Abstract
The gut microbiota plays a vital role in regulating the host’s physiological functions, including metabolism and immunity. The microbial composition and metabolism are modulated by multiple factors; host sex is an important yet under-explored determinant. To investigate the sex-dependent differences in the gut microbiota within the small and large intestine, sixteen somatic mature Jiangshan black pigs (eight males and eight females) were analyzed. The ileal and colonic microbial community and metabolites were profiled using 16S rRNA gene high-throughput sequencing and gas chromatography. Distinct sex-related discrepancies were observed in both the microbial composition and metabolism of the ileum and colon. In the ileum, compared with the male group, the female group exhibited higher abundances of Unclassified Chloroplast and Pseudomonas but a lower abundance of Romboutsia (adjusted p < 0.05). Functional prediction indicated enrichment in amino acid metabolism pathway in females, with more copy numbers of genes encoding key enzymes for propionate (mmdA) generation and elevated valerate levels (p < 0.05). In the colon, compared with the male group, the female group showed higher abundances of Streptococcus, Phascolarctobacterium, and Prevotella spp. and lower abundances of Eubacterium coprostano-ligenes group, Blautia, Christensenellaceae R-7 group, and Ruminococcus (adjusted p < 0.05). Additionally, the female group had more copies of genes mmdA and LcdA (associated with lactate production), along with higher concentrations of propionate and lactate (p < 0.05). Correlation analysis between microbial metabolites and sex-biased bacteria further revealed that the SCFA concentration positively correlated with Prevotella spp. and negatively correlated with Romboutsia, Christensenellaceae R-7 group, and Blautia. Collectively, these findings highlight the pronounced sex-dependent discrepancies in the microbial composition and metabolism within the small and large intestines of Jiangshan black pigs, providing new insights for precisely modulating the microbiota community and metabolism in pigs according to sex.
1. Introduction
The mammalian gastrointestinal tract harbors a large, complex, and dynamically changing community of microorganisms exceeding 100 trillion microbial cells [1]. Microbial communities and metabolites could directly or indirectly contribute to various physiological functions of the host, such as growth and development, nutrient metabolism, immunity, and signal transmission [2,3], thereby maintaining host health and growth.
The gut microbial community serves as an intricate and heterogeneous ecosystem that is easily influenced by dietary composition, drugs, environment, and host inheritance [4,5,6]. In recent years, a growing number of studies have found that the host sex plays an important role in shaping the gut’s microbial community [7,8]. One human study revealed that there were significant differences in the gut microbial composition between women and men, with women exhibiting higher microbial diversity than men [9]. For instance, the genera Prevotella, Fusobacterium, Megasphaera, and Megamonas were enriched in the gut of men; Akkermansia, Ruminococcus, and Bifidobacterium were enriched in the gut of women [10]. Similar results have also been obtained in mice [11]. Furthermore, compared with normal male mice, sexually mature castrated male mice exhibited a gut microbial composition similar to that of females [12]. In pigs, some studies have revealed that the gut microbial composition exhibited sex-specific characteristics [13,14]. For example, the relative abundances of Escherichia, Roseburia, and Veillonella were higher in the feces of male Duroc pigs, while Bacteroides and Treponema were higher in female Duroc pigs [13]. Moreover, the host sex has significant influences on microbial metabolism [12]. A previous study reported that the metabolic activity of Lactobacillus exhibited a clear sex bias, with higher levels of propionate, butyrate and lactate in females, thus modulating gut health and energy metabolism [15]. The gut microbiota of female and male mice showed significant differences in lipid-metabolism-related genes. Compared with the male mice, the concentrations of total and primary bile acids were higher in the gut of female mice [16], which were closely related to higher relative abundances of Lachnoclostridium and Escherichia Shigella in the duodenum [17]. Sex-related differences are also evident in nitrogenous compound metabolism: Gao et al. reported higher colonic leucine and lower phenylalanine and tyrosine levels in female versus male mice [18]. Despite these findings, the understanding of how host sex influences the microbial composition and metabolism in the gut of pigs remains limited.
Extensive researches have demonstrated that the composition and metabolism of the microbiota in the gastrointestinal tract of pigs varies between the small intestine and large intestine [4,19]. In growing pigs, Firmicutes and Proteobacteria were dominant in the ileum, while Firmicutes and Bacteroidota were dominant in the colon [20]. Previous studies have found that the microbes from the small intestine and large intestine had different roles in amino acid metabolism and carbohydrate metabolism [21,22]. These findings revealed that the microbiota in different intestinal compartments perform unique functional roles. It is widely recognized that the microbiota in the small intestine (104–107 CFU/mL) is substantially smaller than that in the large intestine (1011–1012 CFU/mL) [23], and therefore the microbial community of the small intestine has received relatively limited attention. However, the impact of host sex on the microbiota in different intestinal compartments remains unclear.
Jiangshan black pig, a local Chinese breed pig (from Jiangshan city, Zhejiang Province, China), is known for its rich genetic background and excellent germplasm characteristics, such as roughage resistance, strong maternity, high livability and adaptability, and high disease resistance. Despite these valuable characteristics, few studies have investigated the gut microbiota of the Jiangshan black pig, especially in relation to sex-specific discrepancies. Thus, this study aimed to characterize the differentiation in the composition and metabolism of microbes from both the ileum and colon between the sexes of Jiangshan black pigs using 16S rRNA gene high-throughput sequencing and gas chromatography. The results showed that sex-associated disparities exist in the microbial composition and metabolism in the ileum and colon. These findings provide novel insights into how host sex influences the gut microbial community in different intestinal regions of the Jiangshan black pig and show that sex may be a significant factor in determining the microbial community.
2. Materials and Methods
2.1. Animals and Sampling
A total 16 healthy weaned Jiangshan black pigs aged approximately 50 days were obtained from a commercial farm in Jiangshan City (Zhejiang Province, China). According to sex, the pigs were divided into 2 groups, a female group and a male group (intact boars) (n = 8). All the pigs were housed individually in a metal pen with ad libitum access to feed (a commercial formula diet) and clean water; the diet’s details are listed in Table 1. The pigs were raised until about 200 days of age to ensure somatic maturity (body weight exceeds 50 kg). The final body weight and age of the female pigs were 52.0 ± 4.72 kg and 192 ± 8 days, respectively; those of the male pigs were 59.6 ± 6.09 kg and 205 ± 3 days, respectively. Then, all pigs were slaughtered for sample collection. After slaughtering, the gastrointestinal tract of the pig was removed immediately, and intestinal segments were identified and ligated. The ileum was from the jejunal–ileal junction to the ileal–cecal junction; the colon was from the cecal–colonic junction to the rectal–anal junction. Subsequently, the intestinal segments were cut open longitudinally to collect the digesta using a sterile 5 mL tube and immediately stored at −20 °C for the analysis of the microbial composition and metabolites. A flow chart of the experimentation is shown in Figure 1.

Table 1.
Composition of diet and nutrient levels.
Figure 1.
A flow chart of the experimental design in this study.
