Fungal Community Responses to Natural Humus Amendment Potentially Facilitate the Enhancement of Saline–Alkali Soil Multifunctionality
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Site, Design, and Sampling
2.2. Determination of Soil and Plant Properties
2.3. Quantification of Enzzyme Activities and Nutrient Limitation
2.4. Quantitative Calculation of Soil Multifunctionality
2.5. Microbial Community Composition Revealed by Illumina Sequencing
2.6. Statistical Analyses
3. Results
3.1. Soil Physicochemical Properties
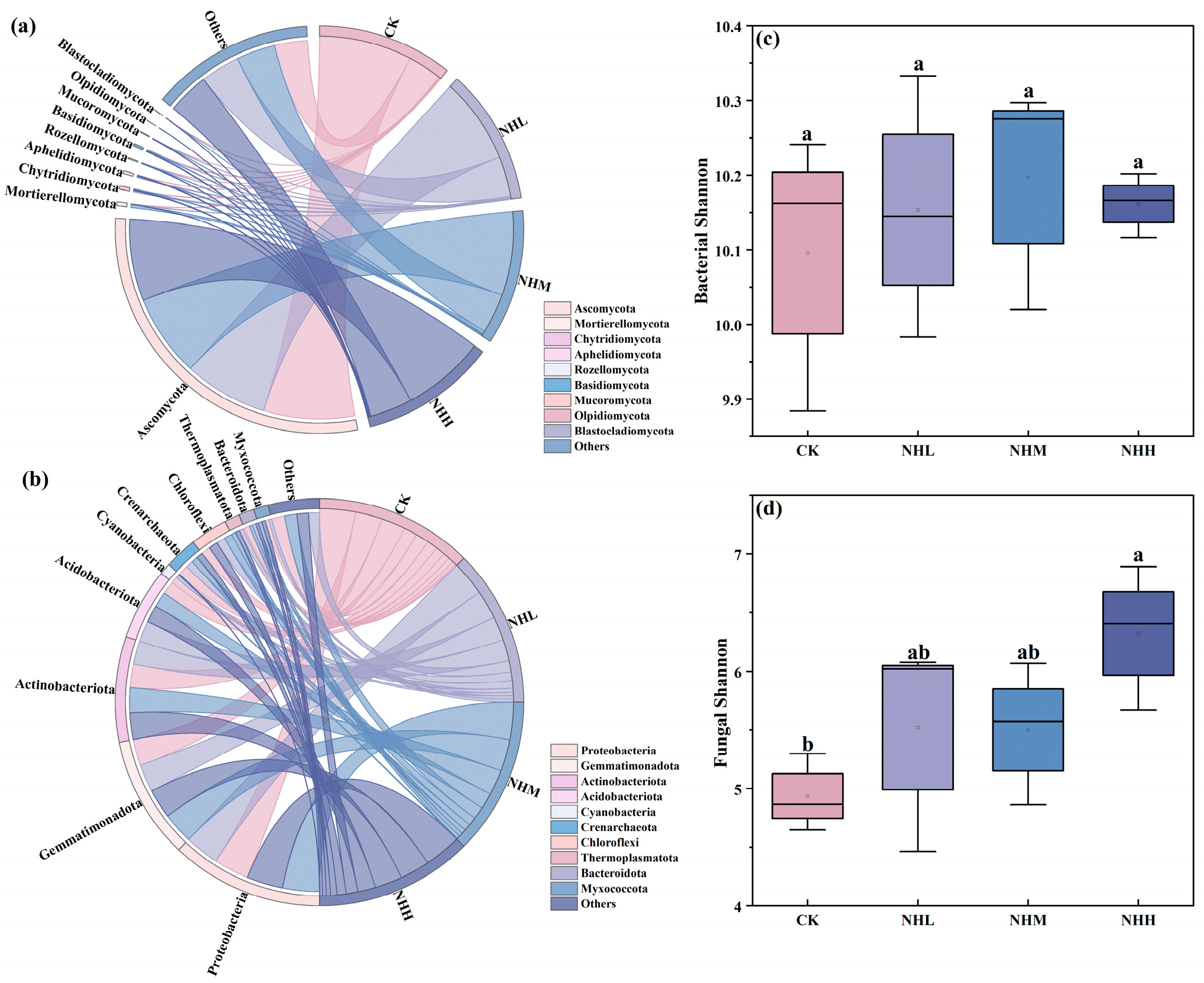
3.2. Soil Microbial Community Composition and Life History Strategies
3.3. Soil Extracellular Enzyme Activities and Microbial Nutrient Limitation
3.4. Soil Multifunctionality and Corn Biomass: Linkages and Key Drivers
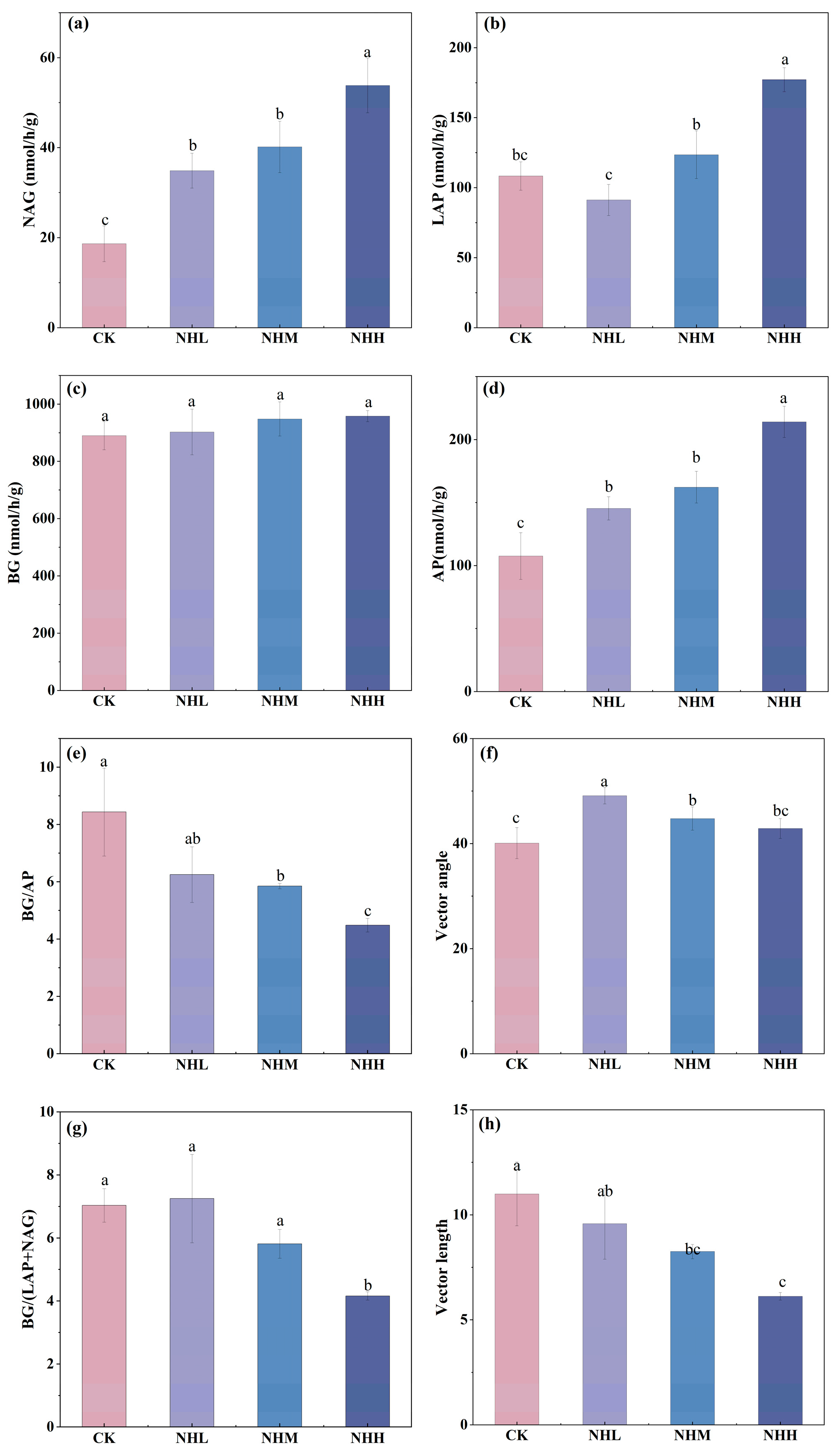

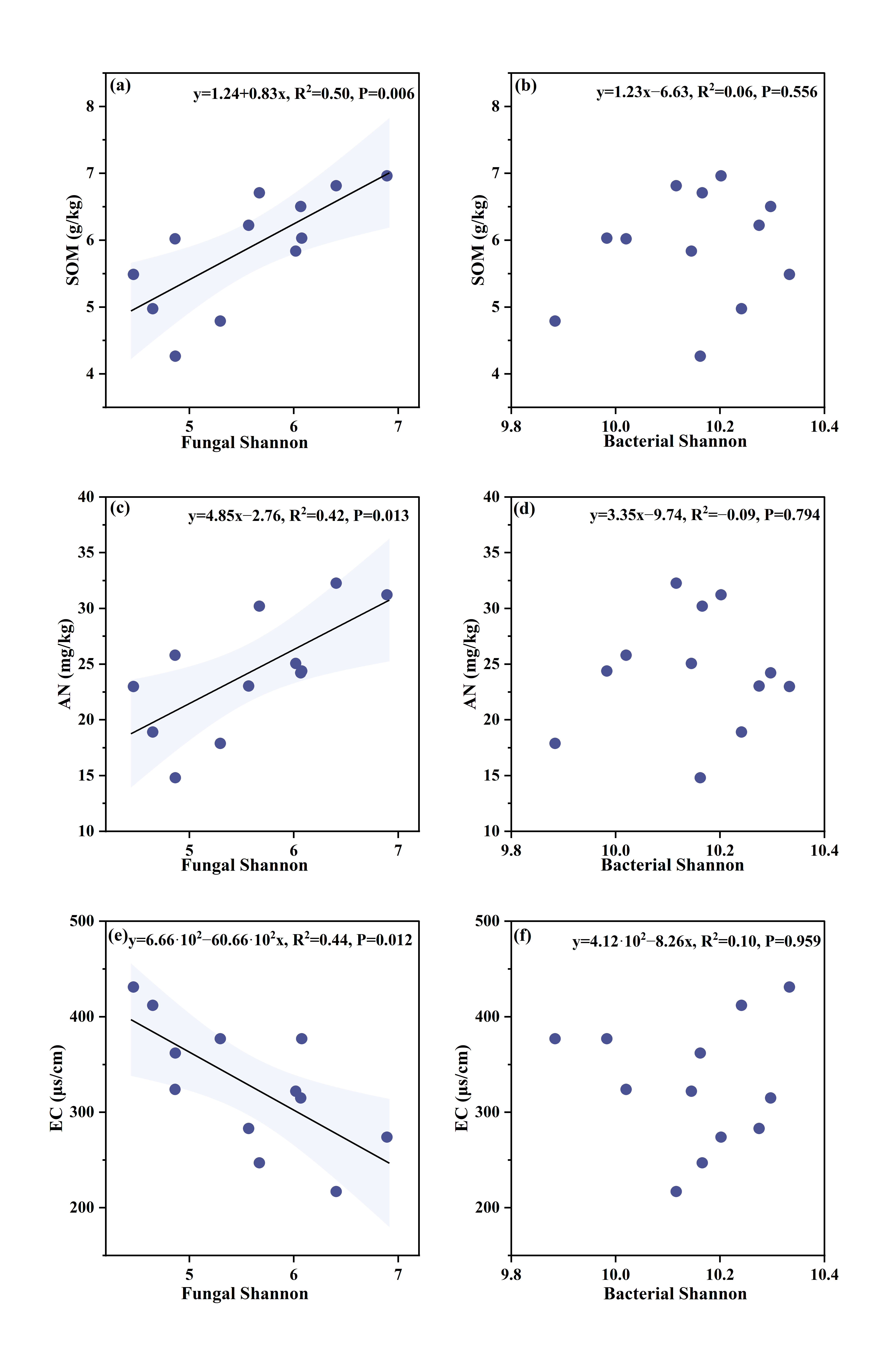
4. Discussion
4.1. The Addition of Natural Humus Increases SMF in Saline–Alkali Soils Primarily Through Fungal Communities
4.2. As the Amount of Added Natural Humus Increased, Microorganisms Gradually Became N Limited, Yet the Soil Microbial Community Maintained a High SMF by Adjusting Its Life History Strategies
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SMF | Soil multifunctionality |
| SOM | Soil organic matter |
| AN | Alkali-hydrolyzable nitrogen |
| EC | Electrical conductivity |
| SWC | Soil moisture content |
| SBD | Soil bulk density |
| AGB | Above-ground corn biomass |
| WR | Below-ground corn biomass |
| BG | β-glucosidase |
| LAP | L-leucine aminopeptidase |
| AP | Alkaline phosphatase |
| NAG | N-acetyl-β-glucosaminidase |
| VA | Vector angle |
| VL | Vector length |
| NHL | 7.5 t/ha natural humus |
| NHM | 15 t/ha natural humus |
| NHH | 30 t/ha natural humus |
| CK | 0 t/ha natural humus |
| MWD | Mass median diameter |
| SWA | Unstable aggregate index |
| TP | Total porosity |
Appendix A

| Phylum (Subphylum) | r-K Selection | References | |
|---|---|---|---|
| Bacteria | Proteobacteria | r-selection | [41,42,43] |
| Gemmatimonadota | K-selection | [44,45,46] | |
| Actinobacteriota | K-selection | [46,47,48] | |
| Acidobacteriota | K-selection | [19,41,49,50,51] | |
| Cyanobacteria | K-selection | [52] | |
| Crenarchaeota | K-selection | [53] | |
| Chloroflexi | K-selection | [46,47,48] | |
