Decoding the Gut Microbiome in Primary Sjögren’s Syndrome and Primary Biliary Cholangitis: Shared Dysbiosis, Distinct Patterns, and Associations with Clinical Features
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Group
2.2. DNA Extraction, 16S rRNA Gene Amplicon Sequencing and Sequence Analysis
2.3. Bioinformatics and Statistical Analysis
2.4. Statistical Analyses
3. Results
3.1. Clinical Indicators of PBC, pSS, and HCs
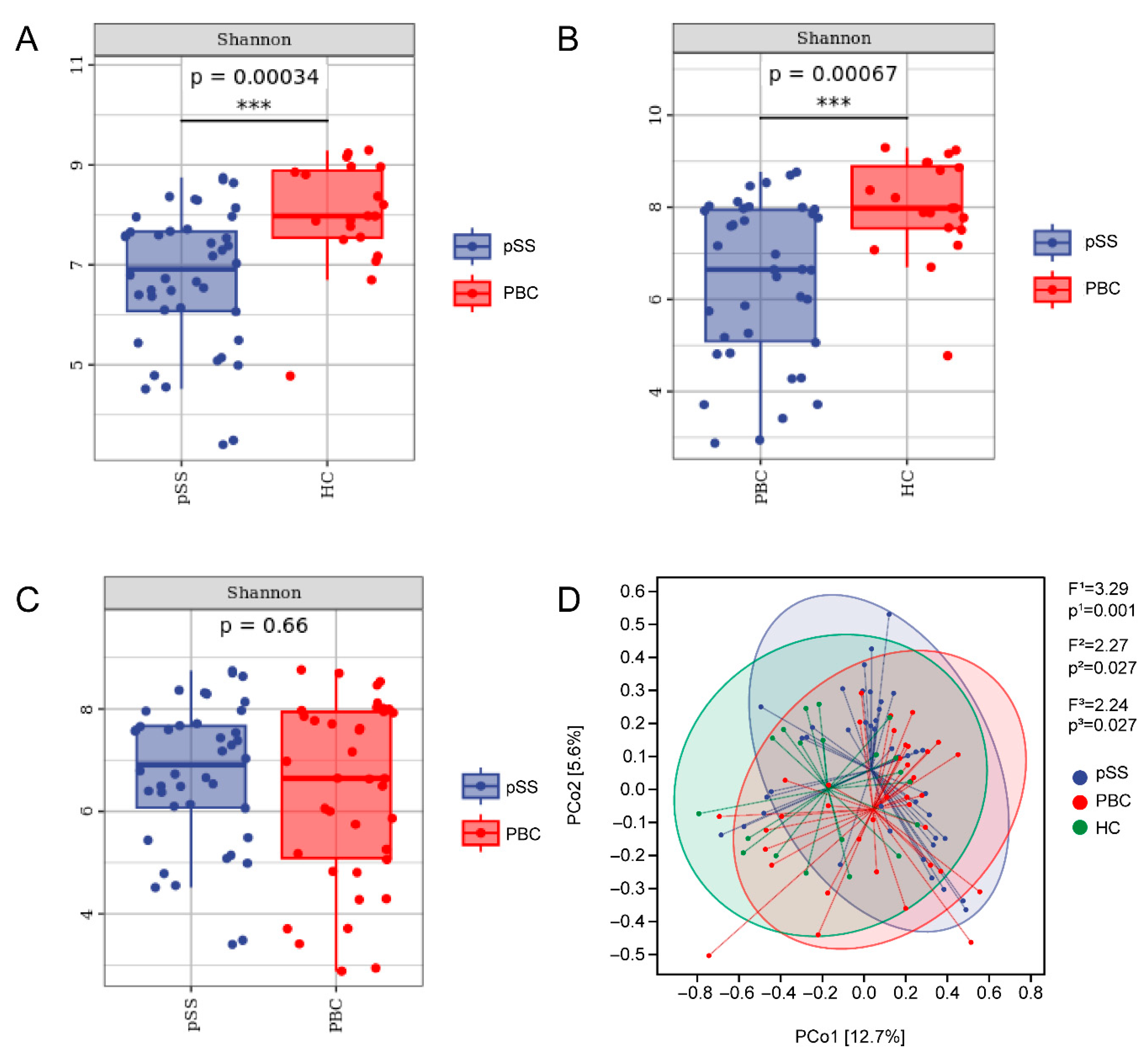
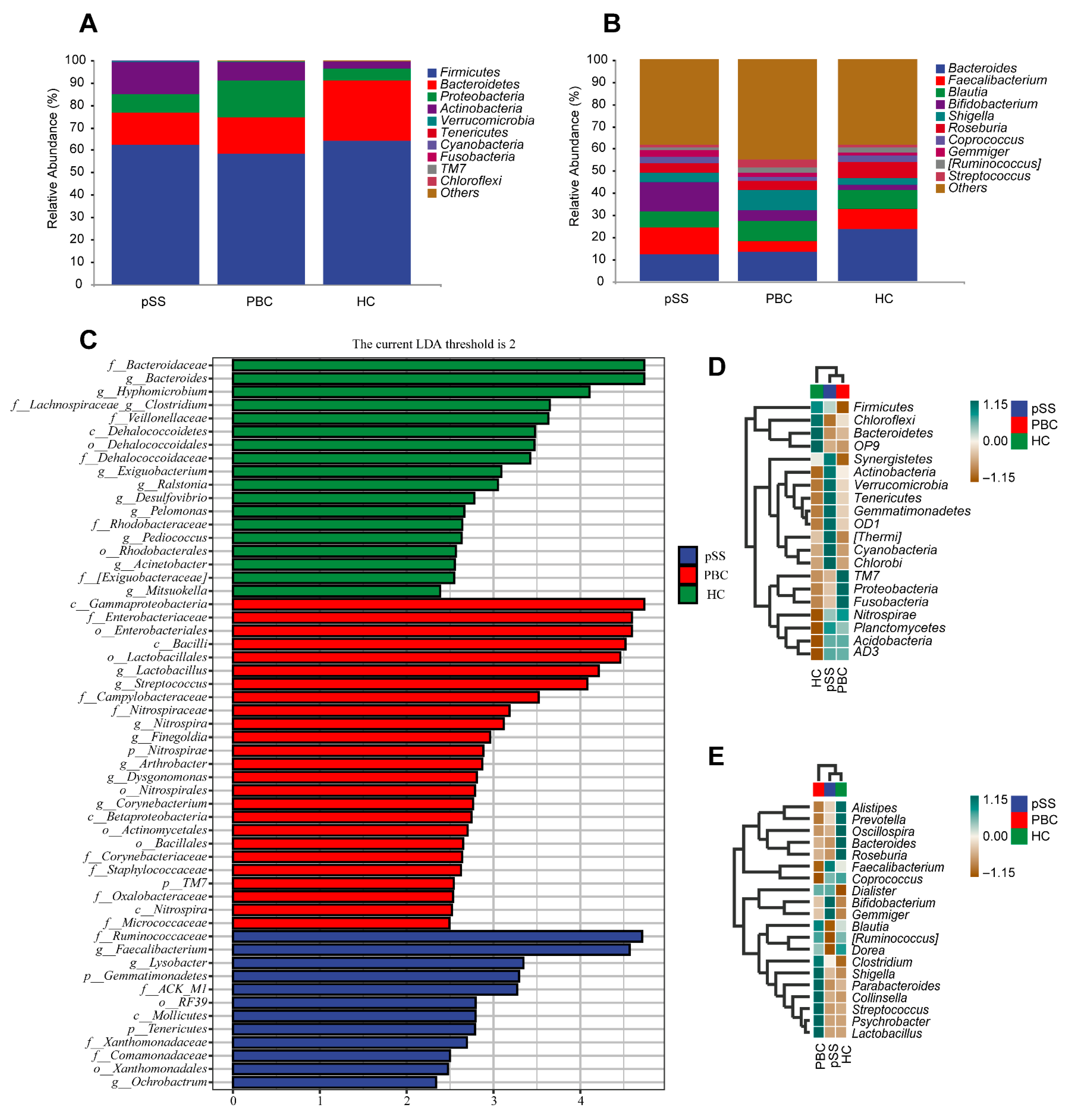
3.2. Differences in Gut Microbiome Composition
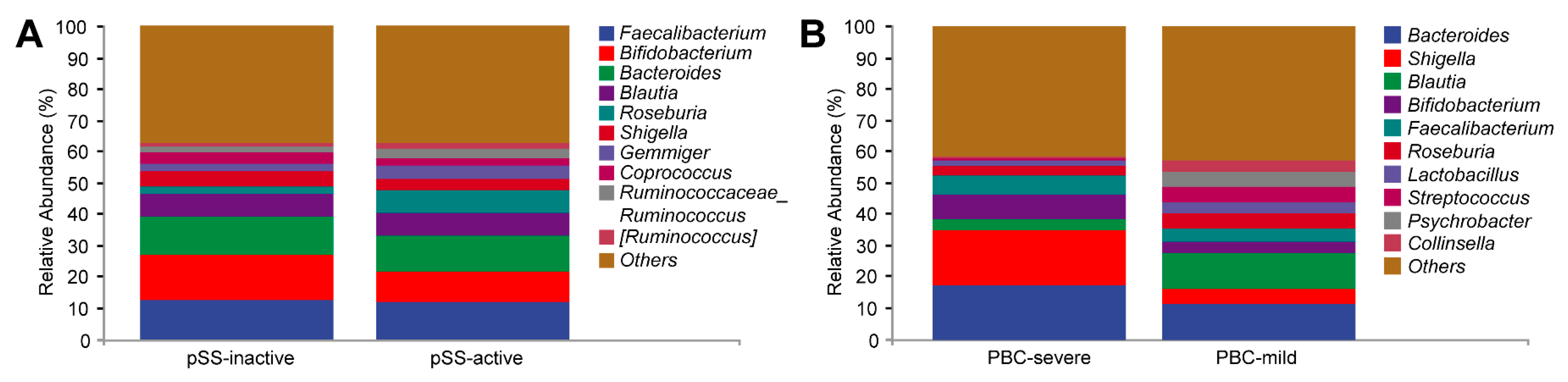
3.3. Serological Chemistry and Gut Microbiome Comparison Based on Disease Severity in PBC and pSS Patients
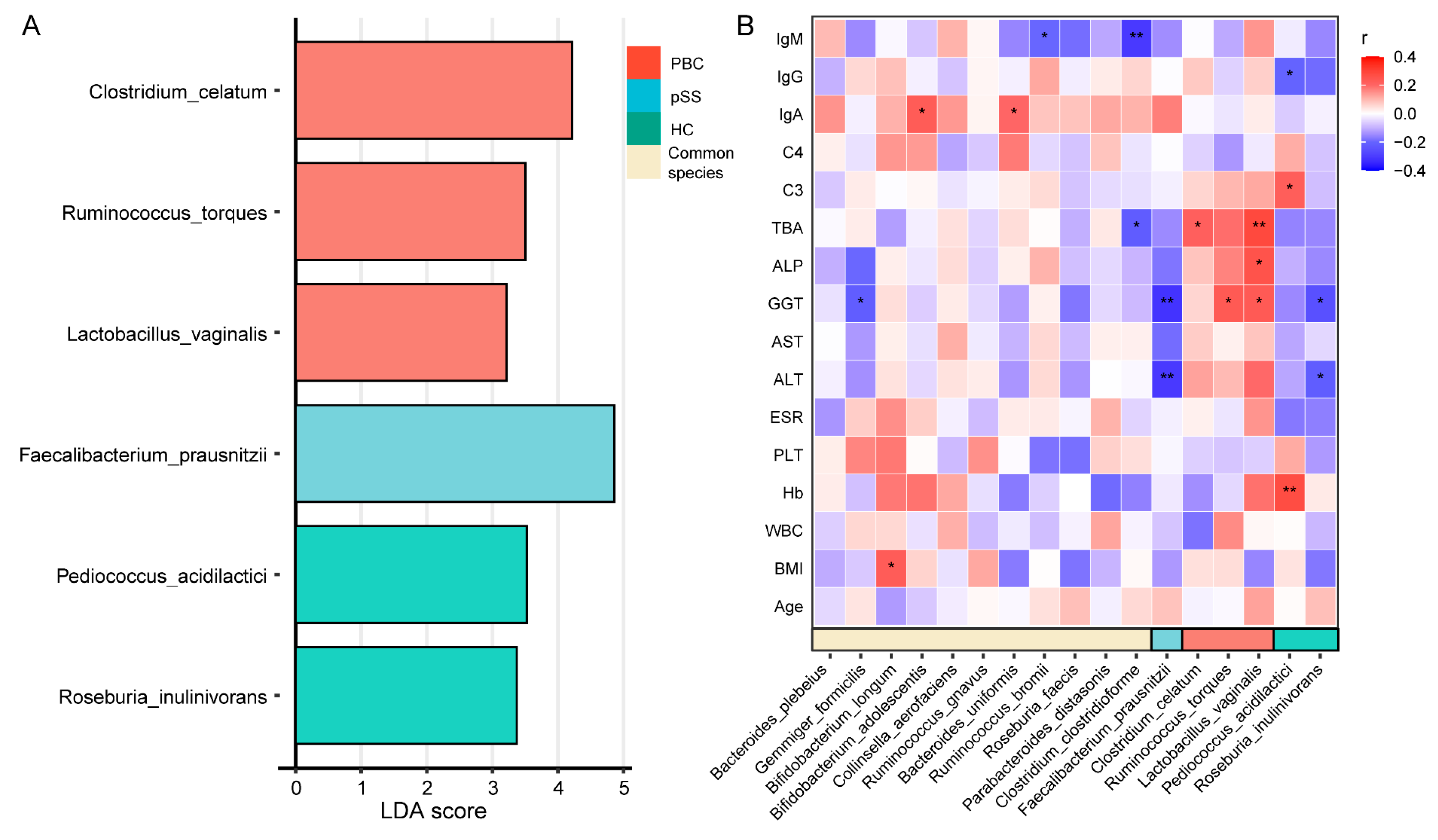
3.4. Characteristic Species of the PBC, pSS, and HC Groups and Their Correlation with Clinical Indicators
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PBC | Primary biliary cholangitis |
| pSS | Primary Sjögren’s syndrome |
| HC | Healthy control |
| ALP | Alkaline phosphatase |
| ULN | Upper limit of normal |
| ACR | American College of Rheumatology |
| PCoA | Principal coordinate analysis |
| SD | Standard deviation |
| ALT | Alanine aminotransferase |
| AST | Aspartate aminotransferase |
| GGT | Gamma-glutamyl transpeptidase |
| TBA | Total bile acid |
| IgM | Immunoglobulin M |
| IgG | Immunoglobulin G |