2.2. DNA Extraction, Illumina MiSeq Sequencing, and Data Processing
Total microbial genomic DNA was extracted from 0.5 g of ileal or colonic digesta samples according to a previous study by Dai et al. [24]. Briefly, the samples were mixed with sterile cetyltrimethyl ammonium bromide (CTAB) buffer using bead-beating for splitting the cell wall of the bacteria. The DNA was extracted by the phenol–chloroform method and finally suspended in TE buffer. The quantity of DNA was measured using a NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE, USA) and stored at −80 °C until further processing.
The V3 to V4 regions of the bacterial 16S rRNA gene were amplified using a universal forward primer, 341F (5′-CCTAYGGGRBGCASCAG-3′), and a reverse primer, 806R (5′-GGACGGACTACNNGGGTATCTAAT-3′). The amplicons were purified using a DNA Gel Extraction Kit according to the manufacturer’s instructions (Axygen Biosciences, Union City, CA, USA). Purified amplicons were pooled in equimolar and paired-end-sequenced on an Illumina MiSeq PE250 platform according to the standard protocols [25].
The raw sequence data from 16S rRNA gene MiSeq sequencing were analyzed using QIIME 2 (version 2020.2). Briefly, raw reads were qualified, denoised, classified, and counted using DADA2 to generate an amplicon sequence variant (ASV) table [26]. The ASVs with a minimum of four reads and present in more than one sample were retained. The Silva 138 database was used to annotate the ASVs [27]. The α-diversity (Chao1 richness estimators, Shannon diversity indices) and principal coordinate analysis (PCoA) based on the Bray–Curtis distance metrics were assessed using MOTHUR software (version 1.48.0) [28]. Significant and differential ASVs between 2 groups were determined by linear discriminant analysis (LDA) effect size (LDA score > 2 as discriminant taxa) [29].
2.3. Functional Prediction of Microbial Metagenomes
To predict the functional capacity of the gut microbiome, metagenomics function based on 16S rRNA gene was predicted with PICRUSt [30]. The gene categories were predicted at levels 2 and 3 of the Kyoto Encyclopedia of Genes and Genomes (KEGG) ontology groups [31]. Then, the predicted KEGG ontologies were classified into each KEGG pathway, the relative abundance of which was calculated. Significant differences in key KEGG pathways were analyzed between the groups.
2.4. Quantifying Copy Numbers of Functional Bacterial Groups and Genes by qPCR
To further reflect the effects of sex on microbial community and function in the ileum and colon, the copy numbers of functional bacterial groups and genes encoding key enzymes involved in propionate and butyrate generation and lactate utilization were quantified by a real-time PCR assay on a QuantStudio5 Flex detection system (Thermo Fisher Inc., Waltham, MA, USA) according to the manufacturer’s instructions using a SYBR Green Pro Taq HS Premix kit (Cat. AG11718, Accurate Biology Co., Ltd., Changsha, China). The detailed procedures, including reaction mixtures, PCR conditions, clone library constructions, and standard curve preparation, were performed according to previous methods [32]. The bacterial groups included total bacteria, Firmicutes, Bacteroidota, Lactobacillus, Bifidobacterium, Streptococcus, Coprococcus eutactus, Faecalibacterium prausnitzii, Megasphaera elsdenii, and Escherichia coli. And, the functional genes, including methylmalonyl-CoA decarboxylase (mmdA) and butyryl-CoA:acetate CoA-transferase (BCoAT), were used for quantifying propionate- and butyrate-producing enzymes, respectively; lactoyl-CoA dehydratase (LcdA) was used for quantifying lactate-utilizing enzymes. All the primers used in this study are listed in Table 2.

Table 2.
The sequence of primers in this study.
2.5. Determination of Microbial Metabolites
Short-chain fatty acids (SCFAs) in the ileal and colonic digesta samples were detected by gas chromatography according to a previous method [42]. Briefly, the digesta samples were weighted into a sterile centrifuge tube, double distilled water was added, and the samples were vortexed and centrifuged. Then, the supernatant was mixed with 25% (w/v) metaphosphoric acid. After allowing to stand overnight at −20 °C, the supernatant was centrifuged and filtered with a 0.22 μm filter for measurement. Lactate was measured using a commercial kit according to the manufacturer’s instructions (Nanjing Jiancheng Biological Engineering Institute, Nanjing, China). The concentration of NH3-N and microbial protein (MCP) in the digesta was assayed by spectrophotometer colorimetry according to the methods in previous studies [43,44].
2.6. Statistical Analysis
All data were analyzed by using SPSS 20.0 (SPSS Inc., Chicago, IL, USA). The statistical differences in bacterial abundance and the KEGG pathways were analyzed using the Mann–Whitney U test. To avoid type I errors during microbiota analysis, the p-value was adjusted with the Benjamini–Hochberg false discovery rate (FDR) multiple-testing correction; q (adjusted p-value) < 0.05 was regarded as statistically significant. The data of copy numbers of functional bacterial groups; genes; microbial metabolites, including SCFAs, lactate, NH3-N, and MCP, were analyzed with Student’s t-test to detect significant differences between the two groups. A p-value < 0.05 was considered statistically significant. All data were visualized using GraphPad Prism version 10.0 (GRAPHPAD Software, San Diego, CA, USA).
3. Results
Throughout the whole experiment, none of the pigs had diarrhea or other health problems (no pigs received medication).
3.1. The Effect of Sex on the Microbial Community in the Ileum and Colon
The effects of sex on the ileal and colonic microbial composition were revealed by 16S rRNA high-throughput sequencing. In total, 805,056 reads were obtained after the quality control, with an average of 25,158 reads per sample. The rarefaction curve of the individual samples based on the ASV numbers showed we had sufficient sequences for further analysis (Figure 2A). The alpha diversity of the microbial community, including Chao1 and Shannon indices, did not differ significantly between the female group and male group in either the ileum or colon (Figure 2B,C). For the beta diversity of the microbial community, principal coordinate analysis (PCoA) showed a clear distinct between the ileum and colon (Figure 2D) and between the female group and male group in the ileum and colon (Figure 2E,F), suggesting that the sex had a significant influence on the microbial structure in the various intestinal segments.
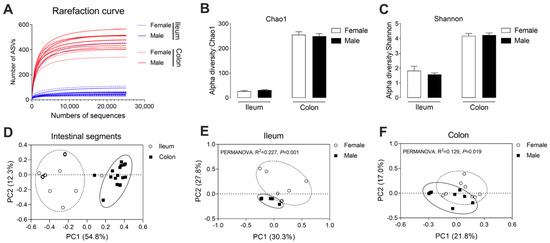
Figure 2.
The microbial community structure in the ileum and colon. (A) The rarefaction curve; (B,C) the alpha diversity of the microbial community in the gut: Chao 1 index, Shannon index. Data are presented as mean ± SEM, n = 8; (D) PCoA of the microbiota in different intestinal segments; (E,F) PCoA of the microbiota in the ileum and colon, respectively.
