Exploring the Role of Rhizobacteria in Sorghum bicolor Adaptation to Combined Drought and Heat Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Planting Materials, Experimental Design, Study Sites, and Crop Management
2.2. Data Collection
2.2.1. Rhizospheric Soil Sampling for 16S rRNA Amplicon Sequencing
2.2.2. Grain Yield Performance
2.3. Data Analysis
2.3.1. 16S rRNA Sequence Analysis
2.3.2. Bacteria Diversity Analysis
2.3.3. Rhizobacteria Incidence of Occurrence and Their Impact on Sorghum bicolor GY Performance
2.3.4. Integrating Microbial Diversity Indices to Select Superior Sorghum bicolor Genotypes for Production Under CDHS Conditions
3. Results
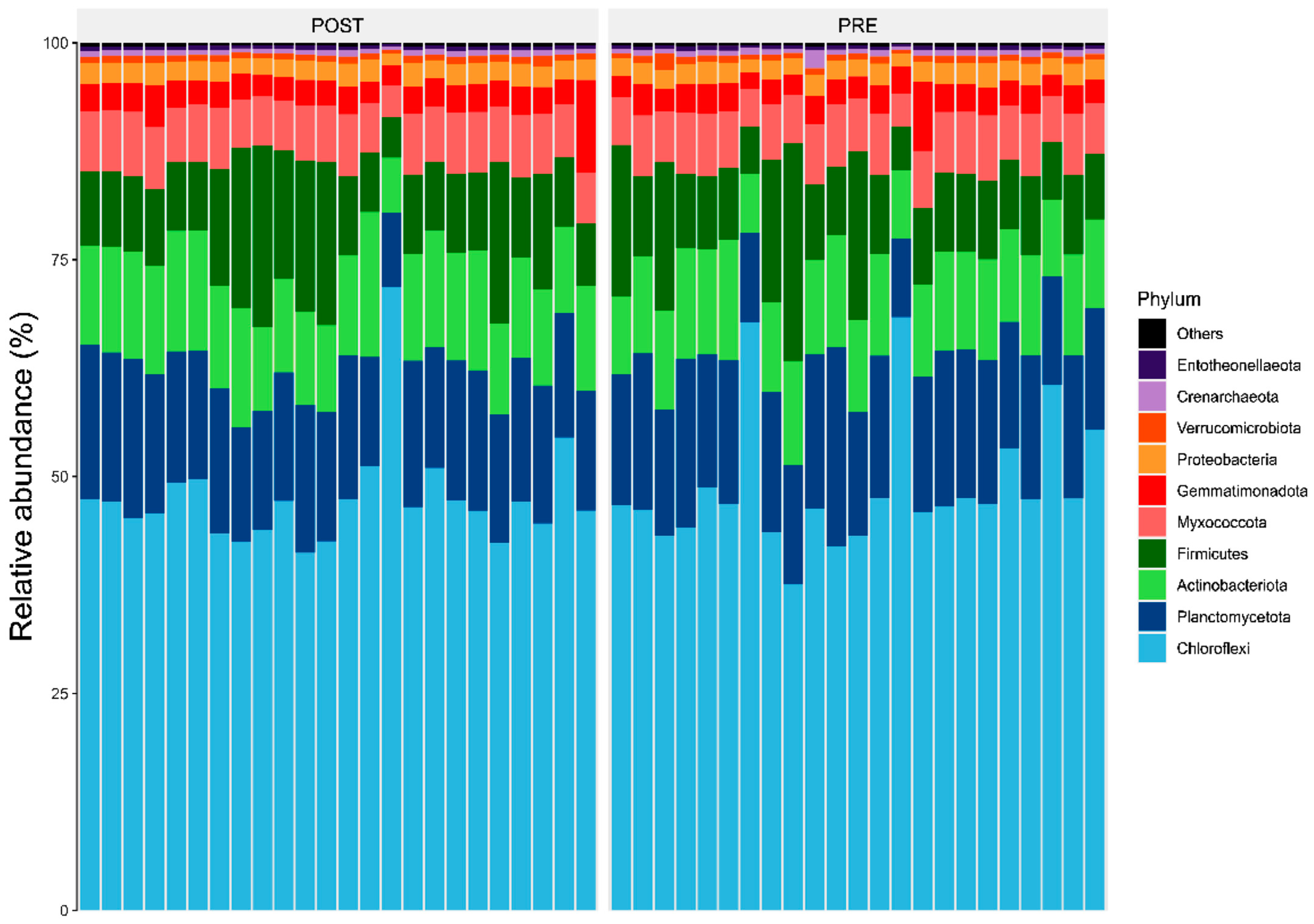
3.1. Bacterial Diversity and Community Composition
3.2. Rhizobacteria Incidence of Occurrence and Their Impact on Sorghum bicolor GY Performance Under CDHS Conditions
3.3. Integrating Microbial Diversity Indices to Select Superior Sorghum bicolor Genotypes for Production Under CDHS Conditions
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MLR | Multiple Linear Regression |
| ANOVA | Analysis of Variance |
| MTSI | Multiple Trait Selection Index |
| ICRISAT | International Crops Research Institute for the Semi-Arid Tropics |
| CIMMYT | International Maize and Wheat Improvement Center |
| RCBD | Randomized Complete Block Design |
| CDHS | Combined Drought and Heat Stress |
| RCZ | Research Council of Zimbabwe |
| AIC | Akaike Information Criterion |
| BIC | Bayesian Information Criterion |
| DADA2 | Divisive Amplicon Denoising Algorithm 2 |
| PERMANOVA | Permutational Multivariate Analysis of Variance |
| out | Operational Taxonomic Unit |
References
- Karlova, R.; Boer, D.; Hayes, S.; Testerink, C. Root plasticity under abiotic stress. Plant Physiol. 2021, 187, 1057–1070. [Google Scholar] [CrossRef]
- Marines Marli, G.K. Current challenges in plant breeding to achieve zero hunger and overcome biotic and abiotic stresses induced by the global climate changes: A review. J. Plant Sci. Phytopathol. 2021, 5, 53–57. [Google Scholar] [CrossRef]
- Munir, N.; Hanif, M.; Abideen, Z.; Sohail, M.; El-Keblawy, A.; Radicetti, E.; Mancinelli, R.; Haider, G. Mechanisms and Strategies of Plant Microbiome Interactions to Mitigate Abiotic Stresses. Agronomy 2022, 12, 2069. [Google Scholar] [CrossRef]
- Esmaeili, N.; Shen, G.; Zhang, H. Genetic manipulation for abiotic stress resistance traits in crops. Front. Plant Sci. 2022, 13, 1011985. [Google Scholar] [CrossRef] [PubMed]
- Dutta, C.; Sarma, R.N. Role of Root Traits and Root Phenotyping in Drought Tolerance. Int. J. Environ. Clim. Chang. 2022, 12, 2300–2309. [Google Scholar] [CrossRef]