| Bacteroidota | r-selection | [46,48] | |
| Myxococcota | r-selection | [54] | |
| Fungi | Ascomycota | r-selection | [48,55,56,57] |
| Mortierellomycota | r-selection | [58] | |
| Chytridiomycota | r-selection | [52] | |
| Aphelidiomycota | K-selection | [52] | |
| Rozellomycota | K-selection | [55,56,57] | |
| Basidiomycota | K-selection | [53] | |
| Mucoromycota | K-selection | [53] |
References
- Cui, Q.; Xia, J.; Yang, H.; Liu, J.; Shao, P. Biochar and effective microorganisms promote sesbania cannabina growth and soil quality in the coastal saline-alkali soil of the yellow river delta. Sci. Total Environ. 2021, 756, 143801. [Google Scholar] [CrossRef] [PubMed]
- Tirabadi, M.S.M.; Banihabib, M.E.; Randhir, T.O. SWAT-S: A SWAT-salinity module for watershed-scale modeling of natural salinity. Environ. Model. Softw. 2021, 135, 104906. [Google Scholar] [CrossRef]
- Yensen, N.P.; Biel, K.Y. Soil remediation via salt-conduction and the hypotheses of halosynthesis and photoprotection. In Ecophysiology of High Salinity Tolerant Plants; Springer: Berlin/Heidelberg, Germany, 2006; pp. 313–344. [Google Scholar]
- Rengasamy, P.; Olsson, K.A. Sodicity and soil structure. Soil Res. 1991, 29, 935–952. [Google Scholar] [CrossRef]
- Qadir, M.; Oster, J. Vegetative bioremediation of calcareous sodic soils: History, mechanisms, and evaluation. Irrig. Sci. 2002, 21, 91–101. [Google Scholar] [CrossRef]
- Creamer, R.E.; Barel, J.M.; Bongiorno, G.; Zwetsloot, M.J. The life of soils: Integrating the who and how of multifunctionality. Soil Biol. Biochem. 2022, 166, 108561. [Google Scholar] [CrossRef]
- Zhao, M.; Liu, S.; Sun, Y.; Chen, Y. Soil multifunctionality is associated with soil water content and fungal richness in Robinia pseudoacacia plantations. Plant Soil 2024, 502, 467–480. [Google Scholar] [CrossRef]
- Hu, W.; Ran, J.; Dong, L.; Du, Q.; Ji, M.; Yao, S.; Sun, Y.; Gong, C.; Hou, Q.; Deng, J.; et al. Aridity-driven shift in biodiversity–soil multifunctionality relationships. Nat. Commun. 2021, 12, 5350. [Google Scholar] [CrossRef]