For the composition of the microbiota, Firmicutes (91.6%) and Proteobacteria (7.3%) were the two dominant phyla in the ileum, and Firmicutes (73.7%) and Bacteroidota (18.1%) were the two dominant phyla in the colon (Figure 3A). At the genus level, Romboutsia, Terrisporobacter, and Lactobacillus were the three dominant genera in the ileum (Figure 3B). Further differential analysis revealed that the relative abundances of Romboutsia and Burkholderia–Caballeronia–Paraburkholderia (BCP) were higher, while Unclassified Chloroplast and Pseudomonas were lower in the male group than in the female group (q < 0.05, Figure 4A). In the colon, Terrisporobacter, Lactobacillus, and Escherchia_Shigella were the three dominant genera (Figure 3B); and compared with the female group, the male group showed higher abundances in the genera unclassified Lachnospiraceae, Eubacterium coprostano-ligenes group, Christensenellaceae R-7 group, Blautia, Clostridium sensu stricto 1, Ruminococcus, Agathobacter, and Coprococcus, with lower abundances in genera Streptococcus, Phascolarctobacterium, Alloprevotella, Prevotellaceae UCG-003, Prevotella_9, Prevotella, Butyricicoccaceae UCG-009, and Desulfovibrio (q < 0.05, Figure 4B).
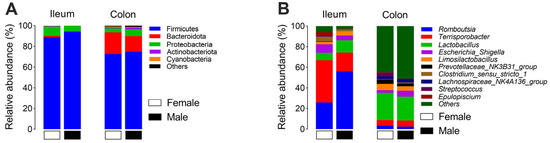
Figure 3.
The relative abundance of microbial composition at the phylum (A) and genus (B) levels in both the ileum and colon.
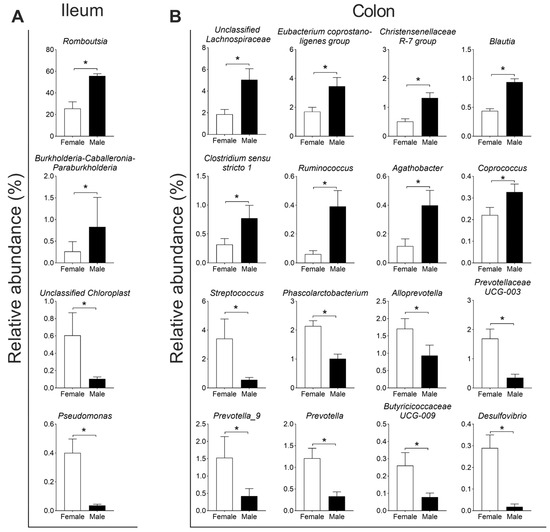
Figure 4.
The significantly different genera in the ileum (A) and colon (B). Data are presented as mean ± SEM, n = 8, * q < 0.05.
At the ASV level, in the ileum, nine ASVs showed differential abundances between the female group and male group, including Campylobacter iguaniorum, Pseudomonas ceruminis, Clostridium perfringens, Potamosiphon australiensis, and Terrisporobacter petrolearius, which were higher in the female group; Burkholderia vietnamiensis, Lactobacillus amylovorus, and Romboutsia timonesis were higher in the male group (Figure 5A). In the colon, a total of twelve ASVs were different in abundance between the groups, with Prevotella copri, Phascolarctobacterium succinatutens, Streptococcus alactolyticus, Coprococcus comes, Ruminococcus callidus and Agathobacter ruminis being higher in the female group; Prevotella oris, Desulfovibrio piger, Paraprevotella clara, Gehongia tenuis, Clostridium geopurificans, and Eubacterium coprostanoligenes were higher in the male group (Figure 5B). These results indicated that sex could serve as one important factor influencing the composition of the gut microbiota.
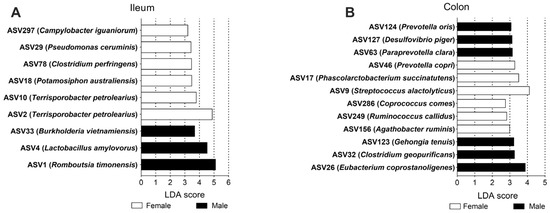
Figure 5.
The significantly different ASVs based on LDA in the ileum (A) and colon (B). ASV: amplicon sequence variants; LDA: linear discriminant analysis.
3.2. The Effect of Sex on the Microbial Function in the Ileum and Colon Based on Metagenomics Prediction
To reveal the differentiation in the microbial function between the female group and male group in the ileum and colon, microbial function was predicted with the KEGG database. Partial least-squares discriminant analysis (PLS-DA) demonstrated significant differences in microbial function between the ileum and colon (Figure 6A). Moreover, the microbial function between the female group and male group was also significantly different in both the ileum and colon (Figure 6B,C).
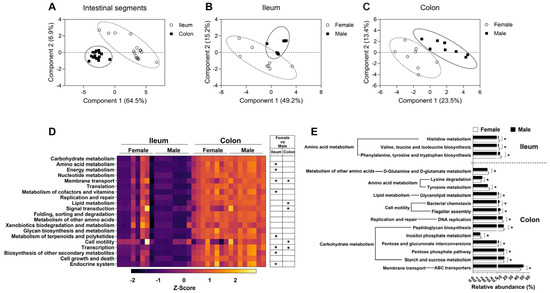
Figure 6.
The microbial functions based on the metagenomic prediction results. (A) PLS-DA of the microbial function in intestinal segments; (B,C) PLS-DA of the microbial function in ileum and colon, respectively; (D) the heatmap of microbial functional metabolism-related pathways based on KEGG database at level 2 KOs; (E) significantly different bacterial metabolic pathways at level 3 KOs. Data are presented as mean ± SEM, n = 8, * q < 0.05.
At KEGG level 2 of KOs, the top 20 functional pathways with relative abundance differences between the ileum and colon were visualized with a heatmap (Figure 6D). In the ileum, the relative abundances of KOs predicted to be involved in amino acid metabolism, energy metabolism, membrane transport, metabolism of cofactors and vitamins, metabolism of terpenoids and polyketides, and transcription and endocrine system were higher in the female group than in the male group (q < 0.05, Figure 6D). In the colon, the relative abundances of KOs predicted to be involved in membrane transport, lipid metabolism, signal transduction, cell motility and transcription were higher in the male group than in the female group (q < 0.05, Figure 6D).
At KEGG level 3 of KOs, the microbial functional pathways were further analyzed, as shown in Figure 6E. In the ileum, the relative abundances of KOs predicted to be involved in histidine metabolism; valine, leucine, and isoleucine biosynthesis; phenylalanine, tyrosine, and tryptophan biosynthesis were higher in the female group than in the male group (q < 0.05, Figure 6E). In the colon, the relative abundances of KOs predicted to be involved in D-glutamine and D-glutamate metabolism, tyrosine metabolism, peptidoglycan biosynthesis, and inositol phosphate metabolism were higher in the female group; lysine degradation, glycerolipid metabolism, bacterial chemotaxis, flagellar assembly, pentose and glucuronate interconversions, pentose phosphate pathway, starch and sucrose metabolism and ABC transporters was higher in the male group (q < 0.05, Figure 6E). These findings suggested that sex also exerted an influence on the function of the gut microbiota.
