Abstract
Long-term fertilization profoundly influences soil biochemical processes and microbial functionality, yet the coupling mechanisms between soil enzyme activities and functional genes in nutrient cycling remain unclear. This study investigated the effects of different fertilization regimes—nitrogen alone (N), nitrogen–phosphorus–potassium fertilizer (NPK), organic fertilizer (M), and combined organic–inorganic fertilizer (MNPK)—on soil properties, enzyme activities, N- and P-cycling-related functional gene abundances, and faba bean (Vicia faba L.) yield in a 45-year ongoing field experiment in subtropical eastern China. Results showed that long-term fertilization significantly affected soil pH, electrical conductivity, nutrient contents, and crop yield. Organic fertilizer addition (M and MNPK) markedly improved soil organic matter, total and available nutrients, and enhanced faba bean grain yield by 75.07–92.79% compared with NPK, whereas NPK had limited benefits on total and available soil nutrients compared with N-only application. Soil enzyme activity analysis revealed that the MNPK treatment achieved the highest urease and neutral protease activities, while acid and alkaline protease activities responded inconsistently. Phosphorus-related enzymes (acid, neutral, and alkaline phosphatases) were strongly stimulated by organic inputs, reflecting enhanced P mineralization potential. Functional gene analysis showed that N-fixation and assimilatory nitrate reduction genes increased under M and MNPK, while N assimilation, N mineralization, anammox, nitrification, denitrification, and dissimilatory nitrate reduction genes were enriched under N treatment. Phosphate uptake and transport genes were upregulated under NPK, M, and MNPK, whereas inorganic P solubilization genes were highest under N. Significant positive correlations were observed among soil enzyme activities, nutrient contents, and faba bean yield, whereas acid and alkaline protease activities showed opposite trends. The relative abundances of N- and P-cycling functional genes exhibited distinct yet coordinated relationships with soil fertility indicators and enzyme activities. These findings provide mechanistic insights into the long-term regulation of soil–microbe interactions and nutrient cycling, offering a scientific basis for sustainable fertilization strategies in agroecosystems.
1. Introduction
Soil enzymes constitute indispensable and dynamic components within the soil ecosystem, originating from soil microorganisms, plant roots, soil fauna, and plant residues [1,2,3]. They play a pivotal role in the mineralization and degradation of soil organic substrates, thereby driving nutrient mobilization and transformation [4,5]. They effectively regulate the kinetics of soil nutrient cycles [6], exerting a significant influence on soil fertility and crop productivity. Soil enzyme activity serves as a critical indicator for assessing soil fertility, reflecting the intensity and temporal dynamics of various nutrient metabolic processes in soils [7,8]. Compared with soil physicochemical properties, soil enzyme activity is highly dynamic and particularly sensitive to environmental management practices. Notably, agronomic interventions such as tillage regimes [9,10], crop rotation systems [11,12], and fertilization strategies [13,14] have been shown to strongly influence soil enzyme activity.
Enhancing soil fertility represents a fundamental pathway for increasing crop growth potential, yield, and achieving high nutritional value [15]. While fertilization serves as the primary measure for improving soil fertility, long-term indiscriminate application of chemical fertilizers can diminish soil fertility. Such yield-oriented fertilization strategies, pursued at environmental cost, exert detrimental effects on soil enzyme activities [16,17]. Long-term field experiments have consistently demonstrated that continuous application of N, P, and K fertilizers suppresses soil enzyme activity [18,19]. Conversely, rational fertilization strategies can effectively enhance soil enzyme activity, thereby promoting soil metabolism processes, altering soil nutrient forms, increasing soil fertility, and improving soil properties [20,21,22]. Consequently, exploring optimal fertilization management practices is particularly important.
Organic fertilizers, as mature amendments, substantially increase soil organic matter (SOM) content when incorporated into soil. This not only improves soil physicochemical properties but also provides favorable environmental conditions and energy substrates for microbial growth [23,24,25]. SOM, a key determinant of soil enzyme activities, is decomposed by microorganisms into a diverse range of products that supply essential nutrients and energy to both microbes and plants, and induce the production of soil urease, phosphatase, and protease, thereby modulating enzymatic activity [26,27,28]. Long-term field experiments conducted by Zhang et al. [1] revealed that sustained manure application elevated activities of soil enzymes such as acid phosphatase and significantly raised soil total P content. Similarly, in a 12-year tobacco-maize rotation trial, Jiang et al. [29] found that adding organic fertilizer increased crop yields, soil acid phosphatase activity, and overall soil nutrient status. A five-year study on winter wheat likewise verified that organic fertilizer application enhanced the activities of soil urease and protease, with crop yield and soil organic carbon being significantly positively correlated with urease and protease activities [30]. Adequate soil organic nutrient pools are generally associated with elevated soil enzyme activity, thereby accelerating SOM mineralization rates and boosting microbial metabolic vigor. Moreover, the chemical decomposition of SOM is a continuous, long-term process that helps maintain a dynamic balance of nutrient supply, thereby enhancing the ecological sustainability of soil nutrient cycling [31,32].
Soil enzymes play an important role in plant nutrient cycling and utilization efficiency [33]. However, the extent of their activities ultimately depends on the metabolic potential of the soil microbial community [22]. Functional genes, as the molecular basis of microbial metabolic functions, directly regulate enzyme synthesis and expression, and their distribution and expression profiles of these functional genes are strongly shaped by soil environmental conditions [34,35,36]. Long-term fertilization, by altering soil physicochemical properties, microbial community composition and structure, can directly or indirectly regulate the relative abundances of functional genes involved in N and P cycling [37,38,39]. For instance, long-term application of chemical fertilizers alone may reduce the abundance of N-fixation functional genes and organic-phosphorus mineralization functional genes as a consequence of soil acidification and nutrient imbalance [40,41], whereas organic fertilizer inputs can increase the abundance of ammonia-oxidizing and phosphatase functional genes by enhancing carbon source availability and improving soil environment [42,43]. Elucidating the mechanistic linkages between shifts in functional gene abundance and corresponding changes in soil enzyme activity is essential for revealing the underlying pathways through which fertilization practices regulate soil nutrient cycling [44]. Meanwhile, soil enzymes, which mediate SOM decomposition and nutrient mineralization, have been used as sensitive bioindicators for assessing the effectiveness of agricultural management practices [45].
Although many studies have examined the effects of long-term fertilization on soil microbial communities or enzyme activities separately, few have systematically integrated multi-enzyme activities with the functional gene profiles involved in N- and P-cycling to elucidate their coordinated regulatory mechanisms. Moreover, the quantitative relationships among soil chemical properties, enzyme activities, and microbial functional potentials under contrasting fertilization regimes remain largely unexplored, particularly in multi-decadal field systems. Therefore, this study aimed to fill these knowledge gaps by investigating the relationships between soil enzyme activities and the relative abundances of N- and P-cycling functional genes under long-term fertilization in a 45-year ongoing field experiment. Specifically, we sought to address three key questions: (1) how soil enzyme activities respond to different long-term fertilization treatments; (2) how microbial functional genes encoding nitrogen- and phosphorus-cycling enzymes change under these treatments; and (3) how soil enzyme activities are correlated with the relative abundances of these functional genes, soil properties, and faba bean yield.
2. Materials and Methods
2.1. Experimental Site
The long-term field trial was located at the experimental base of the Jiangsu Yanjiang Institute of Agricultural Sciences in Xueyao Town, Rugao City, Jiangsu Province, China (120°37′ E, 32°07′ N). This area experiences a subtropical monsoon climate, characterized by mild temperatures (mean annual temperature of 16.2 °C) and abundant rainfall averaging 1250 mm per year. The soil at the site is a fluvo-aquic type developed from alluvial deposits of the Yangtze River and exhibits a sandy-loam texture. When the experiment was initiated in 1979, the topsoil (0–15 cm) contained 14.4 g kg−1 of soil organic matter, 133 mg kg−1 of alkali-hydrolyzable nitrogen, 29.0 mg kg−1 of available phosphorus, 64.0 mg kg−1 of available potassium, and had a pH of 7.86.
2.2. Experimental Design
The long-term experiment has been continuously maintained since 1979 under a six-season crop rotation system within a three-year cycle: rice (Oryza sativa L.), faba bean (Vicia faba L.), maize (Zea mays L.), barley (Hordeum vulgare L.), cotton (Gossypium spp.), and wheat (Triticum aestivum L.). In June 2022, after the wheat harvest, the crop rotation system was adjusted to wheat–soybean–faba bean–maize–barley–rice, while the long-term fertilization treatments and management regimes were maintained consistently. The current study was based on soil and plant samples collected in May 2023 from this ongoing long-term fertilization trial.
The experiment included four fertilization treatments: nitrogen alone (N), NPK fertilizer (NPK), organic fertilizer alone (M), and NPK plus organic fertilizer (MNPK). A randomized complete block design was used, with a plot area of 16.8 m2 and four replicates. For faba bean, 45 kg N ha−1 was applied as urea, while for other crops a rate of 225 kg N ha−1 was used. All crops received 24.56 kg P ha−1 as superphosphate and 139.00 kg K ha−1 as potassium chloride. In the M and MNPK treatments, organic fertilizer (pig manure) was applied at 18 t ha−1. The pig manure had a pH of 7.6, organic matter 312.2 g kg−1, N 5.7 g kg−1, P 3.0 g kg−1, and K 3.6 g kg−1. Phosphorus, potassium, and pig manure were supplied before planting as base fertilization. Nitrogen was applied as topdressing for faba bean, and split (40% basal, 60% topdressing) for the other crops. At grain maturity, all crops were manually harvested, and no straw was returned to the field.
2.3. Soil Sampling
Soil sampling was conducted on 4 May 2023 during the maturation stage of faba bean. In each plot, five soil cores (0–20 cm) were collected using a hollow auger and subsequently mixed thoroughly to produce one representative composite sample. The collected soils were immediately sealed in sterile bags and transported to the laboratory in insulated containers packed with ice. Upon arrival, the fresh soil was gently sieved through a 2 mm mesh to eliminate fine roots and surface debris. A portion of the fresh soil was directly used for measuring soil moisture and for extracting inorganic nitrogen. Approximately 50 g of fresh soil from each sample was frozen at −80 °C for DNA analysis. The remaining soil was air-dried in a shaded environment, finely ground, and sieved through 0.85 mm and 0.149 mm meshes for subsequent chemical determinations.