| C3 | Complement component 3 |
| IgA | Immunoglobulin A |
| C4 | Complement component 4 |
| PLT | Platelets |
| PCoA | Principle Coordinate Analysis |
| LEfSe | Linear Discriminant Analysis Effect Size |
| Hb | Hemoglobin |
| AIDs | Autoimmune diseases |
References
- Selmi, C.; Meroni, P.L.; Gershwin, M.E. Primary biliary cirrhosis and Sjögren’s syndrome: Autoimmune epithelitis. J. Autoimmun. 2012, 39, 34–42. [Google Scholar] [CrossRef]
- Talwalkar, J.A.; Lindor, K.D. Primary biliary cirrhosis. Lancet 2003, 362, 53–61. [Google Scholar] [CrossRef]
- Cristansson, J. Corneal changes in a case of hepatitis. Acta Ophthalmol. 1954, 32, 161–164. [Google Scholar] [CrossRef]
- Skopouli, F.N.; Barbatis, C.; Moutsopoulos, H.M. Liver involvement in primary Sjögren’s syndrome. Br. J. Rheumatol. 1994, 33, 745–748. [Google Scholar] [CrossRef] [PubMed]
- De Luca, F.; Shoenfeld, Y. The microbiome in autoimmune diseases. Clin. Exp. Immunol. 2019, 195, 74–85. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.; Fang, D.; Shi, D.; Chen, D.; Yan, R.; Zhu, Y.; Chen, Y.; Shao, L.; Guo, F.; Wu, W.; et al. Alterations and correlations of the gut microbiome, metabolism and immunity in patients with primary biliary cirrhosis. Environ. Microbiol. 2016, 18, 2272–2286. [Google Scholar] [CrossRef]