3.3. The Effect of Sex on the Functional Bacterial Groups and Functional Genes in the Ileum and Colon
Regarding 16S rRNA high-throughput sequencing, only the relative abundance of microbes was considered when analyzing the microbial composition. Therefore, qPCR was further performed to quantify the changes in the major functional bacterial groups between the female and male pigs. Firstly, the numbers of total bacteria did not differ significantly between the two groups in either the ileum or colon (p > 0.05, Figure 7A). At the taxonomy level, both in the ileum and colon, there were no differences in the numbers of the dominant phyla (Firmicutes and Bacteroidota), common genera (Lactobacillus, Bifidobacterium and Streptococcus), and butyrate-producing species (Coprococcus eutactus, Faecalibacterium prausnitzii) or harmful species (Escherichia coli) between the two groups (p > 0.05, Figure 7A). However, the numbers of Megasphaera elsdenii were significantly lower in the male group than in the female group in the colon (p < 0.05, Figure 7A).
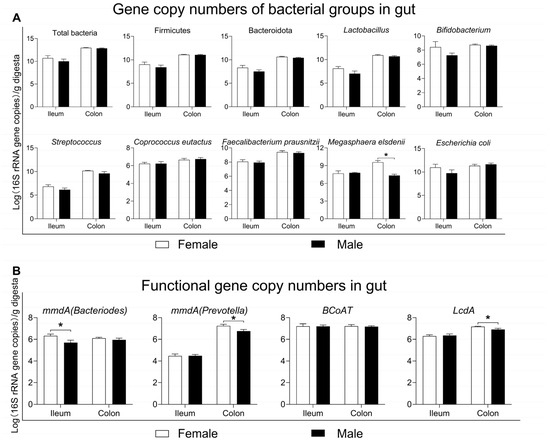
Figure 7.
Quantification of function bacterial groups and genes in the gut.: (A) 16S rRNA gene copy numbers of bacterial groups in the ileum and colon; (B) copy numbers of functional genes encoding key enzymes involved in SCFA generation (mmdA and BCoAT) and lactate utilization (LcdA). Data are presented as mean ± SEM, n = 8, * p < 0.05. mmdA: methylmalonyl-CoA decarboxylase; BCoAT: butyryl-CoA:acetate CoA-transferase; LcdA: lactoyl-CoA dehydratase.
In addition to measuring the numbers of bacterial groups in the gut, the copy numbers of functional genes encoding key enzymes involved in SCFA biosynthesis were also assayed, as shown in Figure 7B. In the ileum, the copy numbers of the functional gene mmdA (Bacteriodes), involved in propionate production, were lower in the male group than in the female group (p < 0.05, Figure 7B). In the colon, the copy numbers of functional genes mmdA (Prevotella) and LcdA, involved in propionate production and lactate utilization, respectively, were also lower in the male group than in the female group (p < 0.05, Figure 7B), hinting that the female group possessed a greater metabolic potential for propionate production by the gut microbiota.
3.4. The Effect of Sex on the Microbial Metabolites in the Ileal and Colonic Digesta
Undigested nutrients in the diet could serve as nutritional substrates to be fermented by gut microbes, producing a variety of metabolites, such as SCFAs, lactate, NH3-N, and MCP. In the ileum, the concentration of valerate was higher in the female group than in the male group (p < 0.05, Figure 8A), whereas no significant differences were observed in other SCFAs, lactate, NH3-N, or MCP between the two groups (p > 0.05, Figure 8A–E). In the colon, compared with the female group, the concentrations of propionate, isobutyrate, valerate, lactate, and NH3-N were lower in the male group (p < 0.05, Figure 8B–D). There was no difference between the two groups in the MCP concentration (p > 0.05, Figure 8E).
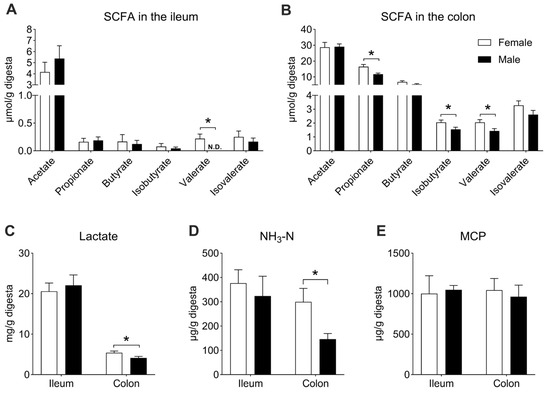
Figure 8.
The concentrations of microbial metabolites in the gut. (A) SCFA concentration in the ileum; (B) SCFA concentration in the colon; (C) lactate concentration in the ileum and colon; (D) NH3-N concentration in the ileum and colon; (E) MCP concentration in the ileum and colon. Data are presented as mean ± SEM, n = 8, * p < 0.05. “N.D.” indicates not detected.
To further elucidate the relationship between the gut microbial community and microbial metabolites, Spearman’s correlation analysis was performed based on the relative abundance of bacterial groups and microbial metabolite concentrations. In the ileum, the relative abundances of Romboutsia and Unclassified Chloroplast negatively correlated with valerate and MCP concentrations, respectively (Figure 9A). In contrast, the relative abundance of Unclassified Chloroplast positively correlated with butyrate, isobutyrate, and isovalerate (Figure 9A). In the colon, the relative abundance of Streptococcus negatively correlated with acetate concentration, while the relative abundances of Christensenellaceae R-7 group and Blautia negatively correlated with propionate concentration. In contrast, Alloprevotella, Prevotellaceae UCG-003, Prevotella_9, Prevotella, and Desulfovibrio were positively correlated with propionate concentration (Figure 9B). The relative abundance of Prevotellaceae UCG-003 positively correlated with butyrate content, whereas the relative abundances of Blautia, Clostridium sensu stricto 1, and Coprococcus negatively correlated with isobutyrate content. Meanwhile, Prevotellaceae UCG-003, Prevotella, Butyricicoccaceae UCG-009, and Desulfovibrio were positively correlated with isobutyrate content. Additionally, the relative abundances of Blautia and Clostridium sensu stricto 1 negatively correlated with valerate content, while Alloprevotella, Prevotellaceae UCG-003, and Prevotella were positively correlated with valerate content (Figure 9B). The relative abundance of Coprococcus negatively correlated with isovalerate concentration, while Prevotellaceae UCG-003, Prevotella, Butyricicoccaceae UCG-009, and Desulfovibrio were positively correlated with it (Figure 9B). Finally, the relative abundance of Unclassified Lachnospiraceae was negatively correlated with lactate concentration; the relative abundance of Blautia negatively correlated with NH3-N concentration, while Prevotellaceae UCG-003, Prevotella_9, and Prevotella were positively correlated with NH3-N concentration (Figure 9B).