- Delmotte, S.; Brunel, C.; Castanier, L.; Fevrier, A.; Brauman, A.; Versini, A. Organic fertilization improves soil multifunctionality in sugarcane agroecosystems. Agronomy 2024, 14, 2475. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhang, C.; Wang, H.; Li, F.; Pan, H.; Yang, Q.; Li, J.; Zhang, J. The effects of natural humus material amendment on soil organic matter and integrated fertility in the black soil of Northeast China: Preliminary results. Agron. J. 2023, 13, 794. [Google Scholar] [CrossRef]
- Zheng, Y.Y.; Zhang, J.B.; Tan, J.; Zhang, C.Z.; Yu, Z.H. Chemical Composition and Structure of Humus Relative to Sources. Acta Pedol. Sin. 2019, 56, 386–397. [Google Scholar]
- Cui, J.; Zhu, R.; Wang, X.; Xu, X.; Ai, C.; He, P.; Liang, G.Q.; Zhou, W.; Zhu, P. Effect of high soil C/N ratio and nitrogen limitation caused by the long-term combined organic-inorganic fertilization on the soil microbial community structure and its dominated SOC decomposition. J. Environ. Manag. 2022, 303, 114155. [Google Scholar] [CrossRef]
- Qiu, L.; Zhang, Q.; Zhu, H.; Reich, P.B.; Banerjee, S.; van der Heijden, M.G.; Sadowsky, M.J.; Ishii, S.; Shao, M.; Liu, B.; et al. Erosion reduces soil microbial diversity, network complexity and multifunctionality. ISME J. 2021, 15, 2474–2489. [Google Scholar] [CrossRef] [PubMed]
- Allison, S.D.; Vitousek, P.M. Responses of extracellular enzymes to simple and complex nutrient inputs. Soil Biol. Biochem. 2005, 37, 937–944. [Google Scholar] [CrossRef]
- Khan, M.F. Microbial Remediation of Agrochemical-Contaminated Soils: Enzymatic Mechanisms, Quorum Sensing, and Emerging Opportunities. IEAM 2025, vjaf167. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.F.; Rama, M.; Murphy, C.D. Biodegradation of fluorinated β-triketone herbicide tembotrione by a bacterial–fungal consortium. Biocatal. Agric. Biotechnol. 2025, 70, 103828. [Google Scholar] [CrossRef]
- Delgado-Baquerizo, M.; Oliverio, A.M.; Brewer, T.E.; Benavent-González, A.; Eldridge, D.J.; Bardgett, R.D.; Maestre, F.T.; Singh, B.K.; Fierer, N. A global atlas of the dominant bacteria found in soil. Science 2018, 359, 320–325. [Google Scholar] [CrossRef]
- Wang, Y.F.; Chen, P.; Wang, F.H.; Han, W.X.; Qiao, M.; Dong, W.X.; Hu, C.S.; Zhu, D.; Chu, H.Y.; Zhu, Y.G. The ecological clusters of soil organisms drive the ecosystem multifunctionality under long-term fertilization. Environ. Int. 2022, 161, 107133. [Google Scholar] [CrossRef]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an ecological classification of soil bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef]