- Tang, R.; Wei, Y.; Li, Y.; Chen, W.; Chen, H.; Wang, Q.; Yang, F.; Miao, Q.; Xiao, X.; Zhang, H.; et al. Gut microbial profile is altered in primary biliary cholangitis and partially restored after UDCA therapy. Gut 2018, 67, 534–541. [Google Scholar] [CrossRef] [PubMed]
- Mandl, T.; Marsal, J.; Olsson, P.; Ohlsson, B.; Andréasson, K. Severe intestinal dysbiosis is prevalent in primary Sjögren’s syndrome and is associated with systemic disease activity. Arthritis Res. Ther. 2017, 19, 237. [Google Scholar] [CrossRef]
- Heathcote, E.J. Management of primary biliary cirrhosis. The American Association for the Study of Liver Diseases practice guidelines. Hepatology 2000, 31, 1005–1013. [Google Scholar] [CrossRef]
- Shiboski, S.C.; Shiboski, C.H.; Criswell, L.A.; Baer, A.N.; Challacombe, S.; Lanfranchi, H.; Schiodt, M.; Umehara, H.; Vivino, F.; Zhao, Y.; et al. American College of Rheumatology classification criteria for Sjogren’s syndrome: A data-driven, expert consensus approach in the Sjogren’s International Collaborative Clinical Alliance cohort. Arthritis Care Res. 2012, 64, 475–487. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef] [PubMed]
- Ramette, A. Multivariate analyses in microbial ecology. FEMS Microbiol. Ecol. 2007, 62, 142–160. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H.; Mitra, S.; Ruscheweyh, H.-J.; Weber, N.; Schuster, S.C. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011, 21, 1552–1560. [Google Scholar] [CrossRef]