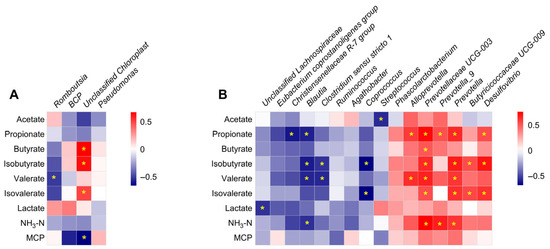
Figure 9.
Correlation analysis of significantly different relative abundances of bacteria at the genus level and microbial metabolites concentrations in the ileum (A) and colon (B). * p < 0.05.
4. Discussion
The gut microbiota has been reported to contribute significantly to host metabolism and immune functions, thereby maintaining intestinal homeostasis and host health. The differences in the intestinal microbial composition and metabolism mainly depend on host genotype, diet, and environment [6,45,46]. In addition to these factors, sex has recently been recognized as an important contributor to microbiota variability [7]. In this study, we characterized the sex-specific discrepancies in microbial composition and metabolism in different intestinal compartments of Jiangshan black pigs. The results showed that the sex-related differences in the microbial composition and metabolism were significant in the ileum and colon of Jiangshan black pigs. Some sex-biased bacteria were identified. Meanwhile, the correlation between the sex-biased bacteria and microbial metabolites was investigated. Taken together, these findings enhance our understanding of the influences of sex on the microbial community in the different intestinal compartments of Jiangshan black pigs.
The alpha-diversity indices are mainly used to evaluate the variation in microbial diversity. In the present study, we found no differences in alpha-diversity indices (e.g., Chao1, Shannon) between the female and male groups in either the ileum or colon, suggesting that sex exerted minimal influences on the diversity of the microbiota in the intestinal tract. These results differed from those of previous studies on humans and mice, which reported lower gut microbial diversity in males [11,47]. Similarly, in commercial purebred Duroc pigs, entire boars also had a lower fecal microbiota diversity than gilts [14]. These discrepancies may reflect differences in age and the relatively small sample size used in our study [48], which also may have been influenced by diet composition and genetic background [49].
In this study, PCoA revealed the variation in microbial composition between groups, consistent with prior observations in pigs [14]. Meanwhile, many sex-biased bacteria in the gut of Jiangshan black pig were identified in this study. Specifically, in the ileum, the abundances of the genera Romboutsia and Pseudomonas were higher in the male and female Jiangshan black pigs, respectively. In the colon, the male Jiangshan black pigs showed higher abundances od Unclassified Lachnospiraceae, Eubacterium coprostano-ligenes group, Christensenellaceae R-7 group, Clostridium_sensu_stricto 1, and Ruminococcus and lower abundances of Streptococcus, Phascolarctobacterium, Prevotella spp., and Butyricicoccaceae UCG-009. These results were also supported by a previous study on pigs [50]. In mice, Lactobacillus, Roseburia, Eubacterium, and Coprococcus were reported to be the representative sex-biased bacteria [11]; Eubacterium, Blautia and Treponema were identified as sex-biased bacteria in humans [51]. The sex-biased bacteria might be attributed to the differences in sex hormones, such as testosterone and estradiol [7]. It was reported that estrogen inhibited Escherichia coli growth in the intestine of rats [52], while this variation was not observed in the present study. Some previous studies showed that, compared with boars, the composition of gut microbiota in castrated male pigs was comparable to that of female pigs [14,50]. Moreover, the circulating testosterone concentration in female mice increased after removing the microbiota, which decreased the concentration in male mice, indicating a bidirectional interaction between male sex hormone levels and microbiota [12].
Functional shifts often accompany compositional changes. The metagenomic functional prediction revealed differences in the microbiota’s gene functions in Jiangshan black pigs between the sexes based on the KEGG database. For example, the metabolic pathways of the gut microbiota enriched in female Jiangshan black pigs were related to amino acid metabolism, such as histidine metabolism; valine, leucine, and isoleucine biosynthesis; phenylalanine, tyrosine, and tryptophan biosynthesis; and tyrosine metabolism. This pattern suggests a stronger microbial capacity for amino acid metabolism in female than in male Jiangshan black pigs. These results might be attributed to increased Pseudomonas in the gut of the female pigs. Pseudomonas is one member of the Proteobacteria. And Proteobacteria are regarded as the main type of bacteria involved in protein utilization in the gut, which contribute to amino acid metabolism [53]. For the male Jiangshan black pigs, the metabolic pathways enriched in the gut microbiota were related to carbohydrate metabolism (such as pentose and glucuronate interconversions, pentose phosphate pathway, starch and sucrose metabolism) and membrane transport (ABC transporters). These results indicated that the microbiota in the gut of male Jiangshan black pigs had a stronger ability to use carbohydrates in the diet, which might have been associated with the increases in Eubacterium, Ruminococcus, and Christensenellaceae R-7 group. Eubacterium and Ruminococcus were the primary fiber-degrading bacteria in the gut, promoting the fermentation and utilization of carbohydrates in the diet [54,55]. A previous study showed that Ruminococcus gnavus possessed a large number of genes encoding starch- and sucrose-degrading enzymes [56]. Moreover, Christensenellaceae R-7 group belongs to the Christensenellaceae, which was reported to be a marker of gut health [57]. A study on humans further showed that Christensenellaceae was inversely related to host body mass index (BMI), total cholesterol, and low-density lipoprotein, and positively correlated with fiber fermentation and protein catabolism [58]. In summary, although direct performance data were not collected, the observed enrichment in fiber-degrading bacteria in males suggests positive impacts on feed efficiency, especially roughage, which should be confirmed through growth or nutrient utilization studies.
The microbial metabolites are the key indicators reflecting microbiota metabolism activity, such as SCFAs and lactate. In the present study, higher concentrations of propionate, isobutyrate and valerate were found in the gut of the female Jiangshan black pigs, hinting that the bacteria in female pigs exhibited a stronger capacity for SCFA production. These results might have been due to the increases in Prevotella spp., Phascolarctobacterium, and Butyricicoccaceae UCG-009, as Prevotella spp. are well-known propionate-producing bacteria in the intestinal tract [59]. Correlation analysis further confirmed these results (as shown in Figure 9). Phascolarctobacterium can also utilize succinate to yield propionate [59]. Furthermore, in this study, the gut microbiota in the female Jiangshan black pigs expressed higher copy numbers of functional genes encoding mmdA, involved in propionate production [60]. This finding further demonstrated that the microbiota possessed a stronger capacity to produce propionate in the gut of female Jiangshan black pigs. Lactate, another important microbial metabolite, is mainly produced by lactate-producing bacteria. In this study, the female Jiangshan black pigs showed a higher level of lactate in the gut, which was mainly associated with an increase in Prevotella_9 and Streptococcus [61,62]. Additionally, lactate, as a substrate, could be converted to propionate by lactate-utilizing bacteria, like Megasphaera elsdenii [63]. M. elsdenii, a specific lactate-utilizing bacteria, can convert lactate to propionate via the acrylate metabolic pathway [63]. Meanwhile, the functional gene LcdA expressed by lactate-utilizing bacteria plays a vital role in the process of converting lactate to propionate [64]. Interestingly, we found higher copy numbers of M. elsdenii and LcdA in the gut of female Jiangshan black pigs, which suggested that the gut microbiota promoted the lactate-to-propionate conversion. NH3-N, an amino acid metabolism product, is mainly produced by the microbiota through the deamination process. In this study, female Jiangshan black pigs had a higher concentration of NH3-N than male pigs, which might have been related to the increase in Streptococcus in the gut. Previous studies have shown that Streptococcus is the major amino-acid-utilizing bacteria in the gut [53]. In addition, a significant positive correlation between Prevotella spp. and NH3-N concentration was observed in this study, suggesting that Prevotella spp. played an important role in the process of metabolizing amino acids to produce NH3-N. Taken together, sex elicited differential effects on the ileal and colonic microbial community and metabolism in Jiangshan black pigs.