2.4. The Determinations of Soil Properties, Crop Yield, and Soil Enzyme Activities
The measured soil indicators included pH, electrical conductivity (EC), soil organic matter (SOM), total carbon (TC), total nitrogen (TN), total phosphorus (TP), ammonium-N (NH4+-N), nitrate-N (NO3−-N), available phosphorus (AP), and available potassium (AK). Soil pH and EC were measured using soil suspensions prepared with water at ratios of 1:2.5 (w/v) and 1:5 (w/v), respectively, and analyzed using a pH meter and a portable conductivity meter (Mettler-Toledo, Shanghai, China). SOM was quantified following the dichromate oxidation procedure specified in Chinese Standard GB9834-88 [46]. Total carbon and nitrogen were determined by high-temperature combustion method with an elemental analyzer (Vario EL CUBE, Elementar, Langenselbold, Germany). TP was analyzed after acid digestion using the molybdenum–antimony colorimetric method. For NH4+-N and NO3−-N, 5 g of fresh soil was shaken for 1 h with 1 M KCl (soil:solution = 1:5), after which the filtrates were quantified using a continuous-flow analyzer (San++, Skalar, Netherlands). Available P was extracted with 0.5 M NaHCO3 by shaking at 180 rpm for 30 min and determined by UV spectrophotometry based on the molybdenum blue method. AK was extracted with 1 M ammonium acetate by shaking at 120 rpm for 30 min and measured with a flame photometer. At the faba bean maturity stage, whole plants from each plot were manually harvested, air dried, and then threshed to determine grain yield.The activities of soil urease, acid protease, neutral protease, alkaline protease, acid phosphatase, neutral phosphatase, and alkaline phosphatase were quantified with soil enzyme assay kits (Solarbio Science & Technology Co., Beijing, China). For urease, one activity unit corresponded to the enzymatic generation of 1 μg NH3-N per g of soil per day. Soil protease activity (acidic, neutral, or alkaline) was expressed as the amount of enzyme producing 1 μmol tyrosine per gram soil per day. One unit of acid, neutral, or alkaline phosphatase activity was defined as the amount of enzyme releasing 1 nmol phenol per g of soil per day at 37 °C.
2.5. Functional Gene Screening
Based on the 16S rRNA gene amplicon sequencing data, the functional potential of soil bacterial communities was predicted using Tax4Fun2 [47]. It should be noted that Tax4Fun2 provides inferred rather than directly measured functional profiles, representing the potential functional capacity of microbial communities rather than actual gene expression. Functional annotations in Tax4Fun2 rely on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, and the predictions are reported as relative abundances. Following previous studies [44,48,49], a total of 151 KEGG functional pathways were selected and categorized into 14 metabolic pathways associated with N and P cycling. Details are provided in Table A1.
2.6. Statistical Analysis
Data analysis was carried out using Microsoft Excel, with results expressed as mean ± standard deviation. Differences among treatments were evaluated using one-way ANOVA in SPSS 27.0, followed by Duncan’s multiple range test to determine statistically significant variations. Pearson correlation analysis was conducted in R (4.3.1) via the cor package to examine the relationships among soil properties, faba bean yield, the relative abundances of N- and P-cycling functional genes, and soil enzyme activities. The correlation results were visualized with the corrplot package, while bar charts and boxplots were generated using Origin 2025.
3. Results
3.1. The Effects of Long-Term Fertilization Regimes on Soil Properties and Faba Bean Yields
Table 1 demonstrates that long-term fertilization regimes significantly influenced soil properties and faba bean yield. Compared with sole N or organic fertilizer application, NPK and MNPK treatments markedly decreased soil pH, with the lowest value under MNPK. MNPK also significantly increased soil EC, while no significant variation was detected among the remaining treatments. The use of organic fertilizer, either applied independently (M) or jointly with mineral fertilizers (MNPK), substantially enhanced soil fertility indices, including SOM, TC, TN, TP, AP, and AK, compared with chemical fertilizer treatments. In contrast, balanced NPK fertilization provided only marginal improvements over sole N, without statistical significance. No significant treatment effects were observed for NO3−-N or NH4+-N contents. Faba bean yield increased by 1097.32% under NPK compared with sole N, and was further promoted by organic fertilizer treatments, with M and MNPK increasing grain yield by 75.07% and 92.79% over NPK, respectively, although the difference between M and MNPK was not significant.

Table 1.
Soil characteristics and faba bean yields under different fertilization treatments.
3.2. Effects of Long-Term Fertilization Treatments on Soil N-Related Enzyme Activities
As shown in Figure 1, compared with sole N application, all other fertilization treatments significantly increased soil urease activity to varying extents (Figure 1A), following the order of MNPK > M > NPK > N, with respective increases of 83.21%, 58.04%, and 23.24%. The mean activities of soil acid protease, neutral protease, and alkaline protease under long-term fertilization treatments were 2.5354 U/g, 0.3951 U/g, and 0.3976 U/g, respectively (Figure 1B–D). Specifically, compared with N-only treatment, NPK significantly enhanced soil acid protease activity by 23.26%. However, acid protease activity under both M and MNPK treatments was significantly lower than under NPK, with no significant difference observed between N and MNPK (Figure 1B). For soil neutral protease activity, no significant differences were observed between N and NPK treatments, whereas organic fertilizer application significantly increased neutral protease activity, with the highest activity under MNPK (Figure 1C). In contrast, soil alkaline protease activity showed no significant differences among N, NPK, and M treatments, but was significantly reduced under the MNPK treatment (Figure 1D).
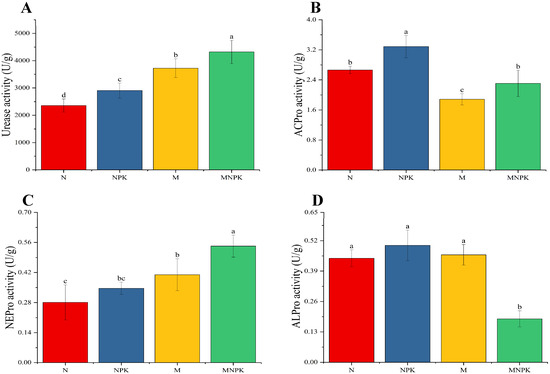
Figure 1.
Activity of soil N-related enzymes under long-term fertilization treatments. (A) Soil urease (Urease); (B) Soil acid protease (ACPro); (C) Soil neutral protease (NEPro); (D) Soil alkaline protease (ALPro). Error bars represent standard deviations, and different lowercase letters above the bars indicate significant differences among treatments at p < 0.05.
3.3. Effects of Long-Term Fertilization Regimes on Soil P-Related Enzyme Activities
Under long-term fertilization regimes, soil acid, neutral, and alkaline phosphatase activities exhibited similar variation patterns (Figure 2). Application of organic fertilizer alone or in combination with inorganic fertilizers significantly enhanced the activities of acid, neutral, and alkaline phosphatases compared with chemical fertilizer treatments, corresponding to 1.81~2.46-fold, 5.01~5.89-fold, and 1.58~1.67-fold increases relative to N and NPK treatments, respectively. Except for alkaline phosphatase, where NPK treatment showed significantly higher activity than N treatment, no significant differences were found between N and NPK treatments for either acid or neutral phosphatase activities (Figure 2B).
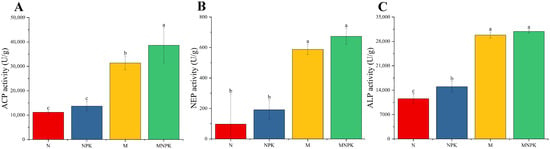
Figure 2.
Activity of soil P-related enzymes under long-term fertilization treatments. (A) Soil acid phosphatase (ACP); (B) Soil neutral phosphatase (NEP); (C) Soil alkaline phosphatase (ALP). Different lowercase letters above the error bars indicate statistically significant differences in enzyme activities among treatments (p < 0.05).
3.4. Effects of Long-Term Fertilization Regimes on the Functional Diversity of Soil Bacterial Communities
The relative abundances of N-cycling functional genes exhibited distinct response patterns under the four long-term fertilization treatments (Figure 3). Among them, the abundance of N fixation genes was highest under MNPK, significantly exceeding that under NPK, but no significant differences were observed compared with N or M (Figure 3A). Genes associated with N mineralization showed the highest abundance under N treatment and the lowest under M, while NPK and MNPK treatments did not differ significantly from N (Figure 3C). The N-only treatment markedly increased the abundance of nitrification genes, with values significantly higher than under NPK and M. Although MNPK was also lower than N, the difference between MNPK and N was not statistically significant (Figure 3E). Organic fertilizer application significantly enhanced the abundance of assimilatory nitrate reduction genes compared with chemical fertilizer treatments; however, no significant differences were detected between M and MNPK or between N and NPK (Figure 3G). Functional genes related to N assimilation, anammox, denitrification, and dissimilatory nitrate reduction displayed similar patterns, with the highest abundances under N treatment, significantly exceeding those of the other treatments, while NPK, M, and MNPK showed no significant differences (Figure 3B,D,F,H).
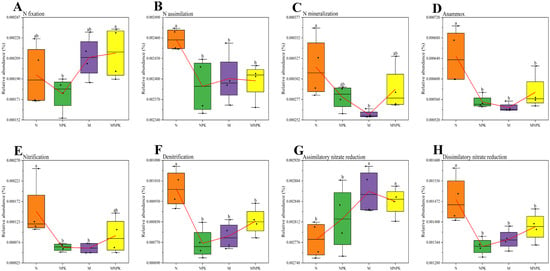
Figure 3.
Enzyme-encoding genes related to nitrogen cycling in soil bacterial communities under long-term fertilization regimes. (A) N fixation; (B) N assimilation; (C) N mineralization; (D) Anammox; (E) Nitrification; (F) Denitrification; (G) Assimilatory nitrate reduction; (H) Dissimilatory nitrate reduction. Red lines represent mean values. Different lowercase letters above the error bars indicate significant differences in soil enzyme activity among treatments (p < 0.05, Duncan’s test).
Among the enzyme-coding genes related to the P-cycling (Figure 4), the relative abundances of genes associated with polyphosphate polymerization and P starvation response regulation exhibited no significant differences across different treatments (Figure 4A,D). The relative abundance of genes involved in polyphosphate degradation was highest under the N, M, and MNPK treatments than that under the balanced NPK fertilization (Figure 4B). Genes related to phosphate uptake and transport exhibited the lowest relative abundance under the N treatment, significantly lower than those under NPK, M, and MNPK treatments, whereas no significant differences were observed among the latter three treatments (Figure 4C). Compared with the N treatment, the relative abundances of inorganic P solubilizing genes were markedly lower in the NPK, M, and MNPK treatments, while no significant differences were found among these three treatments (Figure 4E). The relative abundance of genes involved in organic P mineralization was highest in the NPK treatment, showing significant differences compared with the N and MNPK treatments, but not with the M treatment (Figure 4F).
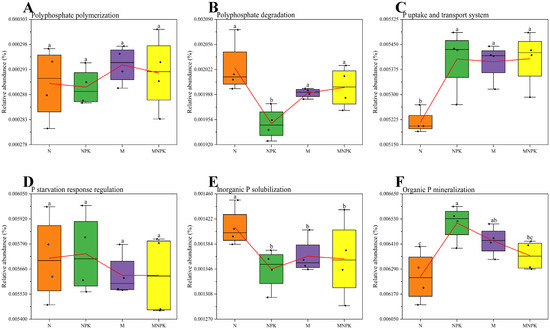
Figure 4.