- Asnicar, F.; Weingart, G.; Tickle, T.L.; Huttenhower, C.; Segata, N. Compact graphical representation of phylogenetic data and metadata with GraPhlAn. PeerJ 2015, 3, e1029. [Google Scholar] [CrossRef]
- Zaura, E.; Keijser, B.J.F.; Huse, S.M.; Crielaard, W. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol. 2009, 9, 12. [Google Scholar] [CrossRef]
- Petersen, C.; Round, J.L. Defining dysbiosis and its influence on host immunity and disease. Cell. Microbiol. 2014, 16, 1024–1033. [Google Scholar] [CrossRef]
- Zaheer, M.; Wang, C.; Bian, F.; Yu, Z.; Hernandez, H.; de Souza, R.G.; Simmons, K.T.; Schady, D.; Swennes, A.G.; Pflugfelder, S.C.; et al. Protective role of commensal bacteria in Sjögren Syndrome. J. Autoimmun. 2018, 93, 45–56. [Google Scholar] [CrossRef]
- de Paiva, C.S.; Jones, D.B.; Stern, M.E.; Bian, F.; Moore, Q.L.; Corbiere, S.; Streckfus, C.F.; Hutchinson, D.S.; Ajami, N.J.; Petrosino, J.F.; et al. Altered Mucosal Microbiome Diversity and Disease Severity in Sjogren Syndrome. Sci. Rep. 2016, 6, 23561. [Google Scholar] [CrossRef]
- Shin, N.R.; Whon, T.W.; Bae, J.W. Proteobacteria: Microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef]
- Nie, Q.; Luo, X.; Wang, K.; Ding, Y.; Jia, S.; Zhao, Q.; Li, M.; Zhang, J.; Zhuo, Y.; Lin, J.; et al. Gut symbionts alleviate MASH through a secondary bile acid biosynthetic pathway. Cell 2024, 187, 2717–2734.e33. [Google Scholar] [CrossRef]