5. Conclusions
This study systematically investigated the sex-related differences in the gut microbiota composition and metabolism of Jiangshan black pigs. The results revealed that there were distinct sex-associated differences in the microbial structure and composition in the ileum and colon. In microbial metabolism, female Jiangshan black pigs displayed a greater ability to enhance amino acid metabolism and SCFA generation; male Jiangshan black pigs showed stronger carbohydrate metabolism and transport capacities. These findings provide important guidance for the precision feeding of Jiangshan black pigs, suggesting that low-protein diets may be more suitable for sows, while high-fiber diets could benefit boars. Collectively, the present study highlights the sex discrepancies in the microbial community and metabolism in the small and large intestines of Jiangshan black pigs, providing new insights for precisely modulating the gut microbiota community and metabolism according to sex.
Author Contributions
Conceptualization, Y.Z. and X.L.; animal experiment, X.W. and D.S.; analysis, Y.Z., X.W. and P.W.; data curation, Y.Z. and X.W.; writing—original draft preparation, Y.Z.; writing—review and editing, H.W.; funding acquisition, H.W. and Y.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by grants from the Research and Development Fund of Zhejiang A&F University (2023LFR006) and the Open Project of Key Laboratory of Animal Molecular Nutrition (Zhejiang University), Ministry of Education (KLMAN202307).
Institutional Review Board Statement
The experiment in this study was performed with protocols approved by the Animal Care and Use Committee of Zhejiang Agricultural and Forestry University, Hangzhou, China, in compliance with the regulations for the Administration of Affairs Concerning Experimental Animals of China (approval No. ZAFUAC2022034; approval date: 5 October 2021).
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in this study are included in the article. Further inquiries can be directed to the corresponding authors.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this study:
| SCFA | Short chain fatty acid |
| MCP | Microbial protein |
| ASV | Amplicon sequence variants |
| LDA | linear discriminant analysis |
| FDR | False discovery rate |
| KEGG | Kyoto encyclopedia of genes and genomes |
| PCoA | Principal coordinate analysis |
| PLS-DA | Partial least squares discriminant analysis |
| PCR | Polymerase chain reaction |
| mmdA | Methylmalonyl-CoA decarboxylase |
| BCoAT | Butyryl-CoA:acetate CoA-transferase |
| LcdA | Lactoyl-CoA dehydratase |
References
- Flint, H.J.; Duncan, S.H.; Scott, K.P.; Louis, P. Interactions and competition within the microbial community of the human colon: Links between diet and health. Environ. Microbiol. 2007, 9, 1101–1111. [Google Scholar] [CrossRef]
- Flint, H.J.; Scott, K.P.; Louis, P.; Duncan, S.H. The role of the gut microbiota in nutrition and health. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 577–589. [Google Scholar] [CrossRef] [PubMed]
- Kayama, H.; Okumura, R.; Takeda, K. Interaction Between the Microbiota, Epithelia, and Immune Cells in the Intestine. Annu. Rev. Immunol. 2020, 38, 23–48. [Google Scholar] [CrossRef] [PubMed]
- Looft, T.; Allen, H.K.; Cantarel, B.L.; Levine, U.Y.; Bayles, D.O.; Alt, D.P.; Henrissat, B.; Stanton, T.B. Bacteria, phages and pigs: The effects of in-feed antibiotics on the microbiome at different gut locations. ISME J. 2014, 8, 1566–1576. [Google Scholar] [CrossRef] [PubMed]
- Louis, P.; Scott, K.P.; Duncan, S.H.; Flint, H.J. Understanding the effects of diet on bacterial metabolism in the large intestine. J. Appl. Microbiol. 2007, 102, 1197–1208. [Google Scholar] [CrossRef]
- Guo, X.; Li, J.; Xu, J.; Zhang, L.; Huang, C.; Nie, Y.; Zhou, Y. Gut microbiota and epigenetic inheritance: Implications for the development of IBD. Gut Microbes 2025, 17, 2490207. [Google Scholar] [CrossRef]
- Valeri, F.; Endres, K. How biological sex of the host shapes its gut microbiota. Front. Neuroendocrinol. 2021, 61, 100912. [Google Scholar] [CrossRef]
- Yoon, K.; Kim, N. Roles of Sex Hormones and Gender in the Gut Microbiota. J. Neurogastroenterol. Motil. 2021, 27, 314–325. [Google Scholar] [CrossRef]
- Sinha, T.; Vila, A.V.; Garmaeva, S.; Jankipersadsing, S.A.; Imhann, F.; Collij, V.; Bonder, M.J.; Jiang, X.; Gurry, T.; Alm, E.J.; et al. Analysis of 1135 gut metagenomes identifies sex-specific resistome profiles. Gut Microbes 2019, 10, 358–366. [Google Scholar] [CrossRef]
- Takagi, T.; Naito, Y.; Inoue, R.; Kashiwagi, S.; Uchiyama, K.; Mizushima, K.; Tsuchiya, S.; Dohi, O.; Yoshida, N.; Kamada, K.; et al. Differences in gut microbiota associated with age, sex, and stool consistency in healthy Japanese subjects. J. Gastroenterol. 2019, 54, 53–63, Erratum in J. Gastroenterol. 2019, 54, 96–98. [Google Scholar] [CrossRef]
- Elderman, M.; Hugenholtz, F.; Belzer, C.; Boekschoten, M.; van Beek, A.; de Haan, B.; Savelkoul, H.; de Vos, P.; Faas, M. Sex and strain dependent differences in mucosal immunology and microbiota composition in mice. Biol. Sex Differ. 2018, 9, 26. [Google Scholar] [CrossRef] [PubMed]