Enzyme-encoding genes related to P-cycling in soil bacterial communities under long-term fertilization treatments. (A) Polyphosphate polymerization; (B) Polyphosphate degradation; (C) P uptake and transport; (D) P starvation response regulation; (E) Inorganic P solubilization; (F) Organic P mineralization. The red connecting lines represent mean values across replicates, and different lowercase letters above the short horizontal bars denote significant differences in gene abundances among treatments (p < 0.05, Duncan’s test).
3.5. Correlation Analysis of Soil Properties and Faba Bean Yield with Soil Enzyme Activities
This study revealed the correlation patterns among soil properties, soil enzyme activities, and faba bean yield under long-term fertilization regimes through correlation analysis (Figure 5). The results showed that, under different long-term fertilization treatments, the activities of soil acid phosphatase, neutral phosphatase, alkaline phosphatase, urease, and neutral protease were all highly positively correlated with each other. The activity of soil acid protease showed a significant negative relationship with soil acid phosphatase, neutral phosphatase, and alkaline phosphatase activities, whereas soil alkaline protease activity was significantly negatively correlated with soil acid phosphatase, neutral phosphatase, alkaline phosphatase, urease, and neutral protease activities.
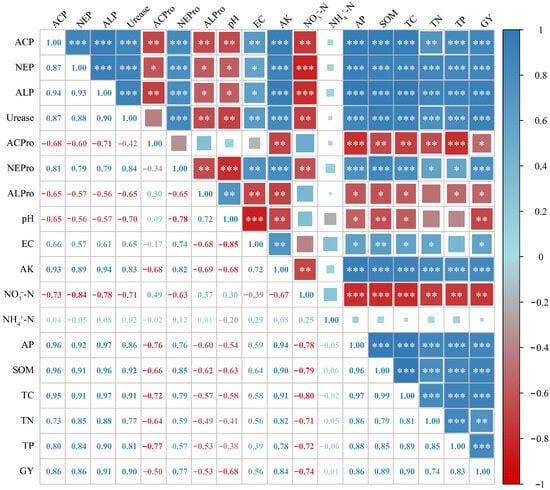
Figure 5.
Correlation analysis of soil properties and faba bean yield with soil enzyme activities. Pearson correlation coefficient, * p < 0.05; ** p < 0.01; *** p < 0.001.
Significant correlations were also observed between soil enzyme activities and various soil properties. The activities of soil acid phosphatase, neutral phosphatase, alkaline phosphatase, urease, and neutral protease showed consistent trends of correlation with soil properties and faba bean yield, being significantly or highly positively correlated with EC, AK, AP, SOM, TC, TN, TP, and GY, while significantly negatively correlated with pH and NO3−-N, and showing no significant correlation with NH4+-N. In contrast, the activities of soil acid protease and alkaline protease generally exhibited opposite trends, being significantly negatively correlated with EC, AK, AP, SOM, TC, TN, TP, and GY (except for soil acid protease vs. EC and soil alkaline protease vs. TN, which showed no significant correlations). Soil alkaline protease activity was significantly positively correlated with pH, whereas soil acid protease activity showed no significant correlation with pH, and both soil acid and alkaline protease activities were not significantly correlated with NH4+-N and NO3−-N.
3.6. Correlation Analysis Between Soil Properties and Faba Bean Yield with the Relative Abundance of Soil N- and P-Related Functional Genes
Correlation analysis (Figure 6) revealed distinct functional partitioning in the associations between soil properties, faba bean grain yield (GY), and the abundances of N- and P-cycling functional genes.
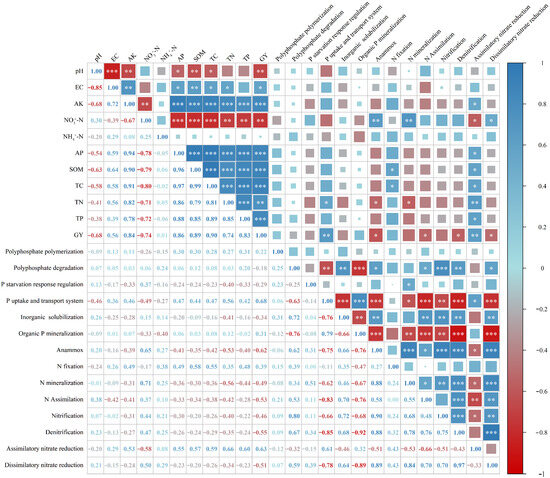
Figure 6.
Correlation analysis of soil properties and faba bean grain yield with the relative abundances of N- and P-cycling functional genes. Note: Correlation analysis (Pearson correlation coefficient, * p < 0.05; ** p < 0.01; *** p < 0.001) revealed distinct functional partitioning in the associations between soil properties, faba bean grain yield (GY), and the abundances of N- and P-cycling functional genes.
Regarding soil P-cycling genes, only genes related to the phosphate uptake and transport system were significantly correlated with TN and GY, whereas other P-cycling functional genes showed weak and non-significant correlations with soil properties and GY. For N-cycling genes, anammox genes were positively correlated with NO3−-N but significantly negatively correlated with TN and GY. N fixation genes were positively correlated with SOM and TC. N mineralization genes were positively correlated with NO3−-N but negatively correlated with TN. N assimilation and denitrification genes were all significantly negatively correlated with GY. Assimilatory nitrate reduction genes showed significant positive correlations with SOM, TC, TN, TP, AP, AK, and GY, but were negatively correlated with NO3−-N. In contrast, dissimilatory nitrate reduction genes were significantly positively correlated with NO3−-N and negatively correlated with GY. Soil pH, EC, and NH4+-N were not significantly correlated with any N- and P-cycling functional genes.
3.7. Correlation Analysis Between Soil Enzyme Activities and the Relative Abundance of Soil N- and P-Related Functional Genes
Correlation analysis (Figure 7) indicated that soil neutral phosphatase, alkaline phosphatase, and urease activities were significantly positively correlated with phosphate uptake and transport functional genes. Anammox genes were negatively correlated with neutral phosphatase activity. N fixation genes showed significant positive correlations with soil acid phosphatase, alkaline phosphatase, urease, and neutral protease activities. Assimilatory nitrate reduction genes were significantly positively correlated with soil acid phosphatase, neutral phosphatase, alkaline phosphatase, and urease activities. Moreover, N- and P-cycling functional genes exhibited clear synergistic and antagonistic relationships.
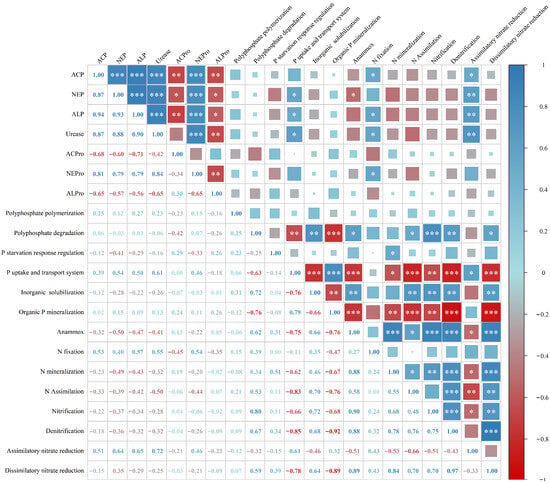
Figure 7.
Correlation analysis between soil enzyme activities and the relative abundance of N- and P-related functional genes. Pearson correlation coefficient, * p < 0.05; ** p < 0.01; *** p < 0.001.
Polyphosphate polymerization genes were not significantly correlated with any N-cycling functional genes. In contrast, polyphosphate degradation genes were significantly positively correlated with anammox, nitrogen assimilation, nitrification, denitrification, and dissimilatory nitrate reduction genes. P starvation response regulation genes showed significant positive correlations with N mineralization genes. Phosphate uptake and transport system genes exhibited strong negative correlations with multiple N transformation pathways, including anammox, nitrogen mineralization, nitrogen assimilation, nitrification, denitrification, and dissimilatory nitrate reduction. In addition, phosphate uptake and transport functional genes were significantly positively correlated with assimilatory nitrate reduction genes.
Similarly, organic P mineralization genes were significantly negatively correlated with anammox, N mineralization, N assimilation, nitrification, denitrification, and dissimilatory nitrate reduction pathways. In contrast, inorganic P solubilization genes were generally significantly positively correlated with these N transformation genes, including anammox, N assimilation, nitrification, denitrification, and dissimilatory nitrate reduction, showing consistent directional patterns.
4. Discussion
4.1. Relationships Between Long-Term Fertilization Regimes and N- and P-Related Soil Enzyme Activities
The primary function of soil urease is to hydrolyze urea into carbon dioxide and ammonia [50], and its activity can reflect the soil nitrogen supply level [51]. Soil urease activity is regulated by multiple environmental factors, such as soil texture, land use type, soil organic carbon content, and total nitrogen content [52,53,54]. Under the sole N treatment, the soil is simultaneously constrained by insufficient carbon sources and limitations of P and K, resulting in the lowest soil urease activity. In contrast, NPK application markedly promotes faba bean root growth and biomass, thereby supplying microbes with a certain amount of carbon via root exudates [55,56]. The M and MNPK treatments further increase carbon supply and nutrient availability to microbes, leading to the highest soil urease activity [57,58,59], which is consistent with the observed significant positive correlations between soil urease activity and SOM, TC, and both total and available nutrient contents. The trend of soil neutral protease is consistent with that of soil urease. However, soil acidic and alkaline proteases exhibited higher activities under the N and NPK treatments. These results suggested that under N and NPK treatments, where soils remained carbon-limited due to the absence of organic inputs, microorganisms appeared to rely more heavily on soil acid and alkaline proteases to decompose recalcitrant protein substrates and obtain N. In contrast, soil neutral protease activity increased substantially under M and MNPK treatments, reflecting the greater availability of labile carbon that supports the decomposition and release of labile organic nitrogen from organic fertilizer.
Soil phosphatase activity is an important indicator for assessing the direction and intensity of soil phosphorus biotransformation [60,61]. The organic fertilizer additions significantly enhanced the activities of soil acid, neutral, and alkaline phosphatases, consistent with previous studies [62,63]. The possible reasons include that the application of organic fertilizer introduces large amounts of labile organic carbon and organic phosphorus, thereby increasing microbial biomass and metabolic activity and supplying hydrolyzable organic-P substrates [64,65]. In addition, the increases in faba bean root biomass and root exudation elevate both root-derived phosphatases and microbial extracellular phosphatases [66,67]. The significant positive correlations between soil phosphatases and SOM and other nutrient indicators indicated that the enrichment of SOM and multiple nutrients markedly enhances microbial and root metabolic activity and extracellular enzyme secretion, thereby increasing phosphatase activity and promoting the mineralization and turnover of organic phosphorus in soil.