- Chen, Z.; Chen, H.; Huang, W.; Guo, X.; Yu, L.; Shan, J.; Deng, X.; Liu, J.; Li, W.; Shen, W.; et al. Bacteroides fragilis alleviates necrotizing enterocolitis through restoring bile acid metabolism balance using bile salt hydrolase and inhibiting FXR-NLRP3 signaling pathway. Gut Microbes 2024, 16, 2379566. [Google Scholar] [CrossRef]
- Quraishi, M.N.; Cheesbrough, J.; Rimmer, P.; Mullish, B.H.; Sharma, N.; Efstathiou, E.; Acharjee, A.; Gkoutus, G.; Patel, A.; Marchesi, J.R.; et al. Open label vancomycin in primary sclerosing cholangitis-inflammatory bowel disease: Improved colonic disease activity and associations with changes in host-microbiome-metabolomic signatures. J. Crohn’s Colitis 2025, 19, jjae189. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wright, K.; Davis, J.M.; Jeraldo, P.; Marietta, E.V.; Murray, J.; Nelson, H.; Matteson, E.L.; Taneja, V. An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med. 2016, 8, 43. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Yang, M.; Liu, J.; Zhou, X.; Li, F.; Yang, B. Diversity analysis of intestinal microbiota in Graves disease patients. J. Pr. Med. 2020, 36, 767–773. Available online: https://d.wanfangdata.com.cn/periodical/syyxzz202006012 (accessed on 12 May 2020).