- Keneni, G.; Bekele, E.; Imtiaz, M. Challenges Associated with Crop Breeding for Adaptation to Drought-Prone Environments. Ethiop. J. Agric. Sci. 2017, 27, 1–24. [Google Scholar]
- Liu, H.; Su, Y.; Ye, C.; Zuo, D.; Wang, L.; Mei, X.; Deng, W.; Liu, Y.; Huang, H.; Hao, J.; et al. Nucleotides enriched under heat stress recruit beneficial rhizomicrobes to protect plants from heat and root-rot stresses. Microbiome 2025, 13, 160. [Google Scholar] [CrossRef] [PubMed]
- Uroz, S.; Courty, P.E.; Oger, P. Plant Symbionts Are Engineers of the Plant-Associated Microbiome. Trends Plant Sci. 2019, 24, 905–916. [Google Scholar] [CrossRef]
- Lopes, M.A. Rethinking plant breeding and seed systems in the era of exponential changes. Cienc. E Agrotecnol. 2023, 47, e0001R23. [Google Scholar] [CrossRef]
- Rizaludin, M.S.; Stopnisek, N.; Raaijmakers, J.M.; Garbeva, P. The chemistry of stress: Understanding the ‘cry for help’ of plant roots. Metabolites 2021, 11, 357. [Google Scholar] [CrossRef]
- Liu, F.; Hewezi, T.; Lebeis, S.L.; Pantalone, V.; Grewal, P.S.; Staton, M.E. Soil indigenous microbiome and plant genotypes cooperatively modify soybean rhizosphere microbiome assembly. BMC Microbiol. 2019, 19, 201. [Google Scholar] [CrossRef]
- Valliere, J.M.; Dixon, K.W.; Nevill, P.G.; Zhong, H. Preparing for the worst: Utilizing stress-tolerant soil microbial communities to aid ecological restoration in the Anthropocene. Ecol. Solut. Evid. 2020, 1, e12027. [Google Scholar] [CrossRef]
- Lamaoui, M.; Jemo, M.; Datla, R.; Bekkaoui, F. Heat and Drought Stresses in Crops and Approaches for Their Mitigation. Front. Chem. 2018, 6, 26. [Google Scholar] [CrossRef] [PubMed]
- Schrader, L.; Trautner, J.; Tebbe, C.C. Identifying environmental factors affecting the microbial community composition on outdoor structural timber. Appl. Microbiol. Biotechnol. 2024, 108, 254. [Google Scholar] [CrossRef]
- Zverev, A.; Kichko, A.; Shapkin, V.; Pinaev, A.; Provorov, N.; Andronov, E. Does plant diversity determine the diversity of the rhizosphere microbial community? bioRxiv 2021. [Google Scholar] [CrossRef]
- Trivedi, P.; Batista, B.D.; Bazany, K.E.; Singh, B.K. Plant–microbiome interactions under a changing world: Responses, consequences and perspectives. New Phytol. 2022, 234, 1951–1959. [Google Scholar] [CrossRef]
- Korenblum, E.; Massalha, H.; Aharoni, A. Plant–microbe interactions in the rhizosphere via a circular metabolic economy. Plant Cell 2022, 34, 3168–3182. [Google Scholar] [CrossRef]