4.2. Responses of N-Cycling Enzyme-Encoding Genes to Different Fertilization Treatments
In this study, the higher relative abundance of nitrogen fixation functional genes from MNPK treatment likely implied the incorporation of organic fertilizer substantially increased soil SOM and TC contents, thereby providing essential carbon sources and energy substrates for diazotrophic microorganisms. Correlation analysis also revealed that the abundance of nitrogen fixation genes was significantly positively correlated with SOM and TC, underscoring the pivotal role of carbon availability in supporting the nitrogen fixation process. Moreover, the combined application of organic and inorganic fertilizers can optimize the soil C:N ratio, which further promotes the proliferation and activity of nitrogen-fixing microorganisms [68,69].
Functional genes related to N assimilation, mineralization, anammox, nitrification, denitrification, and dissimilatory nitrate reduction showed most abundance under N-only treatment. Sole nitrogen fertilization often results in limited available carbon and phosphorus (C, P co-limitation) and a reduced rhizosphere carbon flow due to less root biomass and exudates, which substantially restricts microbial growth and energy metabolism. Under such nutrient-impoverished conditions, microorganisms rely more on inorganic nitrogen assimilation, organic nitrogen mineralization, and inorganic nitrogen transformation processes to maintain nitrogen acquisition and basic metabolic activity, thereby enriching the associated functional genes [70,71,72]. In contrast, multi-nutrient sufficiency and enhanced rhizosphere carbon inputs expand microbial metabolic niches and functional gene diversity, reducing the relative investment in inorganic nitrogen uptake and transformation pathways and thus lowering the proportional abundance of these genes, despite possible greater microbial biomass and activity [73,74,75,76]. This is also consistent with long-term fertilization studies showing that N-only treatment leads to substantially greater nitrogen losses and lower nitrogen use efficiency than balanced or manure-based fertilization systems via leaching as NO3− and gas volatilization or emissions as NH3, N2O, and N2 [77,78,79]. Such elevated N-loss pressures under N-only conditions can further stimulate microbial nitrogen transformation processes, leading to higher abundances of nitrification, denitrification, anammox functional genes. Faba bean yield was negatively associated with N-assimilation, anammox, denitrification, and dissimilatory nitrate reduction genes, but positively with assimilatory nitrate reduction genes. These contrasting correlations indicate that nitrogen transformation pathways associated with microbial nutrient stress (e.g., assimilation, dissimilatory nitrate reduction, anammox, denitrification) dominate in low-fertility soils and coincide with reduced crop performance, whereas assimilatory nitrate reduction increases under fertile, carbon-enriched soils that better support plant nitrogen uptake and higher yields.
4.3. Responses of P-Cycling Enzyme-Encoding Genes to Different Fertilization Treatments
The relative abundance of polyphosphate degradation genes was lowest under the NPK treatment, consistent with previous findings [80,81]. This can be primarily attributed to the direct input of soluble phosphorus fertilizers in the NPK treatment, which effectively alleviated soil phosphorus limitation and substantially reduced the dependence on intracellular polyphosphate degradation pathways. In contrast, the N-only treatment necessitated the activation of microbial polyphosphate degradation to release phosphorus as a compensatory strategy to secure essential P for basic metabolic needs, thereby stimulating its expression [82,83]. Under the M and MNPK treatments, the highest biomass and yield indicate greater plant demand for bioavailable P, likely increasing reliance on microbial P-releasing processes. Therefore, polyphosphate degradation pathways are still necessary to supplement more phosphorus under organic fertilization treatments [84].
Microorganisms drive soil phosphorus cycling by secreting organic acids and phosphatases, which were responsible for solubilizing inorganic P and releasing P from organic matter, respectively [85,86,87]. The generation of extracellular enzymes requires substantial resources, including carbon and nutrient inputs for constructing the enzyme molecules, as well as considerable metabolic energy to support protein synthesis and subsequent secretion [31,88]. For organic P mineralization functional genes, the NPK treatment showed the highest relative abundance, followed by M and MNPK, and lowest under N treatment. Acid phosphatase is predominantly derived from plants and fungi, while bacteria are the primary producers of alkaline phosphatase [89]. Phosphatase-driven organic P mineralization accounts for nearly 90% of total organic P turnover in soils [90,91]. These results are also consistent with our measured soil phosphatase activities, especially alkaline phosphatase activity, which was lowest under the N treatment and significantly higher under NPK, M, and MNPK. Because extracellular enzymes are short-lived, microorganisms must obtain a sufficient return on their investment to sustain growth, particularly under resource-limited conditions [31]. The severe carbon and nutrient limitation under the N treatment restricts microbial metabolic, making the energy-intensive mineralization process less favorable and lowering organic P-mineralizing gene abundance. Although organic P-mineralizing genes were slightly more abundant under NPK than under organic-added treatments, organic fertilization supported greater soil microbial biomass and still resulted in significantly higher soil phosphatase activities. The functional genes involved in inorganic P solubilization exhibited the highest abundance under the N-only treatment, significantly exceeding the other treatments. These results may reflect that inorganic P solubilization microbes are favored under low-P conditions, whereas high available phosphorus due to long-term P fertilization suppresses their abundance and activity through negative feedback regulation [92]. The NPK treatment directly supplied P and stimulated crop growth and nutrient demand, whereas M and MNPK increased SOM and nutrient levels, inducing the expression of P uptake and transport genes. This upregulation subsequently promoted plant phosphorus uptake, transport and utilization, ultimately leading to a significant increase in both biomass and grain yield of faba bean [87,93].
5. Conclusions
Long-term fertilization markedly reshaped soil nutrient status, enzyme activities, and microbial functional potential. Compared with chemical fertilization, organic fertilization significantly improved SOM, total and available nutrients, and achieved the highest faba bean yield. Urease, phosphatases, and neutral protease were key enzymatic indicators positively associated with soil fertility and productivity. Long-term fertilization also altered the abundance and coordination of N- and P-cycling functional genes. Genes related to assimilatory nitrate reduction and phosphate transport were strongly linked to higher soil nutrient availability and yield, while those involved in denitrification and anammox exhibited negative associations, reflecting microbial functional differentiation under different nutrient regimes. Overall, balanced application of organic and inorganic fertilizers effectively enhanced microbial nutrient cycling and soil biochemical functioning, offering an effective pathway for improving soil fertility and sustaining crop production in long-term agroecosystems.
It should be noted that soil sampling was conducted during a single crop season to ensure consistency across treatments. Although this approach effectively reflects the cumulative effects of long-term fertilization, it may not capture potential seasonal variations in microbial activity and enzyme dynamics. Future studies involving multi-seasonal sampling would provide further insight into temporal fluctuations of microbial functions under long-term fertilization regimes. Moreover, although Tax4Fun2 effectively predicts microbial functional potential, it does not directly reflect the active gene expression or enzymatic activity. Therefore, the observed relationships between predicted functional genes and soil enzyme activities should be interpreted as indicative of potential associations. Future studies employing metagenomic or metatranscriptomic analyses would provide more direct evidence of microbial functional dynamics under long-term fertilization treatments.
Author Contributions
Conceptualization, K.W. and A.H.; methodology, D.X., S.J. and N.Z.; software, S.J., M.Y. and E.Z.; validation, Y.W., F.D. and X.G.; formal analysis, D.X. and S.J.; investigation, S.J., Y.M. and K.W.; data curation, S.J., F.D. and X.G.; writing—original draft preparation, D.X. and S.J.; writing—review and editing, X.W., Y.M., L.W., K.W. and A.H.; visualization, D.X., S.J. and N.Z.; supervision, K.W. and A.H.; project administration, M.Y. and E.Z.; funding acquisition, X.W., A.H. and L.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Earmarked Fund for China Agriculture Research System (CARS-08-Z10), the Youth Fund of Natural Science Foundation of Nantong City (JC2023027), Jiangsu Province Seed Industry Revitalization Project Foundation (JBGS (2021)056), the Innovative and Entrepreneurial Talent Program of Jiangsu Province (JSSCRC2022469).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original data presented in the study are openly available in NCBI Sequence Read Archive database with the accession number PRJNA1118028.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| N | nitrogen |
| P | phosphorus |
| K | potassium |
| NPK | combined N, P, and K fertilization |
| M | organic fertilizer alone |
| MNPK | NPK plus organic fertilizer |
| EC | electrical conductivity |
| TN | total nitrogen |
| TP | total phosphorus |
| TC | total carbon |
| SOM | soil organic matter |
| AP | available phosphorus |
| AK | available potassium |
| GY | grain yield |
| Urease | soil urease |
| ACPro | soil acid protease |
| NEPro | soil neutral protease |
| ALPro | soil alkaline protease |
| ACP | soil acid phosphatase |
| NEP | soil neutral phosphatase |
| ALP | soil alkaline phosphatase |
Appendix A

Table A1.
Soil bacterial KEGG pathways that related to nitrogen and phosphorus cycles.
Table A1.
Soil bacterial KEGG pathways that related to nitrogen and phosphorus cycles.