- Nouailles, G.; Dorhoi, A.; Koch, M.; Zerrahn, J.; Weiner, J.; Faé, K.C.; Arrey, F.; Kuhlmann, S.; Bandermann, S.; Loewe, D.; et al. CXCL5-secreting pulmonary epithelial cells drive destructive neutrophilic inflammation in tuberculosis. J. Clin. Investig. 2014, 124, 1268–1282. [Google Scholar] [CrossRef]
- Ferrari, M.L.; Malardé, V.; Grassart, A.; Salavessa, L.; Nigro, G.; Descorps-Declere, S.; Rohde, J.R.; Schnupf, P.; Masson, V.; Arras, G.; et al. Shigella promotes major alteration of gut epithelial physiology and tissue invasion by shutting off host intracellular transport. Proc. Natl. Acad. Sci. USA 2019, 116, 13582–13591. [Google Scholar] [CrossRef]
- Gui, W.-Y.; Yin, J.-G.; Liao, J.-C.; Luo, H.-Z.; You, Q.; Gong, J.-H.; Xiang, J.; Zou, J.-D.; Li, C.-Y. Integrated analysis of metabolome, lipidome, and gut microbiome reveals the immunomodulation of Astragali radix in healthy human subjects. Chin. Med. 2024, 19, 174. [Google Scholar] [CrossRef]
- Han, H.S.; Hwang, S.; Choi, S.Y.; Hitayezu, E.; Humphrey, M.A.; Enkhbayar, A.; Song, D.; Kim, M.; Park, J.; Park, Y.; et al. Roseburia intestinalis-derived extracellular vesicles ameliorate colitis by modulating intestinal barrier, microbiome, and inflammatory responses. J. Extracell. Vesicles 2024, 13, e12487. [Google Scholar] [CrossRef]
- Hu, J.; Wang, C.; Huang, X.; Yi, S.; Pan, S.; Zhang, Y.; Yuan, G.; Cao, Q.; Ye, X.; Li, H. Gut microbiota-mediated secondary bile acids regulate dendritic cells to attenuate autoimmune uveitis through TGR5 signaling. Cell Rep. 2021, 36, 109726. [Google Scholar] [CrossRef]
- Guan, B.; Tong, J.; Hao, H.; Yang, Z.; Chen, K.; Xu, H.; Wang, A. Bile acid coordinates microbiota homeostasis and systemic immunometabolism in cardiometabolic diseases. Acta Pharm. Sin. B 2022, 12, 2129–2149. [Google Scholar] [CrossRef]
- Jiao, N.; Baker, S.S.; Chapa-Rodriguez, A.; Liu, W.; Nugent, C.A.; Tsompana, M.; Mastrandrea, L.; Buck, M.J.; Baker, R.D.; Genco, R.J.; et al. Suppressed hepatic bile acid signalling despite elevated production of primary and secondary bile acids in NAFLD. Gut 2018, 67, 1881–1891. [Google Scholar] [CrossRef]
- Schaus, S.R.; Pereira, G.V.; Luis, A.S.; Madlambayan, E.; Terrapon, N.; Ostrowski, M.P.; Jin, C.; Henrissat, B.; Hansson, G.C.; Martens, E.C. Ruminococcus torques is a keystone degrader of intestinal mucin glycoprotein, releasing oligosaccharides used by Bacteroides thetaiotaomicron. mBio 2004, 15, e0003924. [Google Scholar] [CrossRef]
- Thangamani, S.; Monasky, R.; Lee, J.K.; Antharam, V.; HogenEsch, H.; Hazbun, T.R.; Jin, Y.; Gu, H.; Guo, G.L. Bile Acid Regulates the Colonization and Dissemination of Candida albicans from the Gastrointestinal Tract by Controlling Host Defense System and Microbiota. J. Fungi 2021, 7, 1030. [Google Scholar] [CrossRef]
- Wang, H.; Gong, J.; Chen, J.; Zhang, W.; Sun, Y.; Sun, D. Intestinal microbiota and biliary system diseases. Front. Cell Infect. Microbiol. 2024, 14, 1362933. [Google Scholar] [CrossRef]