- Erbilgin, O.; Bowen, B.P.; Kosina, S.M.; Jenkins, S.; Lau, R.K.; Northen, T.R. Dynamic substrate preferences predict metabolic properties of a simple microbial consortium. BMC Bioinf. 2017, 18, 57. [Google Scholar] [CrossRef]
- Oshiki, M.; Satoh, H.; Okabe, S. Ecology and physiology of anaerobic ammonium oxidizing bacteria. Environ. Microbiol. 2016, 18, 2784–2796. [Google Scholar] [CrossRef]
- Moorhead, D.L.; Sinsabaugh, R.L. A theoretical model of litter decay and microbial interaction. Ecol. Monogr. 2006, 76, 151–174. [Google Scholar] [CrossRef]
- Bao, S. Soil Agrochemical Analysis, 3rd ed.; China Agriculture Press: Beijing, China, 2000. [Google Scholar]
- Shao, M.A.; Wang, Q.J.; Huang, M.B. Soil Physics; Higher Education Press: Beijing, China, 2006; pp. 37–38. [Google Scholar]
- Lu, R.K. Analytical Methods for Soil and Agricultural Chemistry; China Agricultural Science and Technology Press: Beijing, China, 1999; Volume 107–108, p. 269. [Google Scholar]
- Tang, F.K.; Cui, M.; Lu, Q.; Liu, Y.G.; Guo, H.Y.; Zhou, J.X. Effects of vegetation restoration on the aggregate stability and distribution of aggregate-associated organic carbon in a typical karst gorge region. Solid Earth 2016, 7, 141–151. [Google Scholar] [CrossRef]
- Liu, C.; Li, Z.; Fu, S.; Ding, L.; Wu, G. Influence of soil aggregate characteristics on the sediment transport capacity of overland flow. Geoderma 2020, 369, 114338. [Google Scholar] [CrossRef]
- Zhu, L.; Tian, G.; Zhang, L.; Zhao, X.; Wang, J.; Wang, C.; Wang, P. Biochar application affects dynamics of soil microbial biomass and maize grain yield. ScienceAsia 2022, 48, 673–680. [Google Scholar] [CrossRef]
- Guo, Y.; Huang, G.; Guo, Q.; Peng, C.; Liu, Y.; Zhang, M.; Li, Z.; Zhou, Y.; Duan, L. Increase in root density induced by coro-natine improves maize drought resistance in North China. Crop J. 2023, 11, 278–290. [Google Scholar] [CrossRef]
- Moorhead, D.L.; Sinsabaugh, R.L.; Hill, B.H.; Weintraub, M.N. Vector analysis of ecoenzyme activities reveal constraints on coupled C, N and P dynamics. Soil Biol. Biochem. 2016, 93, 1–7. [Google Scholar] [CrossRef]
- Hooper, D.U.; Vitousek, P.M. Effects of Plant Composition and Diversity on Nutrient Cycling. Ecol. Monogr. 1998, 68, 121–149. [Google Scholar] [CrossRef]