| Nutrient Cycle | Function | KEGG Pathway | Corresponding Gene | Description |
|---|---|---|---|---|
| Nitrogen | Anammox | K00368 | nirK | nitrite reductase (NO-forming) [EC:1.7.2.1] |
| K01428 | ureC | urease subunit alpha [EC:3.5.1.5] | ||
| K01429 | ureB | urease subunit beta [EC:3.5.1.5] | ||
| K01430 | ureA | urease subunit gamma [EC:3.5.1.5] | ||
| K10535 | hao | hydroxylamine dehydrogenase [EC:1.7.2.6] | ||
| N fixation | K00531 | anfG | nitrogenase delta subunit [EC:1.18.6.1] | |
| K02586 | nifD | nitrogenase molybdenum-iron protein alpha chain [EC:1.18.6.1] | ||
| K02588 | nifH | nitrogenase iron protein NifH [EC:1.18.6.1] | ||
| K02591 | nifK | nitrogenase molybdenum-iron protein beta chain [EC:1.18.6.1] | ||
| K02592 | nifN | nitrogenase molybdenum-iron protein NifN | ||
| K02593 | nifT | nitrogen fixation protein NifT | ||
| K02594 | nifV | homocitrate synthase NifV [EC:2.3.3.14] | ||
| K02595 | nifW | nitrogenase-stabilizing/protective protein | ||
| K02596 | nifX | nitrogen fixation protein NifX | ||
| N mineralization | K00260 | gudB | glutamate dehydrogenase [EC:1.4.1.2] | |
| K00261 | GLUD1_2 | glutamate dehydrogenase (NAD(P)+) [EC:1.4.1.3] | ||
| K00262 | gdhA | glutamate dehydrogenase (NADP+) [EC:1.4.1.4] | ||
| N assimilation | K00265 | gltB | glutamate synthase (NADPH/NADH) large chain [EC:1.4.1.13 1.4.1.14] | |
| K00266 | gltD | glutamate synthase (NADPH/NADH) small chain [EC:1.4.1.13 1.4.1.14] | ||
| K00284 | GLU | glutamate synthase (ferredoxin) [EC:1.4.7.1] | ||
| K00459 | nmo | nitronate monooxygenase [EC:1.13.12.16] | ||
| K01424 | ansB | L-asparaginase [EC:3.5.1.1] | ||
| K01425 | glsA | glutaminase [EC:3.5.1.2] | ||
| K01953 | asnB | asparagine synthase (glutamine-hydrolysing) [EC:6.3.5.4] | ||
| Nitrification | K10535 | hao | hydroxylamine dehydrogenase [EC:1.7.2.6] | |
| K10944 | pmoA-amoA | methane/ammonia monooxygenase subunit A [EC:1.14.18.3 1.14.99.39] | ||
| K10945 | pmoB-amoB | methane/ammonia monooxygenase subunit B | ||
| K10946 | pmoC-amoC | methane/ammonia monooxygenase subunit C | ||
| Denitrification | K00368 | nirK | nitrite reductase (NO-forming) [EC:1.7.2.1] | |
| K00370 | narG, narZ, nxrA | nitrate reductase/nitrite oxidoreductase, alpha subunit [EC:1.7.5.1 1.7.99.-] | ||
| K00371 | narH, narY, nxrB | nitrate reductase/nitrite oxidoreductase, beta subunit [EC:1.7.5.1 1.7.99.-] | ||
| K00374 | narI, narV | nitrate reductase gamma subunit [EC:1.7.5.1 1.7.99.-] | ||
| K00376 | nosZ | nitrous-oxide reductase [EC:1.7.2.4] | ||
| K02305 | norC | nitric oxide reductase subunit C | ||
| K02567 | napA | periplasmic nitrate reductase NapA [EC:1.7.99.-] | ||
| K02568 | napB | cytochrome c-type protein NapB | ||
| K04561 | norB | nitric oxide reductase subunit B [EC:1.7.2.5] | ||
| K15864 | nirS | nitrite reductase (NO-forming)/hydroxylamine reductase [EC:1.7.2.1 1.7.99.1] | ||
| Assimilatory nitrate reduction | K00265 | gltB | glutamate synthase (NADPH/NADH) large chain [EC:1.4.1.13 1.4.1.14] | |
| K00360 | nasB | assimilatory nitrate reductase electron transfer subunit [EC:1.7.99.-] | ||
| K00366 | nirA | ferredoxin-nitrite reductase [EC:1.7.7.1] | ||
| K00367 | narB | ferredoxin-nitrate reductase [EC:1.7.7.2] | ||
| K00372 | nasA | assimilatory nitrate reductase catalytic subunit [EC:1.7.99.-] | ||
| K01915 | glnA, GLUL | glutamine synthetase [EC:6.3.1.2] | ||
| K02575 | NRT2, narK, nrtP, nasA | MFS transporter, NNP family, nitrate/nitrite transporter | ||
| K15577 | nrtB, nasE, cynB | nitrate/nitrite transport system permease protein | ||
| K15578 | nrtC, nasD | nitrate/nitrite transport system ATP-binding protein [EC:3.6.3.-] | ||
| K15579 | narC | nitrate/nitrite transport system ATP-binding protein | ||
| Dissimilatory nitrate reduction | K00362 | nirB | nitrite reductase (NADH) large subunit [EC:1.7.1.15] | |
| K00363 | nirD | nitrite reductase (NADH) small subunit [EC:1.7.1.15] | ||
| K00370 | narG, narZ, nxrA | nitrate reductase/nitrite oxidoreductase, alpha subunit [EC:1.7.5.1 1.7.99.-] | ||
| K00371 | narH, narY, nxrB | nitrate reductase/nitrite oxidoreductase, beta subunit [EC:1.7.5.1 1.7.99.-] | ||
| K00373 | narJ, narW | nitrate reductase molybdenum cofactor assembly chaperone NarJ/NarW | ||
| K00374 | narI, narV | nitrate reductase gamma subunit [EC:1.7.5.1 1.7.99.-] | ||
| K02567 | napA | periplasmic nitrate reductase NapA [EC:1.7.99.-] | ||
| K02568 | napB | cytochrome c-type protein NapB | ||
| K02569 | napC | cytochrome c-type protein NapC | ||
| K03385 | nrfA | nitrite reductase (cytochrome c-552) [EC:1.7.2.2] | ||
| K04013 | nrfB | cytochrome c-type protein NrfB | ||
| K04014 | nrfC | protein NrfC | ||
| K04015 | nrfD | protein NrfD | ||
| K15576 | nrtA, nasF, cynA | nitrate/nitrite transport system substrate-binding protein | ||
| K15876 | nrfH | cytochrome c nitrite reductase small subunit | ||
| Phosphorus | Polyphosphate polymerization | K00937 | ppk1 | polyphosphate kinase [EC:2.7.4.1] |
| K15986 | ppaC | manganese-dependent inorganic pyrophosphatase [EC:3.6.1.1] | ||
| Polyphosphate degradation | K00858 | ppnK, NADK | NAD+ kinase [EC:2.7.1.23] | |
| K00873 | PK, pyk | pyruvate kinase [EC:2.7.1.40] | ||
| K00886 | ppgK | polyphosphate glucokinase [EC:2.7.1.63] | ||
| K00940 | ndk, NME | nucleoside-diphosphate kinase [EC:2.7.4.6] | ||
| K00951 | relA | GTP pyrophosphokinase [EC:2.7.6.5] | ||
| K01139 | spoT | GTP diphosphokinase/guanosine-3,5-bis(diphosphate) 3-diphosphatase [EC:2.7.6.5 3.1.7.2] | ||
| K03787 | surE | 5-nucleotidase [EC:3.1.3.5] | ||
| K21138 | HDDC3 | guanosine-3,5-bis(diphosphate) 3-pyrophosphohydrolase [EC:3.1.7.2] | ||
| K22468 | ppk2 | polyphosphate kinase [EC:2.7.4.1] | ||
| P starvation response regulation | K02039 | phoU | phosphate transport system protein | |
| K07636 | phoR | two-component system, OmpR family, phosphate regulon sensor histidine kinase PhoR [EC:2.7.13.3] | ||
| K07657 | phoB | two-component system, OmpR family, phosphate regulon response regulator PhoB | ||
| K07658 | phoP | two-component system, OmpR family, alkaline phosphatase synthesis response regulator PhoP | ||
| K10916 | cqsS | two-component system, CAI-1 autoinducer sensor kinase/phosphatase CqsS [EC:2.7.13.3 3.1.3.-] | ||
| P uptake and transport system | K02036 | pstB | phosphate transport system ATP-binding protein [EC:3.6.3.27] | |
| K02037 | pstB | phosphate transport system permease protein | ||
| K02038 | pstA | phosphate transport system permease protein | ||
| K02040 | pstS | phosphate transport system substrate-binding protein | ||
| K02041 | phnC | phosphonate transport system ATP-binding protein [EC:3.6.3.28] | ||
| K02042 | phnE | phosphonate transport system permease protein | ||
| K02043 | phnF | GntR family transcriptional regulator, phosphonate transport system regulatory protein | ||
| K02044 | phnD | phosphonate transport system substrate-binding protein | ||
| K02440 | GLPF | glycerol uptake facilitator protein | ||
| K02443 | glpP | glycerol uptake operon antiterminator | ||
| K02444 | glpR | DeoR family transcriptional regulator, glycerol-3-phosphate regulon repressor | ||
| K02445 | glpT | MFS transporter, OPA family, glycerol-3-phosphate transporter | ||
| K02757 | bglF | PTS system, beta-glucoside-specific IIC component | ||
| K03306 | pit | inorganic phosphate transporter, PiT family | ||
| K03324 | yjbB | phosphate:Na+ symporter | ||
| K05781 | phnK | putative phosphonate transport system ATP-binding protein | ||
| K05813 | ugpB | sn-glycerol 3-phosphate transport system substrate-binding protein | ||
| K05814 | ugpA | sn-glycerol 3-phosphate transport system permease protein | ||
| K05815 | ugpE | sn-glycerol 3-phosphate transport system permease protein | ||
| K05833 | K05833 | putative ABC transport system ATP-binding protein | ||
| K07220 | K07220 | uncharacterized protein | ||
| K07221 | oprO_P | phosphate-selective porin OprO and OprP | ||
| K11082 | phnV | 2-aminoethylphosphonate transport system permease protein | ||
| K16322 | pit | low-affinity inorganic phosphate transporter | ||
| Inorganic P solubilization | K00112 | glpB | glycerol-3-phosphate dehydrogenase subunit B [EC:1.1.5.3] | |
| K00113 | glpC | glycerol-3-phosphate dehydrogenase subunit C [EC:1.1.5.3] | ||
| K00117 | gcd | quinoprotein glucose dehydrogenase [EC:1.1.5.2] | ||
| K01507 | ppa | inorganic pyrophosphatase [EC:3.6.1.1] | ||
| K01524 | ppx | exopolyphosphatase/guanosine-5-triphosphate,3-diphosphate pyrophosphatase [EC:3.6.1.11 3.6.1.40] | ||
| K06136 | pqqB | pyrroloquinoline quinone biosynthesis protein B | ||
| K06137 | pqqC | pyrroloquinoline-quinone synthase [EC:1.3.3.11] | ||
| K06138 | pqqD | pyrroloquinoline quinone biosynthesis protein D | ||
| K06139 | pqqE | pyrroloquinoline quinone biosynthesis protein E | ||
| Organic P mineralization | K00105 | E1.1.3.21 | alpha-glycerophosphate oxidase [EC:1.1.3.21] | |
| K00864 | glpK | glycerol kinase [EC:2.7.1.30] | ||