- Bogdanos, D.; Pusl, T.; Rust, C.; Vergani, D.; Beuers, U. Primary biliary cirrhosis following Lactobacillus vaccination for recurrent vaginitis. J. Hepatol. 2008, 49, 466–473. [Google Scholar] [CrossRef] [PubMed]
- Eade, C.R.; Diaz, C.; Wood, M.P.; Anastos, K.; Patterson, B.K.; Gupta, P.; Cole, A.L.; Cole, A.M. Identification and characterization of bacterial vaginosis-associated pathogens using a comprehensive cervical-vaginal epithelial coculture assay. PLoS ONE 2012, 7, e50106. [Google Scholar] [CrossRef] [PubMed]
- Miquel, S.; Martin, R.; Rossi, O.; Bermudez-Humaran, L.G.; Chatel, J.M.; Sokol, H.; Thomas, M.; Wells, J.M.; Langella, P. Faecalibacterium prausnitzii and human intestinal health. Curr. Opin. Microbiol. 2013, 16, 255–261. [Google Scholar] [CrossRef]
- Jiao, M.; Yan, S.; Shi, Q.; Liu, Y.; Li, Y.; Lv, J.; Ding, S.; Li, A. Alcohol-Related Elevation of Liver Transaminase Is Associated With Gut Microbiota in Male. Front. Med. 2022, 9, 823898. [Google Scholar] [CrossRef]
- Maniat, M.; Salati, A.P.; Zanguee, N.; Mousavi, S.M.; Hoseinifar, S.H. Effects of Dietary Pediococcus acidilactici and Isomaltooligosaccharide on Growth Performance, Immunity, and Antioxidant Defense in Juvenile Common Carp. Aquac. Nutr. 2023, 2023, 1808640. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.V.; Hao, L.; Offermanns, S.; Medzhitov, R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl. Acad. Sci. USA 2014, 111, 2247–2252. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Kaminga, A.C.; Liu, A.; Wen, S.W.; Luo, M.; Luo, J. Gut Microbiota, Glucose, Lipid, and Water-Electrolyte Metabolism in Children With Nonalcoholic Fatty Liver Disease. Front. Cell Infect. Microbiol. 2021, 11, 683743. [Google Scholar] [CrossRef] [PubMed]
| PBC n = 38 | pSS n = 42 | HC n = 20 | p-Value | ||
|---|---|---|---|---|---|
| Age in years, mean ± SD | 46.24 ± 7.74 | 45.43 ± 10.81 | 44.85 ± 8.22 | 0.85 | |
| Gender, n | Male | 2 | 3 | 1 | 1.00 |
| Female | 36 | 39 | 19 | ||
| Height (cm) | 160.45 ± 4.49 | 161.29 ± 3.78 | 160.70 ± 5.30 | 0.69 | |
| Weight (kg) | 57.68 ± 4.66 | 57.57 ± 4.93 | 56.40 ± 5.63 | 0.61 | |
| BMI (kg/m2) | 22.45 ± 2.10 | 22.12 ± 1.53 | 21.82 ± 1.61 | 0.42 |
| pSS | PBC | HC | p a-Value | p b-Value | p c-Value | |
|---|---|---|---|---|---|---|
| WBC (10~9/L) | 4.86 ± 1.62 | 6.19 ± 2.16 | 5.19 ± 1.21 | 0.420 | 0.062 | 0.002 ** |
| Hb (g/L) | 130 ± 12 | 126 ± 14 | 128 ± 13 | 0.679 | 0.512 | 0.188 |
| PLT (10~9/L) | 195 (162, 226) | 153 (128, 242) | 160 (148, 187) | 0.012 * | 0.623 | 0.114 |
| ESR (mm/1 h) | 18 (12, 27) | 14 (11, 23) | 5.55 (4.15, 8.15) | <0.001 *** | <0.001 *** | 0.191 |
| ALT (U/L) | 16 (12.6, 23.1) | 35.7 (26, 56) | 16.2 (14.4, 21) | 0.904 | <0.001 *** | <0.001 *** |
| AST (U/L) | 18.8 (16.6, 25.1) | 34 (23, 42) | 24.8 (19.6, 28.2) | 0.034 * | 0.003 ** | <0.001 *** |
| GGT (U/L) | 18.7 (12.1, 28.6) | 94.9 (38, 136) | 23.5 (14.4, 27) | 0.625 | <0.001 *** | <0.001 *** |
| ALP (U/L) | 55.5 (43.8, 71.7) | 134 (87, 309) | 61.9 (51.8, 75.9) | 0.222 | <0.001 *** | <0.001 *** |