- Markle, J.G.M.; Frank, D.N.; Mortin-Toth, S.; Robertson, C.E.; Feazel, L.M.; Rolle-Kampczyk, U.; von Bergen, M.; McCoy, K.D.; Macpherson, A.J.; Danska, J.S. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 2013, 339, 1084–1088. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Estellé, J.; Kiilerich, P.; Ramayo-Caldas, Y.; Xia, Z.; Feng, Q.; Liang, S.; Pedersen, A.Ø.; Kjeldsen, N.J.; Liu, C.; et al. A reference gene catalogue of the pig gut microbiome. Nat. Microbiol. 2016, 1, 16161. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Gao, J.; Wu, J.; Zhou, Y.; Fu, H.; Ke, S.; Yang, H.; Chen, C.; Huang, L. Host Gender and Androgen Levels Regulate Gut Bacterial Taxa in Pigs Leading to Sex-Biased Serum Metabolite Profiles. Front. Microbiol. 2019, 10, 1359. [Google Scholar] [CrossRef]
- Santos-Marcos, J.A.; Mora-Ortiz, M.; Tena-Sempere, M.; Lopez-Miranda, J.; Camargo, A. Interaction between gut microbiota and sex hormones and their relation to sexual dimorphism in metabolic diseases. Biol. Sex Differ. 2023, 14, 4. [Google Scholar] [CrossRef]
- Baars, A.; Oosting, A.; Lohuis, M.; Koehorst, M.; El Aidy, S.; Hugenholtz, F.; Smidt, H.; Mischke, M.; Boekschoten, M.V.; Verkade, H.J.; et al. Sex differences in lipid metabolism are affected by presence of the gut microbiota. Sci. Rep. 2018, 8, 13426. [Google Scholar] [CrossRef]
- Sisk-Hackworth, L.; Brown, J.; Sau, L.; Levine, A.A.; Tam, L.Y.I.; Ramesh, A.; Shah, R.S.; Kelley-Thackray, E.T.; Wang, S.; Nguyen, A.; et al. Genetic hypogonadal mouse model reveals niche-specific influence of reproductive axis and sex on intestinal microbial communities. Biol. Sex Differ. 2023, 14, 79. [Google Scholar] [CrossRef]
- Gao, H.; Shu, Q.; Chen, J.; Fan, K.; Xu, P.; Zhou, Q.; Li, C.; Zheng, H. Antibiotic Exposure Has Sex-Dependent Effects on the Gut Microbiota and Metabolism of Short-Chain Fatty Acids and Amino Acids in Mice. Msystems 2019, 4, e00048-19. [Google Scholar] [CrossRef]
- Mu, C.; Yang, Y.; Su, Y.; Zoetendal, E.G.; Zhu, W. Differences in Microbiota Membership along the Gastrointestinal Tract of Piglets and Their Differential Alterations Following an Early-Life Antibiotic Intervention. Front. Microbiol. 2017, 8, 797. [Google Scholar] [CrossRef]
- Zhao, W.; Wang, Y.; Liu, S.; Huang, J.; Zhai, Z.; He, C.; Ding, J.; Wang, J.; Wang, H.; Fan, W.; et al. The dynamic distribution of porcine microbiota across different ages and gastrointestinal tract segments. PLoS ONE 2015, 10, e0117441. [Google Scholar] [CrossRef]
- El Kaoutari, A.; Armougom, F.; Gordon, J.I.; Raoult, D.; Henrissat, B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat. Rev. Microbiol. 2013, 11, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.X.; Dai, Z.L.; Zhu, W.Y. Important impacts of intestinal bacteria on utilization of dietary amino acids in pigs. Amino Acids 2014, 46, 2489–2501. [Google Scholar] [CrossRef] [PubMed]
- O’Hara, A.M.; Shanahan, F. The gut flora as a forgotten organ. EMBO Rep. 2006, 7, 688–693. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.L.; Zhang, J.; Wu, G.; Zhu, W.Y. Utilization of amino acids by bacteria from the pig small intestine. Amino Acids 2010, 39, 1201–1215. [Google Scholar] [CrossRef]
- Mu, Y.Y.; Qi, W.P.; Zhang, T.; Zhang, J.Y.; Mao, S.Y. Gene function adjustment for carbohydrate metabolism and enrichment of rumen microbiota with antibiotic resistance genes during subacute rumen acidosis induced by a high-grain diet in lactating dairy cows. J. Dairy Sci. 2021, 104, 2087–2105. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Sato, Y.; Furumichi, M.; Tanabe, M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012, 40, D109–D114. [Google Scholar] [CrossRef]
- Mu, C.; Yang, Y.; Luo, Z.; Zhu, W. Temporal microbiota changes of high-protein diet intake in a rat model. Anaerobe 2017, 47, 218–225. [Google Scholar] [CrossRef]
- Suzuki, M.T.; Taylor, L.T.; DeLong, E.F. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 2000, 66, 4605–4614. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Xia, X.; Tang, R.; Zhou, J.; Zhao, H.; Wang, K. Development of a real-time PCR method for Firmicutes and Bacteroidetes in faeces and its application to quantify intestinal population of obese and lean pigs. Lett. Appl. Microbiol. 2008, 47, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Rinttila, T.; Kassinen, A.; Malinen, E.; Krogius, L.; Palva, A. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J. Appl. Microbiol. 2004, 97, 1166–1177. [Google Scholar] [CrossRef] [PubMed]
- Walter, J.; Hertel, C.; Tannock, G.W.; Lis, C.M.; Munro, K.; Hammes, W.P. Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 2001, 67, 2578–2585. [Google Scholar] [CrossRef]
- Walker, A.W.; Ince, J.; Duncan, S.H.; Webster, L.M.; Holtrop, G.; Ze, X.; Brown, D.; Stares, M.D.; Scott, P.; Bergerat, A.; et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2011, 5, 220–230. [Google Scholar] [CrossRef]
- Reichardt, N.; Vollmer, M.; Holtrop, G.; Farquharson, F.M.; Wefers, D.; Bunzel, M.; Duncan, S.H.; Drew, J.E.; Williams, L.M.; Milligan, G.; et al. Specific substrate-driven changes in human faecal microbiota composition contrast with functional redundancy in short-chain fatty acid production. ISME J. 2018, 12, 610–622. [Google Scholar] [CrossRef]
- Wang, R.F.; Cao, W.W.; Cerniglia, C.E. PCR detection and quantitation of predominant anaerobic bacteria in human and animal fecal samples. Appl. Environ. Microbiol. 1996, 62, 1242–1247. [Google Scholar] [CrossRef]
- Kajihara, Y.; Yoshikawa, S.; Cho, Y.; Ito, T.; Miyamoto, H.; Kodama, H. Preferential isolation of Megasphaera elsdenii from pig feces. Anaerobe 2017, 48, 160–164. [Google Scholar] [CrossRef]