| K00906 | aceK | isocitrate dehydrogenase kinase/phosphatase [EC:2.7.11.5 3.1.3.-] | ||
| K01077 | E3.1.3.1, phoA, phoB | alkaline phosphatase [EC:3.1.3.1] | ||
| K01079 | serB | phosphoserine phosphatase [EC:3.1.3.3] | ||
| K01083 | E3.1.3.8 | 3-phytase [EC:3.1.3.8] | ||
| K01091 | gph | phosphoglycolate phosphatase [EC:3.1.3.18] | ||
| K01092 | IMPA | myo-inositol-1(or 4)-monophosphatase [EC:3.1.3.25] | ||
| K01093 | appA | 4-phytase/acid phosphatase [EC:3.1.3.26 3.1.3.2] | ||
| K01113 | phoD | alkaline phosphatase D [EC:3.1.3.1] | ||
| K01126 | E3.1.4.46, glpQ, ugpQ | glycerophosphoryl diester phosphodiesterase [EC:3.1.4.46] | ||
| K01841 | pepM | phosphoenolpyruvate phosphomutase [EC:5.4.2.9] | ||
| K02043 | phnF | GntR family transcriptional regulator, phosphonate transport system regulatory protein | ||
| K02203 | thrH | phosphoserine/homoserine phosphotransferase [EC:3.1.3.3 2.7.1.39] | ||
| K03270 | kdsC | 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase (KDO 8-P phosphatase) [EC:3.1.3.45] | ||
| K03430 | phnW | 2-aminoethylphosphonate-pyruvate transaminase [EC:2.6.1.37] | ||
| K03788 | aphA | acid phosphatase (class B) [EC:3.1.3.2] | ||
| K05306 | phnX | phosphonoacetaldehyde hydrolase [EC:3.11.1.1] | ||
| K05518 | rsbX | phosphoserine phosphatase RsbX [EC:3.1.3.3] | ||
| K05774 | phnN | ribose 1,5-bisphosphokinase [EC:2.7.4.23] | ||
| K05780 | phnL | alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnL [EC:2.7.8.37] | ||
| K05816 | ugpC | sn-glycerol 3-phosphate transport system ATP-binding protein [EC:3.6.3.20] | ||
| K06162 | phnM | alpha-D-ribose 1-methylphosphonate 5-triphosphate diphosphatase [EC:3.6.1.63] | ||
| K06163 | phnJ | alpha-D-ribose 1-methylphosphonate 5-phosphate C-P lyase [EC:4.7.1.1] | ||
| K06164 | phnI | alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnI [EC:2.7.8.37] | ||
| K06165 | phnH | alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnH [EC:2.7.8.37] | ||
| K06166 | phnG | alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnG [EC:2.7.8.37] | ||
| K06167 | phnP | phosphoribosyl 1,2-cyclic phosphate phosphodiesterase [EC:3.1.4.55] | ||
| K06193 | phnA | protein PhnA | ||
| K07048 | aceK | phosphotriesterase-related protein | ||
| K07175 | PhoH2 | PhoH-like ATPase | ||
| K07315 | rsbU_P | phosphoserine phosphatase RsbU/P [EC:3.1.3.3] | ||
| K08483 | ptsI | phosphotransferase system, enzyme I, PtsI [EC:2.7.3.9] | ||
| K08484 | ptsP | phosphotransferase system, enzyme I, PtsP [EC:2.7.3.9] | ||
| K09474 | phoN | acid phosphatase (class A) [EC:3.1.3.2] | ||
| K09994 | phnO | aminoalkylphosphonate N-acetyltransferase [EC:2.3.1.-] | ||
| K15781 | serB-plsC | putative phosphoserine phosphatase/1-acylglycerol-3-phosphate O-acyltransferase [EC:3.1.3.3 2.3.1.51] | ||
| K16055 | TPS | trehalose 6-phosphate synthase/phosphatase [EC:2.4.1.15 3.1.3.12] |
References
- Zhang, X.Y.; Dong, W.Y.; Dai, X.Q.; Schaeffer, S.; Yang, F.T.; Radosevich, M.; Xu, L.L.; Liu, X.Y.; Sun, X.M. Responses of absolute and specific soil enzyme activities to long term additions of organic and mineral fertilizer. Sci. Total Environ. 2015, 536, 59–67. [Google Scholar] [CrossRef]
- Tabatabai, M.A.; Fu, M. Extraction of enzymes from soils. In Soil Biochemistry; Strotzky, G., Bollag, J.M., Eds.; CRC Press: New York, NY, USA, 1992; Volume 7, pp. 197–227. [Google Scholar] [CrossRef]
- Pausch, J.; Kuzyakov, Y. Carbon input by roots into the soil: Quantification of rhizodeposition from root to ecosystem scale. Glob. Change Biol. 2018, 24, 1–12. [Google Scholar] [CrossRef]
- Burns, R.G. Enzyme activity in soil: Location and a possible role in microbial ecology. Soil Biol. Biochem. 1982, 14, 423–427. [Google Scholar] [CrossRef]
- Sun, X.D.; Ye, Y.Q.; Ma, Q.X.; Guan, Q.W.; Jones, D.L. Variation in enzyme activities involved in carbon and nitrogen cycling in rhizosphere and bulk soil after organic mulching. Rhizosphere 2021, 19, 100376. [Google Scholar] [CrossRef]
- Neemisha; Sharma, S. Soil enzymes and their role in nutrient cycling. In Structure and Functions of Pedosphere, 1st ed.; Giri, B., Kapoor, R., Eds.; Springer Nature: Singapore, 2022; pp. 173–188. [Google Scholar] [CrossRef]
- Roldán, A.; Salinas-García, J.R.; Alguacil, M.M.; Díaz, E.; Caravaca, F. Soil enzyme activities suggest advantages of conservation tillage practices in sorghum cultivation under subtropical conditions. Geoderma 2005, 129, 178–185. [Google Scholar] [CrossRef]
- Jat, M.L.; Chakraborty, D.; Ladha, J.K.; Rana, D.S.; Gathala, M.K.; McDonald, A.; Gerard, B. Conservation agriculture for sustainable intensification in South Asia. Nat. Sustain. 2020, 3, 336–343. [Google Scholar] [CrossRef]
- Jin, K.; Sleutel, S.; Buchan, D.; De Neve, S.; Cai, D.X.; Gabriels, D.; Jin, J.Y. Changes of soil enzyme activities under different tillage practices in the Chinese Loess Plateau. Soil Tillage Res. 2009, 104, 115–120. [Google Scholar] [CrossRef]
- He, L.Y.; Lu, S.X.; Wang, C.G.; Mu, J.; Zhang, Y.L.; Wang, X.D. Changes in soil organic carbon fractions and enzyme activities in response to tillage practices in the Loess Plateau of China. Soil Tillage Res. 2021, 209, 104940. [Google Scholar] [CrossRef]
- Yang, L.; Wang, L.H.; Chu, J.C.; Zhao, H.L.; Zhao, J.; Zang, H.D.; Yang, Y.D.; Zeng, Z.H. Improving soil quality and wheat yield through diversified crop rotations in the North China Plain. Soil Tillage Res. 2024, 244, 106231. [Google Scholar] [CrossRef]
- Li, M.H.; Guo, J.J.; Ren, T.; Luo, G.W.; Shen, Q.R.; Lu, J.W.; Guo, S.W.; Ling, N. Crop rotation history constrains soil biodiversity and multifunctionality relationships. Agric. Ecosyst. Environ. 2021, 319, 107550. [Google Scholar] [CrossRef]
- Muhammad, I.; Yang, L.; Ahmad, S.; Zeeshan, M.; Farooq, S.; Ali, I.; Khan, A.; Zhou, X.B. Irrigation and nitrogen fertilization alter soil bacterial communities, soil enzyme activities, and nutrient availability in maize crop. Front. Microbiol. 2022, 13, 833758. [Google Scholar] [CrossRef]
- Liu, B.; Xia, H.; Jiang, C.C.; Riaz, M.; Yang, L.; Chen, Y.F.; Fan, X.P.; Xia, X.G. 14 year applications of chemical fertilizers and crop straw effects on soil labile organic carbon fractions, enzyme activities and microbial community in rice-wheat rotation of middle China. Sci. Total Environ. 2022, 841, 156608. [Google Scholar] [CrossRef]
- Miao, Y.X.; Stewart, B.A.; Zhang, F.S. Long-term experiments for sustainable nutrient management in China. A review. Agron. Sustain. Dev. 2011, 31, 397–414. [Google Scholar] [CrossRef]
- Bai, Y.C.; Chang, Y.Y.; Hussain, M.; Lu, B.; Zhang, J.P.; Song, X.B.; Lei, X.S.; Pei, D. Soil chemical and microbiological properties are changed by long-term chemical fertilizers that limit ecosystem functioning. Microorganisms 2020, 8, 694. [Google Scholar] [CrossRef]
- Fan, T.L.; Stewart, B.A.; Wang, Y.; Luo, J.J.; Zhou, G.Y. Long-term fertilization effects on grain yield, water-use efficiency and soil fertility in the dryland of Loess Plateau in China. Agric. Ecosyst. Environ. 2005, 106, 313–329. [Google Scholar] [CrossRef]
- Wang, S.X.; Liang, X.Q.; Chen, Y.X.; Luo, Q.X.; Liang, W.S.; Li, S.; Huang, C.L.; Li, Z.Z.; Wan, L.L.; Li, W.; et al. Phosphorus loss potential and phosphatase activity under phosphorus fertilization in long-term paddy wetland agroecosystems. Soil Sci. Soc. Am. J. 2012, 7, 161–167. [Google Scholar] [CrossRef]
- Wang, J.; Fu, X.; Ghimire, R.; Sainju, U.M.; Jia, Y.; Zhao, F.Z. Responses of soil bacterial community and enzyme activity to organic matter components under long-term fertilization on the loess plateau of China. Appl. Soil Ecol. 2021, 166, 103992. [Google Scholar] [CrossRef]
- Ren, J.H.; Liu, X.L.; Yang, W.P.; Yang, X.X.; Li, W.G.; Xia, Q.; Li, J.H.; Gao, Z.Q.; Yang, Z.P. Rhizosphere soil properties, microbial community, and enzyme activities: Short-term responses to partial substitution of chemical fertilizer with organic manure. J. Environ. Manag. 2021, 299, 113650. [Google Scholar] [CrossRef]
- Han, J.Q.; Dong, Y.Y.; Zhang, M. Chemical fertilizer reduction with organic fertilizer effectively improve soil fertility and microbial community from newly cultivated land in the Loess Plateau of China. Appl. Soil Ecol. 2021, 165, 103966. [Google Scholar] [CrossRef]
- Gianfreda, L.; Rao, M.A. Interactions between xenobiotics and microbial and enzymatic soil activity. Crit. Rev. Environ. Sci. Technol. 2008, 38, 269–310. [Google Scholar] [CrossRef]
- Wang, H.X.; Xu, J.L.; Liu, X.J.; Zhang, D.; Li, L.W.; Li, W.; Sheng, L.X. Effects of long-term application of organic fertilizer on improving organic matter content and retarding acidity in red soil from China. Soil Tillage Res. 2019, 195, 104382. [Google Scholar] [CrossRef]
- Wei, W.L.; Yan, Y.; Cao, J.; Christie, P.; Zhang, F.S.; Fan, M.S. Effects of combined application of organic amendments and fertilizers on crop yield and soil organic matter: An integrated analysis of long-term experiments. Agric. Ecosyst. Environ. 2016, 225, 86–92. [Google Scholar] [CrossRef]