| TBA (μmol/L) | 3.5 (2.46, 6.2) | 11 (8, 16) | 3.6 (3.3, 6.85) | 0.484 | <0.001 *** | <0.001 *** |
| C3 (g/L) | 1.10 (0.92, 1.23) | 1.16 (1, 1.36) | 1.2 (1.01, 1.37) | 0.041 * | 0.595 | 0.080 |
| C4 (g/L) | 0.25 (0.18, 0.33) | 0.22 (0.15, 0.26) | 0.25 (0.19, 0.26) | 0.399 | 0.327 | 0.072 |
| IgA (g/L) | 2.58 (1.91, 3.21) | 2.62 (1.75, 3.36) | 2.74 (2.12, 4.05) | 0.641 | 0.707 | 0.904 |
| IgG (g/L) | 15.2 (13.7, 16.8) | 14.2 (11.6, 16.1) | 12.2 (11.2, 13.1) | <0.001 *** | 0.013 * | 0.031 * |
| IgM (g/L) | 1.21 (0.83, 1.58) | 1.57 (0.98, 2.84) | 1.29 (0.71, 1.54) | 0.625 | 0.014 * | 0.022 * |
| Active (n = 12) | Inactive (n = 30) | p-Value | |
|---|---|---|---|
| PLT (10~9/L) | 140 ± 52 | 217 ± 50 | <0.001 *** |
| ESR (mm/1 h) | 13.5 (7.75, 38.5) | 18 (14, 26) | 0.512 |
| AST (U/L) | 20.5 (15.8, 25.2) | 18.6 (17.3, 24.6) | 0.749 |
| C3 (g/L) | 0.89 ± 0.32 | 1.14 ± 0.19 | 0.002 ** |
| IgG (g/L) | 24.5 (17.3, 30.80) | 14.45 (13.6, 15.6) | <0.001 *** |
| Mild (n = 26) | Severe (n = 12) | p-Value | |
|---|---|---|---|
| ESR (mm/1 h) | 13.35 (9, 23) | 16 (11.65, 29.5) | 0.198 |
| ALT (U/L) | 35.2 ± 18.1 | 52.8 ± 22.1 | 0.013 * |
| AST (U/L) | 32.3 ± 13.2 | 44.2 ± 21 | 0.041 * |
| GGT (U/L) | 52.8 (33, 95.8) | 150 (136, 209) | <0.001 ** |
| ALP (U/L) | 102 (69, 134) | 334.5 (310, 398.5) | <0.001 ** |
| TBA (μmol/L) | 9 (7, 12) | 16 (15, 25) | <0.001 ** |
| IgG (g/L) | 12.9 (11.6, 16.1) | 15 (11.07, 16.1) | 0.875 |
| IgM (g/L) | 1.89 (1.02, 3.53) | 1.32 (0.94, 2.25) | 0.307 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zang, B.; Xu, L.; Huang, H.; Liu, Q.; Yao, Y.; Li, J.; Yang, Y.; Zhao, C.; Liu, B.; Liu, B. Decoding the Gut Microbiome in Primary Sjögren’s Syndrome and Primary Biliary Cholangitis: Shared Dysbiosis, Distinct Patterns, and Associations with Clinical Features. Microorganisms 2025, 13, 2668. https://doi.org/10.3390/microorganisms13122668
Zang B, Xu L, Huang H, Liu Q, Yao Y, Li J, Yang Y, Zhao C, Liu B, Liu B. Decoding the Gut Microbiome in Primary Sjögren’s Syndrome and Primary Biliary Cholangitis: Shared Dysbiosis, Distinct Patterns, and Associations with Clinical Features. Microorganisms. 2025; 13(12):2668. https://doi.org/10.3390/microorganisms13122668
Chicago/Turabian StyleZang, Bo, Lishan Xu, Haojie Huang, Qixuan Liu, Yuan Yao, Jiaxiu Li, Yifei Yang, Chenyang Zhao, Bingqian Liu, and Bin Liu. 2025. "Decoding the Gut Microbiome in Primary Sjögren’s Syndrome and Primary Biliary Cholangitis: Shared Dysbiosis, Distinct Patterns, and Associations with Clinical Features" Microorganisms 13, no. 12: 2668. https://doi.org/10.3390/microorganisms13122668
APA StyleZang, B., Xu, L., Huang, H., Liu, Q., Yao, Y., Li, J., Yang, Y., Zhao, C., Liu, B., & Liu, B. (2025). Decoding the Gut Microbiome in Primary Sjögren’s Syndrome and Primary Biliary Cholangitis: Shared Dysbiosis, Distinct Patterns, and Associations with Clinical Features. Microorganisms, 13(12), 2668. https://doi.org/10.3390/microorganisms13122668