- Geml, J.; Pastor, N.; Fernandez, L.; Pacheco, S.; Semenova, T.A.; Becerra, A.G.; Wicaksono, C.Y.; Nouhra, E.R. Large-scale fungal diversity assessment in the Andean Yungas forests reveals strong community turnover among forest types along an altitudinal gradient. Mol. Ecol. 2014, 23, 2452–2472. [Google Scholar] [CrossRef]
- Xu, D.; Yu, X.; Zhang, Y.; Liu, Y.; Chen, C.; Li, L.; Fan, S.; Lu, X.; Zhang, X. Effect of compost as a soil amendment on the structure and function of fungal diversity in saline–alkali soil. Curr. Res. Microb. Sci. 2025, 9, 100405. [Google Scholar] [CrossRef]
- Fan, K.; Delgado-Baquerizo, M.; Guo, X.; Wang, D.; Zhu, Y.G.; Chu, H. Biodiversity of key-stone phylotypes determines crop production in a 4-decade fertilization experiment. ISME J. 2021, 15, 550–561. [Google Scholar] [CrossRef]
- Jiao, S.; Lu, Y.; Wei, G. Soil multitrophic network complexity enhances the link between biodiversity and multifunctionality in agricultural systems. Glob. Change Biol. 2022, 28, 140–153. [Google Scholar] [CrossRef]
- Jiao, P.; Yang, L.; Li, Z.; Liu, C.; Zheng, P.; Tong, D.; Chang, X.; Tang, C.; Xiao, H. Responses of microbial community composition and respiration to soil moisture in eroded soil. Appl. Soil Ecol. 2023, 181, 104662. [Google Scholar] [CrossRef]
- Chen, Y.; Neilson, J.W.; Kushwaha, P.; Maier, R.M.; Barberán, A. Life-history strategies of soil microbial communities in an arid ecosystem. ISME J. 2021, 15, 649–657. [Google Scholar] [CrossRef] [PubMed]
- Mooshammer, M.; Wanek, W.; Zechmeister-Boltenstern, S.; Richter, A. Stoichiometric imbalances between terrestrial decomposer communities and their resources: Mechanisms and implications of microbial adaptations to their resources. Front. Microbiol. 2014, 5, 22. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Amador, P.; Stortenbeker, N.; Wessels, H.J.; Speth, D.R.; Garcia-Heredia, I.; Kartal, B. Enrichment and characterization of a nitric oxide-reducing microbial community in a continuous bioreactor. Nat. Microbiol. 2023, 8, 1574–1586. [Google Scholar] [CrossRef] [PubMed]
- Ghafoor, A.Z.; Javed, H.H.; Karim, H.; Studnicki, M.; Ali, I.; Yue, H.; Xiao, P.; Asghar, M.A.; Brock, C.; Wu, Y. Biological Nitrogen Fixation for Sustainable Agriculture Development Under Climate Change–New Insights From a Meta-Analysis. J. Agron. Crop Sci. 2024, 210, e12754. [Google Scholar] [CrossRef]