- Liu, J.A.; Shu, A.P.; Song, W.F.; Shi, W.C.; Li, M.C.; Zhang, W.X.; Li, Z.Z.; Liu, G.R.; Yuan, F.S.; Zhang, S.X.; et al. Long-term organic fertilizer substitution increases rice yield by improving soil properties and regulating soil bacteria. Geoderma 2021, 404, 115287. [Google Scholar] [CrossRef]
- Geisseler, D.; Horwath, W.R. Regulation of extracellular protease activity in soil in response to different sources and concentrations of nitrogen and carbon. Soil Biol. Biochem. 2008, 40, 3040–3048. [Google Scholar] [CrossRef]
- Xu, H.W.; Qu, Q.; Chen, Y.H.; Liu, G.B.; Xue, S. Responses of soil enzyme activity and soil organic carbon stability over time after cropland abandonment in different vegetation zones of the Loess Plateau of China. Catena 2021, 196, 104812. [Google Scholar] [CrossRef]
- Bhattacharyya, S.S.; Ros, G.H.; Furtak, K.; Iqbal, H.M.N.; Parra-Saldívar, R. Soil carbon sequestration–An interplay between soil microbial community and soil organic matter dynamics. Sci. Total Environ. 2022, 815, 152928. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.L.; Zhang, R.Q.; Zhang, C.Q.; Su, J.E.; Cong, W.F.; Deng, X.P. Long-term organic fertilizer additions elevate soil extracellular enzyme activities and tobacco quality in a tobacco-maize rotation. Front. Plant Sci. 2022, 13, 973639. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.H.; Liu, H.; Wu, J.C.; Zhang, S.S.; Gao, C.M.; Zhang, S.Q.; Tang, D.W.S. Soil enzyme activities, soil physical properties, photosynthetic physical characteristics and water use of winter wheat after long-term straw mulch and organic fertilizer application. Front. Plant Sci. 2023, 14, 1186376. [Google Scholar] [CrossRef]
- Bernard, L.; Basile-Doelsch, I.; Derrien, D.; Fanin, N.; Fontaine, S.; Guenet, B.; Karimi, B.; Marsden, C.; Maron, P.A. Advancing the mechanistic understanding of the priming effect on soil organic matter mineralisation. Funct. Ecol. 2022, 36, 1355–1377. [Google Scholar] [CrossRef]
- Tian, P.; Razavi, B.S.; Zhang, X.C.; Wang, Q.K.; Blagodatskaya, E. Microbial growth and enzyme kinetics in rhizosphere hotspots are modulated by soil organics and nutrient availability. Soil Biol. Biochem. 2020, 141, 107662. [Google Scholar] [CrossRef]
- Xiao, H.; Yang, H.L.; Zhao, M.L.; Monaco, T.A.; Rong, Y.P.; Huang, D.; Song, Q.; Zhao, K.; Wang, D.P. Soil extracellular enzyme activities and the abundance of nitrogen-cycling functional genes responded more to N addition than P addition in an Inner Mongolian meadow steppe. Sci. Total Environ. 2021, 759, 143541. [Google Scholar] [CrossRef] [PubMed]
- Chubukov, V.; Gerosa, L.; Kochanowski, K.; Sauer, U. Coordination of microbial metabolism. Nat. Rev. Microbiol. 2014, 12, 327–340. [Google Scholar] [CrossRef]
- Reeve, J.R.; Schadt, C.W.; Carpenter-Boggs, L.; Kang, S.; Zhou, J.Z.; Reganold, J.P. Effects of soil type and farm management on soil ecological functional genes and microbial activities. ISME J. 2010, 4, 1099–1107. [Google Scholar] [CrossRef]
- Dong, M.H.; Zhou, H.J.; Wang, J.; Yang, J.H.; Lai, J.Z.; Chen, Y.L.; Sun, F.; Ye, X.F.; Wu, Y.J. Responses of soil microbial metabolism, function and soil quality to long-term addition of organic materials with different carbon sources. Biochar 2024, 6, 80. [Google Scholar] [CrossRef]
- Li, W.X.; Wang, C.; Zheng, M.M.; Cai, Z.J.; Wang, B.R.; Shen, R.F. Fertilization strategies affect soil properties and abundance of N-cycling functional genes in an acidic agricultural soil. Appl. Soil Ecol. 2020, 156, 103704. [Google Scholar] [CrossRef]
- Li, Y.L.; Tremblay, J.; Bainard, L.D.; Cade-Menun, B.; Hamel, C. Long-term effects of nitrogen and phosphorus fertilization on soil microbial community structure and function under continuous wheat production. Environ. Microbiol. 2020, 22, 1066–1088. [Google Scholar] [CrossRef]
- Ying, D.; Chen, X.L.; Hou, J.F.; Zhao, F.C.; Li, P. Soil properties and microbial functional attributes drive the response of soil multifunctionality to long-term fertilization management. Appl. Soil Ecol. 2023, 192, 105095. [Google Scholar] [CrossRef]
- Abd-Alla, M.H.; Al-Amri, S.M.; El-Enany, A.W.E. Enhancing rhizobium–legume symbiosis and reducing nitrogen fertilizer use are potential options for mitigating climate change. Agriculture 2023, 13, 2092. [Google Scholar] [CrossRef]
- Chen, X.D.; Jiang, N.; Chen, Z.H.; Tian, J.H.; Sun, N.; Xu, M.G.; Chen, L.J. Response of soil phoD phosphatase gene to long-term combined applications of chemical fertilizers and organic materials. Appl. Soil Ecol. 2017, 119, 197–204. [Google Scholar] [CrossRef]
- Xing, Y.Y.; Xie, Y.X.; Wang, X.K. Enhancing soil health through balanced fertilization: A pathway to sustainable agriculture and food security. Front. Microbiol. 2025, 16, 1536524, Erratum in Front. Microbiol. 2025, 16, 1644143. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.C.; Zhang, Y.J.; Turner, B.L.; He, Y.L.; Chen, X.D.; Che, R.X.; Cui, X.Y.; Liu, X.J.; Jiang, L.; Zhu, J.T. Organic amendments promote soil phosphorus related functional genes and microbial phosphorus cycling. Geoderma 2025, 456, 117247. [Google Scholar] [CrossRef]
- Du, L.; Zhong, H.H.; Guo, X.N.; Li, H.N.; Xia, J.X.; Chen, Q. Nitrogen fertilization and soil nitrogen cycling: Unraveling the links among multiple environmental factors, functional genes, and transformation rates. Sci. Total Environ. 2024, 951, 175561. [Google Scholar] [CrossRef] [PubMed]
- Reardon, C.L.; Klein, A.M.; Melle, C.J.; Hagerty, C.H.; Klarer, E.R.; Machado, S.; Paulitz, T.; Pritchett, L.; Schlatter, D.; Smith, S.F.; et al. Enzyme activities distinguish long-term fertilizer effects under different soil storage methods. Appl. Soil Ecol. 2022, 177, 104518. [Google Scholar] [CrossRef]
- GB9834-88; Method for Determination of Soil Organic Matter. China Standard Press: Beijing, China, 1988.
- Wemheuer, F.; Taylor, J.A.; Daniel, R.; Johnston, E.; Meinicke, P.; Thomas, T.; Wemheuer, B. Tax4Fun2: Prediction of habitat-specific functional profiles and functional redundancy based on 16S rRNA gene sequences. Environ. Microbiome 2020, 15, 11. [Google Scholar] [CrossRef]
- Hu, H.; Wu, X.; Zhao, J.N.; Yang, D.L.; Wang, L.L.; Li, G.; Xiu, W.M. The effects of combined organic and inorganic fertilizer on the bacterial nitrogen cycling functional genes in wheat and maize soils by PICRUSt functional prediction. J. Agro-Environ. Sci. 2021, 40, 144–154. [Google Scholar] [CrossRef]
- Dai, Z.M.; Liu, G.F.; Chen, H.H.; Chen, C.R.; Wang, J.K.; Ai, S.Y.; Wei, D.; Li, D.M.; Ma, B.; Tang, C.X.; et al. Long-term nutrient inputs shift soil microbial functional profiles of phosphorus cycling in diverse agroecosystems. ISME J. 2020, 14, 757–770. [Google Scholar] [CrossRef]
- Krajewska, B.; Ureases, I. Functional, catalytic and kinetic properties: A review. J. Mol. Catal. B Enzym. 2009, 59, 9–21. [Google Scholar] [CrossRef]
- Sigua, G.C.; Stone, K.C.; Bauer, P.J.; Szogi, A.A. Efficacy of supplemental irrigation and nitrogen management on enhancing nitrogen availability and urease activity in soils with sorghum production. Sustainability 2020, 12, 8358. [Google Scholar] [CrossRef]
- Nourbakhsh, F.; Monreal, C.M. Effects of soil properties and trace metals on urease activities of calcareous soils. Biol. Fertil. Soils 2004, 40, 359–362. [Google Scholar] [CrossRef]
- Zhu, Y.; Guo, B.; Liu, C.; Lin, Y.C.; Fu, Q.L.; Li, N.Y.; Li, H. Soil fertility, enzyme activity, and microbial community structure diversity among different soil textures under different land use types in coastal saline soil. J. Soils Sediments 2021, 21, 2240–2252. [Google Scholar] [CrossRef]
- Moghimian, N.; Hosseini, S.M.; Kooch, Y.; Darki, B.Z. Impacts of changes in land use/cover on soil microbial and enzyme activities. Catena 2017, 157, 407–414. [Google Scholar] [CrossRef]
- Yin, H.J.; Li, Y.F.; Xiao, J.; Xu, Z.F.; Cheng, X.Y.; Liu, Q. Enhanced root exudation stimulates soil nitrogen transformations in a subalpine coniferous forest under experimental warming. Glob. Change Biol. 2013, 19, 2158–2167. [Google Scholar] [CrossRef]
- Miao, F.H.; Li, Y.; Cui, S.; Jagadamma, S.; Yang, G.F.; Zhang, Q.P. Soil extracellular enzyme activities under long-term fertilization management in the croplands of China: A meta-analysis. Nutr. Cycl. Agroecosys. 2019, 114, 125–138. [Google Scholar] [CrossRef]
- Ning, C.C.; Gao, P.D.; Wang, B.Q.; Lin, W.P.; Jiang, N.H.; Cai, K.Z. Impacts of chemical fertilizer reduction and organic amendments supplementation on soil nutrient, enzyme activity and heavy metal content. J. Integr. Agric. 2017, 16, 1819–1831. [Google Scholar] [CrossRef]
- Yang, Y.H.; Li, M.J.; Wu, J.C.; Pan, X.Y.; Gao, C.M.; Tang, D.W.S. Impact of combining long-term subsoiling and organic fertilizer on soil microbial biomass carbon and nitrogen, soil enzyme activity, and water use of winter wheat. Front. Plant Sci. 2022, 12, 788651. [Google Scholar] [CrossRef]