- Zhang, Y.; Mu, C.; Liu, S.; Zhu, W. Dietary citrus pectin drives more ileal microbial protein metabolism and stronger fecal carbohydrate fermentation over fructo-oligosaccharide in growing pigs. Anim. Nutr. 2022, 11, 252–263. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yu, K.; Chen, H.; Su, Y.; Zhu, W. Caecal infusion of the short-chain fatty acid propionate affects the microbiota and expression of inflammatory cytokines in the colon in a fistula pig model. Microb. Biotechnol. 2018, 11, 859–868. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Chen, Y.; Moraes, L.E.; Yu, Z.; Zhu, W. Effects of dietary protein sources and nisin on rumen fermentation, nutrient digestion, plasma metabolites, nitrogen utilization, and growth performance in growing lambs. J. Anim. Sci. 2018, 96, 1929–1938. [Google Scholar] [CrossRef] [PubMed]
- Makkar, H.P.; Sharma, O.P.; Dawra, R.K.; Negi, S.S. Simple determination of microbial protein in rumen liquor. J. Dairy Sci. 1982, 65, 2170–2173. [Google Scholar] [CrossRef]
- Michalak, L.; Gaby, J.C.; Lagos, L.; La Rosa, S.L.; Hvidsten, T.R.; Tétard-Jones, C.; Willats, W.G.T.; Terrapon, N.; Lombard, V.; Henrissat, B.; et al. Microbiota-directed fibre activates both targeted and secondary metabolic shifts in the distal gut. Nat. Commun. 2020, 11, 5773. [Google Scholar] [CrossRef]
- Han, Y.; Liu, X.; Jia, Q.; Xu, J.; Shi, J.; Li, X.; Xie, G.; Zhao, X.; He, K. Longitudinal multi-omics analysis uncovers the altered landscape of gut microbiota and plasma metabolome in response to high altitude. Microbiome 2024, 12, 70. [Google Scholar] [CrossRef]
- Gao, X.; Zhang, M.; Xue, J.; Huang, J.; Zhuang, R.; Zhou, X.; Zhang, H.; Fu, Q.; Hao, Y. Body Mass Index Differences in the Gut Microbiota Are Gender Specific. Front. Microbiol. 2018, 9, 1250. [Google Scholar] [CrossRef]
- Han, G.G.; Lee, J.-Y.; Jin, G.-D.; Park, J.; Choi, Y.H.; Kang, S.-K.; Chae, B.J.; Kim, E.B.; Choi, Y.-J. Tracing of the fecal microbiota of commercial pigs at five growth stages from birth to shipment. Sci. Rep. 2018, 8, 6012. [Google Scholar] [CrossRef]
- Org, E.; Mehrabian, M.; Parks, B.W.; Shipkova, P.; Liu, X.; Drake, T.A.; Lusis, A.J. Sex differences and hormonal effects on gut microbiota composition in mice. Gut Microbes 2016, 7, 313–322. [Google Scholar] [CrossRef]
- Zhou, Z.J.; Zheng, W.J.; Shang, W.W.; Du, H.L.; Li, G.L.; Yao, W. How host gender affects the bacterial community in pig feces and its correlation to skatole production. Ann. Microbiol. 2015, 65, 2379–2386. [Google Scholar] [CrossRef]
- Schnorr, S.L.; Candela, M.; Rampelli, S.; Centanni, M.; Consolandi, C.; Basaglia, G.; Turroni, S.; Biagi, E.; Peano, C.; Severgnini, M.; et al. Gut microbiome of the Hadza hunter-gatherers. Nat. Commun. 2014, 5, 3654. [Google Scholar] [CrossRef]
- Yang, Y.; Qu, C.; Liang, S.; Wang, G.; Han, H.; Chen, N.; Wang, X.; Luo, Z.; Zhong, C.; Chen, Y.; et al. Estrogen inhibits the overgrowth of Escherichia coli in the rat intestine under simulated microgravity. Mol. Med. Rep. 2018, 17, 2313–2320. [Google Scholar] [PubMed]
- Davila, A.-M.; Blachier, F.; Gotteland, M.; Andriamihaja, M.; Benetti, P.-H.; Sanz, Y.; Tomé, D. Intestinal luminal nitrogen metabolism: Role of the gut microbiota and consequences for the host. Pharmacol. Res. 2013, 68, 95–107, reprinted in Pharmacol. Res. 2013, 69, 114–126. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.; Lordan, C.; Ross, R.P.; Cotter, P.D. Gut microbes from the phylogenetically diverse genus Eubacterium and their various contributions to gut health. Gut Microbes 2020, 12, 1802866. [Google Scholar] [CrossRef] [PubMed]
- La Reau, A.J.; Suen, G. The Ruminococci: Key symbionts of the gut ecosystem. J. Microbiol. 2018, 56, 199–208. [Google Scholar] [CrossRef]
- Crost, E.H.; Coletto, E.; Bell, A.; Juge, N. Ruminococcus gnavus: Friend or foe for human health. FEMS Microbiol. Rev. 2023, 47, fuad014. [Google Scholar] [CrossRef]
- Mancabelli, L.; Milani, C.; Lugli, G.A.; Turroni, F.; Cocconi, D.; van Sinderen, D.; Ventura, M. Identification of universal gut microbial biomarkers of common human intestinal diseases by meta-analysis. FEMS Microbiol. Ecol. 2017, 93, fix153. [Google Scholar] [CrossRef]
- Waters, J.L.; Ley, R.E. The human gut bacteria Christensenellaceae are widespread, heritable, and associated with health. BMC Biol. 2019, 17, 83. [Google Scholar] [CrossRef]
- Flint, H.J.; Duncan, S.H.; Scott, K.P.; Louis, P. Links between diet, gut microbiota composition and gut metabolism. Proc. Nutr. Soc. 2015, 74, 13–22. [Google Scholar] [CrossRef]
- Walker, A.W.; Duncan, S.H.; Louis, P.; Flint, H.J. Phylogeny, culturing, and metagenomics of the human gut microbiota. Trends Microbiol. 2014, 22, 267–274. [Google Scholar] [CrossRef]
- Bai, Y.; Zhou, X.; Li, N.; Zhao, J.; Ye, H.; Zhang, S.; Yang, H.; Pi, Y.; Tao, S.; Han, D.; et al. In Vitro Fermentation Characteristics and Fiber-Degrading Enzyme Kinetics of Cellulose, Arabinoxylan, beta-Glucan and Glucomannan by Pig Fecal Microbiota. Microorganisms 2021, 9, 1071. [Google Scholar] [CrossRef]
- Shen, J.; Mu, C.; Wang, H.; Huang, Z.; Yu, K.; Zoetendal, E.G.; Zhu, W. Stimulation of Gastric Transit Function Driven by Hydrolyzed Casein Increases Small Intestinal Carbohydrate Availability and Its Microbial Metabolism. Mol. Nutr. Food Res. 2020, 64, e2000250. [Google Scholar] [CrossRef]
- Prabhu, R.; Altman, E.; Eiteman, M.A. Lactate and acrylate metabolism by Megasphaera elsdenii under batch and steady-state conditions. Appl. Environ. Microbiol. 2012, 78, 8564–8570. [Google Scholar] [CrossRef]
- Reichardt, N.; Duncan, S.H.; Young, P.; Belenguer, A.; McWilliam Leitch, C.; Scott, K.P.; Flint, H.J.; Louis, P. Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J. 2014, 8, 1323–1335, Erratum in ISME J. 2014, 8, 1352. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).