- Fierer, N.; Lauber, C.L.; Ramirez, K.S.; Zaneveld, J.; Bradford, M.A.; Knight, R. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 2012, 6, 1007–1017. [Google Scholar] [CrossRef]
- Leff, J.W.; Jones, S.E.; Prober, S.M.; Barberán, A.; Borer, E.T.; Firn, J.L.; Harple, W.S.; Hobbie, S.E.; Hofmockel, K.S.; Fierer, N.; et al. Consistent responses of soil microbial communities to elevated nutrient inputs in grasslands across the globe. Proc. Nat. Acad. Sci. USA 2015, 112, 10967–10972. [Google Scholar] [CrossRef]
- Francioli, D.; Schulz, E.; Lentendu, G.; Wubet, T.; Buscot, F.; Reitz, T. Mineral vs. organic amendments: Microbial community structure, activity and abundance of agriculturally relevant microbes are driven by long-term fertilization strategies. Front. Microbiol. 2016, 7, 1446. [Google Scholar] [CrossRef]
- DeAngelis, K.M.; Pold, G.; Topçuoğlu, B.D.; van Diepen, L.T.; Varney, R.M.; Blanchard, J.L.; Melillo, J.; Frey, S.D. Long-term forest soil warming alters microbial communities in temperate forest soils. Front. Microbiol. 2015, 6, 104. [Google Scholar] [CrossRef]
- Razanamalala, K.; Razafimbelo, T.; Maron, P.A.; Ranjard, L.; Chemidlin, N.; Lelièvre, M.; Dequiedt, S.; Ramaroson, V.H.; Marsden, C.; Bernard, L.; et al. Soil microbial diversity drives the priming effect along climate gradients: A case study in madagascar. ISME J. 2018, 12, 451–462. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zou, J.; Wu, J. Responses of soil bacterial and fungal community structure and functions to different plant species in a mixed forest plantation in a semi-arid region of China. Appl. Soil Ecol. 2024, 198, 105369. [Google Scholar] [CrossRef]
- Liu, T.; Tong, D.; Chen, S.; Ning, C.; Zhang, X.; Filimonenko, E.; Aloufi, A.S.; Cai, W.; Farooq, A.; Yan, W.; et al. Fertilization shapes microbial life strategies, carbon and nitrogen metabolic functions in Camellia oleifera soil. J. Environ. Manag. 2024, 370, 122896. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.; Zhang, W.; Kuzyakov, Y.; Xiao, L.; Xiao, D.; Xu, L.; Chen, H.; Zhao, J.; Wang, K. Linking bacterial life strategies with soil organic matter accrual by karst vegetation restoration. Soil Biol. Biochem. 2023, 177, 108925. [Google Scholar] [CrossRef]