- Ullah, S.; Ali, I.; Yang, M.; Zhao, Q.; Iqbal, A.; Wu, X.Y.; Ahmad, S.; Muhammad, I.; Khan, A.; Adnan, M.; et al. Partial substitution of urea with biochar induced improvements in soil enzymes activity, ammonia-nitrite oxidizers, and nitrogen uptake in the double-cropping rice system. Microorganisms 2023, 11, 527. [Google Scholar] [CrossRef] [PubMed]
- Janes-Bassett, V.; Blackwell, M.S.A.; Blair, G.; Davies, J.; Haygarth, P.M.; Mezeli, M.M.; Stewart, G. A meta-analysis of phosphatase activity in agricultural settings in response to phosphorus deficiency. Soil Biol. Biochem. 2022, 165, 108537. [Google Scholar] [CrossRef]
- Hui, D.F.; Mayes, M.A.; Wang, G.S. Kinetic parameters of phosphatase: A quantitative synthesis. Soil Biol. Biochem. 2013, 65, 105–113. [Google Scholar] [CrossRef]
- Garg, S.; Bahl, G.S. Phosphorus availability to maize as influenced by organic manures and fertilizer P associated phosphatase activity in soils. Bioresour. Technol. 2008, 99, 5773–5777. [Google Scholar] [CrossRef]
- Gao, C.H.; El-Sawah, A.M.; Ali, D.F.I.; Alhaj Hamoud, Y.; Shaghaleh, H.; Sheteiwy, M.S. The integration of bio and organic fertilizers improve plant growth, grain yield, quality and metabolism of hybrid maize (Zea mays L.). Agronomy 2020, 10, 319. [Google Scholar] [CrossRef]
- Yang, Q.Y.; Zhang, M.K. Effect of bio-organic fertilizers partially substituting chemical fertilizers on labile organic carbon and bacterial community of citrus orchard soils. Plant Soil 2023, 483, 255–272. [Google Scholar] [CrossRef]
- Hu, N.J.; Liu, C.Y.; Chen, Q.; Fan, J.D.; Wang, Y.T.; Sun, H.W. Substitution of chemical fertilizer with organic fertilizer can affect soil labile organic carbon fractions and garlic yield by mediating soil aggregate-associated organic carbon. Agronomy 2023, 13, 3062. [Google Scholar] [CrossRef]
- Martins, N.P.; Fuchslueger, L.; Fleischer, K.; Andersen, K.M.; Assis, R.L.; Baccaro, F.B.; Camargo, P.B.; Cordeiro, A.L.; Grandis, A.; Hartley, I.P.; et al. Fine roots stimulate nutrient release during early stages of leaf litter decomposition in a Central Amazon rainforest. Plant Soil 2021, 469, 287–303. [Google Scholar] [CrossRef]
- Furtak, K.; Gawryjołek, K.; Gajda, A.M.; Gałązka, A. Effects of maize and winter wheat grown under different cultivation techniques on biological activity of soil. Plant Soil Environ. 2017, 63, 449–454. [Google Scholar] [CrossRef]
- Gao, P.; Zhang, T.; Lei, X.Y.; Cui, X.W.; Lu, Y.X.; Fan, P.F.; Long, S.P.; Huang, J.; Gao, J.S.; Zhang, Z.H.; et al. Improvement of soil fertility and rice yield after long-term application of cow manure combined with inorganic fertilizers. J. Integr. Agric. 2023, 22, 2221–2232. [Google Scholar] [CrossRef]
- Pan, W.K.; Zhou, J.J.; Tang, S.; Wu, L.H.; Ma, Q.X.; Marsden, K.A.; Chadwick, D.R.; Jones, D.L. Utilisation and transformation of organic and inorganic nitrogen by soil microorganisms and its regulation by excessive carbon and nitrogen availability. Biol. Fertil. Soils 2023, 59, 379–389. [Google Scholar] [CrossRef]
- Luo, M.; Moorhead, D.L.; Ochoa-Hueso, R.; Mueller, C.W.; Ying, S.C.; Chen, J. Nitrogen loading enhances phosphorus limitation in terrestrial ecosystems with implications for soil carbon cycling. Funct. Ecol. 2022, 36, 2845–2858. [Google Scholar] [CrossRef]
- Lv, C.H.; Wang, C.K.; Cai, A.D.; Zhou, Z.H. Global magnitude of rhizosphere effects on soil microbial communities and carbon cycling in natural terrestrial ecosystems. Sci. Total Environ. 2023, 856, 158961. [Google Scholar] [CrossRef]
- Mooshammer, M.; Wanek, W.; Hämmerle, I.; Fuchslueger, L.; Hofhansl, F.; Knoltsch, A.; Schnecker, J.; Takriti, M.; Watzka, M.; Wild, B.; et al. Adjustment of microbial nitrogen use efficiency to carbon:nitrogen imbalances regulates soil nitrogen cycling. Nat. Commun. 2014, 5, 3694. [Google Scholar] [CrossRef]
- Shao, G.D.; Xu, Y.X.; Zhou, J.; Tian, P.; Ai, J.J.; Yang, Y.D.; Zamanian, K.; Zeng, Z.H.; Zang, H.D. Enhanced soil organic carbon stability in rhizosphere through manure application. Soil Tillage Res. 2024, 244, 106223. [Google Scholar] [CrossRef]
- Qiu, S.J.; Ju, X.T.; Ingwersen, J.; Guo, Z.D.; Stange, C.F.; Bisharat, R.; Christie, P.; Zhang, F.S. Role of carbon substrates added in the transformation of surplus nitrate to organic nitrogen in a calcareous soil. Pedosphere 2013, 23, 205–212. [Google Scholar] [CrossRef]
- Zhang, J.B.; Zhu, T.B.; Cai, Z.C.; Qin, S.W.; Müller, C. Effects of long-term repeated mineral and organic fertilizer applications on soil nitrogen transformations. Eur. J. Soil Sci. 2012, 63, 75–85. [Google Scholar] [CrossRef]
- Pan, W.K.; Tang, S.; Zhou, J.J.; Wanek, W.; Gregory, A.S.; Ge, T.; Marsden, K.A.; Chadwick, D.R.; Liang, Y.C.; Wu, L.H.; et al. Long-term manure and mineral fertilisation drive distinct pathways of soil organic nitrogen decomposition: Insights from a 180-year-old study. Soil Biol. Biochem. 2025, 207, 109840. [Google Scholar] [CrossRef]
- Giles, M.; Morley, N.; Baggs, E.M.; Daniell, T.J. Soil nitrate reducing processes–drivers, mechanisms for spatial variation, and significance for nitrous oxide production. Front. Microbiol. 2012, 3, 407. [Google Scholar] [CrossRef]
- Duan, Y.H.; Xu, M.G.; Gao, S.D.; Liu, H.; Huang, S.M.; Wang, B.R. Long-term incorporation of manure with chemical fertilizers reduced total nitrogen loss in rain-fed cropping systems. Sci. Rep. 2016, 6, 33611. [Google Scholar] [CrossRef]
- Duan, Y.H.; Shi, X.J.; Li, S.L.; Sun, X.F.; He, X.H. Nitrogen use efficiency as affected by phosphorus and potassium in long-term rice and wheat experiments. J. Integr. Agric. 2014, 13, 588–596. [Google Scholar] [CrossRef]
- Su, J.Q.; Ding, L.J.; Xue, K.; Yao, H.Y.; Quensen, J.; Bai, S.J.; Wei, W.X.; Wu, J.S.; Zhou, J.Z.; Tiedje, J.M.; et al. Long-term balanced fertilization increases the soil microbial functional diversity in a phosphorus-limited paddy soil. Mol. Ecol. 2015, 24, 136–150. [Google Scholar] [CrossRef]
- Yang, W.; Jiang, Y.; Zhang, J.Q.; Wang, W.; Liu, X.S.; Jin, Y.; Li, S.; Qu, J.J.; Zhu, Y.C. Organic materials promote soil phosphorus cycling: Metagenomic analysis. Agronomy 2025, 15, 1693. [Google Scholar] [CrossRef]
- Deubel, A.; Merbach, W. Influence of microorganisms on phosphorus bioavailability in soils. In Microorganisms in Soils: Roles in Genesis and Functions; Varma, A., Buscot, F., Eds.; Springer Nature: Berlin/Heidelberg, Germany, 2005; Volume 3, pp. 177–191. [Google Scholar] [CrossRef]
- Huang, Y.L.; Lin, J.H.; Tang, C.X.; Xu, J.M. Organic carbon inputs shift the profiles of phosphorus cycling-related genes in maize rhizosphere. Plant Soil 2024, 503, 595–609. [Google Scholar] [CrossRef]
- Shaji, H.; Chandran, V.; Mathew, L. Organic fertilizers as a route to controlled release of nutrients. In Controlled Release Fertilizers for Sustainable Agriculture; Lewu, F.B., Volova, T., Eds.; Elsevier: Amsterdam, The Netherlands, 2021; pp. 231–245. [Google Scholar] [CrossRef]
- Rasul, M.; Yasmin, S.; Yahya, M.; Breitkreuz, C.; Tarkka, M.; Reitz, T. The wheat growth-promoting traits of Ochrobactrum and Pantoea species, responsible for solubilization of different P sources, are ensured by genes encoding enzymes of multiple P-releasing pathways. Microbiol. Res. 2021, 246, 126703. [Google Scholar] [CrossRef]
- Tian, J.; Ge, F.; Zhang, D.Y.; Deng, S.Q.; Liu, X.W. Roles of phosphate solubilizing microorganisms from managing soil phosphorus deficiency to mediating biogeochemical P cycle. Biology 2021, 10, 158. [Google Scholar] [CrossRef] [PubMed]
- Pang, F.; Li, Q.; Solanki, M.K.; Wang, Z.; Xing, Y.X.; Dong, D.F. Soil phosphorus transformation and plant uptake driven by phosphate-solubilizing microorganisms. Front. Microbiol. 2024, 15, 1383813. [Google Scholar] [CrossRef] [PubMed]
- Allison, S.D.; Chacon, S.S.; German, D.P. Substrate concentration constraints on microbial decomposition. Soil Biol. Biochem. 2014, 79, 43–49. [Google Scholar] [CrossRef]
- Fraser, T.D.; Lynch, D.H.; Gaiero, J.; Khosla, K.; Dunfield, K.E. Quantification of bacterial non-specific acid (phoC) and alkaline (phoD) phosphatase genes in bulk and rhizosphere soil from organically managed soybean fields. Appl. Soil Ecol. 2017, 111, 48–56. [Google Scholar] [CrossRef]
- Alori, E.T.; Glick, B.R.; Babalola, O.O. Microbial phosphorus solubilization and its potential for use in sustainable agriculture. Front. Microbiol. 2017, 8, 971. [Google Scholar] [CrossRef]
- Chen, A.; Arai, Y. A review of the reactivity of phosphatase controlled by clays and clay minerals: Implications for understanding phosphorus mineralization in soils. Clays Clay Miner. 2023, 71, 119–142. [Google Scholar] [CrossRef]
- Ramos Cabrera, E.V.; Delgado Espinosa, Z.Y.; Solis Pino, A.F. Use of phosphorus-solubilizing microorganisms as a biotechnological alternative: A review. Microorganisms 2024, 12, 1591. [Google Scholar] [CrossRef]
- Ding, W.; Cong, W.F.; Lambers, H. Plant phosphorus-acquisition and-use strategies affect soil carbon cycling. Trends Ecol. Evol. 2021, 36, 899–906. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).