- Cleveland, C.C.; Nemergut, D.R.; Schmidt, S.K.; Townsend, A.R. Increases in soil respiration following labile carbon additions linked to rapid shifts in soil microbial community composition. Biogeochemistry 2007, 82, 229–240. [Google Scholar] [CrossRef]
- Ramirez, K.S.; Craine, J.M.; Fierer, N. Consistent effects of nitrogen amendments on soil microbial communities and processes across biomes. Glob. Change Biol. 2012, 18, 1918–1927. [Google Scholar] [CrossRef]
- Hungate, B.A.; Mau, R.L.; Schwartz, E.; Caporaso, J.G.; Dijkstra, P.; Van Gestel, N.; Koch, B.J.; Liu, C.M.; McHugh, T.A.; Price, L.B.; et al. Quantitative microbial ecology through stable isotope probing. Appl. Environ. Microbiol. 2015, 81, 7570–7581. [Google Scholar] [CrossRef]
- Du, Y.; Yu, A.; Chi, Y.; Wang, Z.; Han, X.; Liu, K.; Fan, Q.; Hu, X.; Che, R.; Liu, D. Organic carbon decomposition temperature sensitivity positively correlates with the relative abundance of copiotrophic microbial taxa in cropland soils. Appl. Soil Ecol. 2024, 204, 105712. [Google Scholar] [CrossRef]
- Chen, J.; Li, F.; Zhao, X.; Wang, Y.; Zhang, L.; Yan, L.; Yu, L. Change in composition and potential functional genes of microbial communities on carbonatite rinds with different weathering times. Front. Microbiol. 2022, 13, 1024672. [Google Scholar] [CrossRef]
- Wang, H.B.; Liu, X.P.; Shu, Y.C.; Li, G.; Sun, C.L.; Jones, D.L.; Zhu, Y.G.; Lin, X.Y. Molecular composition of exogenous dissolved organic matter regulates dissimilatory iron reduction and carbon emissions in paddy soil. Environ. Sci. Technol. 2025, 59, 12679–12691. [Google Scholar] [CrossRef]
- Ho, A.; Di Lonardo, D.P.; Bodelier, P.L.E. Revisiting life strategy concepts in environmental microbial ecology. FEMS Microbiol. Ecol. 2017, 93, 3. [Google Scholar] [CrossRef]
- Yao, F.; Yang, S.; Wang, Z.; Wang, X.; Ye, J.; Wang, X.; DeBruyn, J.M.; Feng, X.; Jiang, Y.; Li, H. Microbial taxa distribution is associated with ecological trophic cascades along an elevation gradient. Front. Microbiol. 2017, 8, 2071. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yang, S.; Semenov, M.V.; Yao, F.; Ye, J.; Bu, R.; Ma, R.; Lin, J.; Kurganova, I.; Kuzyakov, Y.; et al. Temperature sensitivity of SOM decomposition is linked with a K-selected microbial community. Glob. Change Biol. 2021, 27, 2763–2779. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Liu, P.; Wegner, C.E.; Luo, Y.; Xiao, K.Q.; Cui, Z.; Zhang, F.; Liesack, W.; Peng, J. Deciphering microbial mechanisms underlying soil organic carbon storage in a wheat-maize rotation system. Sci. Total Environ. 2021, 788, 147798. [Google Scholar] [CrossRef] [PubMed]

| Group | Bacterial K/r | Fungal K/r |
|---|---|---|
| CK | 2.026 ± 0.027 a | 0.015 ± 0.003 a |
| NHL | 2.261 ± 0.111 a | 0.018 ± 0.006 a |
| NHM | 2.016 ± 0.204 a | 0.021 ± 0.004 a |
| NHH | 2.093 ± 0.087 a | 0.023 ± 0.005 a |
| Group | SMF | AGB (g/Strain) | WR (g/Strain) |
|---|---|---|---|
| CK | −0.19 ± 0.134 b | 15.16 ± 0.112 a | 0.25 ± 0.015 bc |
| NHL | −0.16 ± 0.093 b | 15.19 ± 0.144 a | 0.24 ± 0.019 c |
| NHM | −0.05 ± 0.192 b | 15.39 ± 0.586 a | 0.29 ± 0.006 ab |
| NHH | 0.41 ± 0.114 a | 15.88 ± 0.612 a | 0.33 ± 0.012 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, X.; Lei, J.; Chu, H.; Liu, Y.; Liu, F.; Li, Y.; Zheng, X.; Zhang, H.; Pan, H.; Zhang, C.; et al. Fungal Community Responses to Natural Humus Amendment Potentially Facilitate the Enhancement of Saline–Alkali Soil Multifunctionality. Microorganisms 2025, 13, 2877. https://doi.org/10.3390/microorganisms13122877
Sun X, Lei J, Chu H, Liu Y, Liu F, Li Y, Zheng X, Zhang H, Pan H, Zhang C, et al. Fungal Community Responses to Natural Humus Amendment Potentially Facilitate the Enhancement of Saline–Alkali Soil Multifunctionality. Microorganisms. 2025; 13(12):2877. https://doi.org/10.3390/microorganisms13122877
Chicago/Turabian StyleSun, Xiaoting, Jing Lei, Hang Chu, Yimin Liu, Fei Liu, Yang Li, Xuejia Zheng, Hui Zhang, Hui Pan, Congzhi Zhang, and et al. 2025. "Fungal Community Responses to Natural Humus Amendment Potentially Facilitate the Enhancement of Saline–Alkali Soil Multifunctionality" Microorganisms 13, no. 12: 2877. https://doi.org/10.3390/microorganisms13122877
APA StyleSun, X., Lei, J., Chu, H., Liu, Y., Liu, F., Li, Y., Zheng, X., Zhang, H., Pan, H., Zhang, C., & Wu, Q. (2025). Fungal Community Responses to Natural Humus Amendment Potentially Facilitate the Enhancement of Saline–Alkali Soil Multifunctionality. Microorganisms, 13(12), 2877. https://doi.org/10.3390/microorganisms13122877