- Inzé, D.; Nelissen, H. The translatability of genetic networks from model to crop species: Lessons from the past and perspectives for the future. New Phytol. 2022, 236, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Grover, M.; Ali, S.Z. Role of microorganisms in adaptation of agriculture crops to abiotic stresses Role of microorganisms in adaptation of agriculture crops to abiotic stresses. World J. Microbiol. Biotechnol. 2011, 27, 1231–1240. [Google Scholar] [CrossRef]
- Velmourougane, K.; Saxena, G.; Prasanna, R. Plant-microbe interactions in the rhizosphere: Mechanisms and their ecological benefits. In Plant-Microbe Interactions in Agro-Ecological Perspectives; Springer: Singapore, 2017; Volume 2, pp. 193–219. [Google Scholar] [CrossRef]
- Matova, P.M.; Kamutando, C.N.; Mutari, B.; Magorokosho, C.; Labuschagne, M. Adaptability and Stability Analysis of Commercial Cultivars, Experimental Hybrids and Lines under Natural Fall Armyworm Infestation in Zimbabwe Using Different Stability Models. Agronomy 2022, 12, 1724. [Google Scholar] [CrossRef]
- Wasimuddin; Malik, H.; Ratovonamana, Y.R.; Rakotondranary, S.J.; Ganzhorn, J.U.; Sommer, S. Anthropogenic Disturbance Impacts Gut Microbiome Homeostasis in a Malagasy Primate. Front. Microbiol. 2022, 13, 911275. [Google Scholar] [CrossRef]
- Callahan, M.B.; Callahan, A.B.; McMurdie, P.; Holmes, S. Package ‘dada2’. 2025. Available online: https://github.com/benjjneb/dada2/issues (accessed on 25 July 2025).
- Merino-Martín, L.; Hernández-Cáceres, D.; Reverchon, F.; Angeles-Alvarez, G.; Zhang, G.; de Segonzac, D.D.; Dezette, D.; Stokes, A. Habitat partitioning of soil microbial communities along an elevation gradient: From plant root to landscape scale. Oikos 2023, 2023, e09034. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, F.; Zhao, C.; Wu, G.; Wang, H.; Cao, X. The Properties and Application of Poisson Distribution. J. Phys. Conf. Ser. 2020, 1550, 032109. [Google Scholar] [CrossRef]
- Trunfio, T.A.; Scala, A.; Giglio, C.; Rossi, G.; Borrelli, A.; Romano, M.; Improta, G. Multiple regression model to analyze the total LOS for patients undergoing laparoscopic appendectomy. BMC Med. Inform. Decis. Mak. 2022, 22, 141. [Google Scholar] [CrossRef]
- Nashrudin, I.S.; Islam, U.; Maulana, N.; Ibrahim, M. Regression Analysis Between Social Media Usage and Number of Posts by Gender Using Minitab. Biomed. Signal Process. 2024. [Google Scholar] [CrossRef]
- Science, P.; Author, T. Evaluating Effect Size in Psychological Research: Sense and Nonsense. Adv. Methods Pract. Psychol. Sci. 2020, 2, 156–168. [Google Scholar] [CrossRef]
- Gazal, A. Smith Hazel Selection Index for the Improvement of Maize Inbred Lines under Water Stress Conditions. Int. J. Pure Appl. Biosci. 2017, 5, 72–81. [Google Scholar] [CrossRef]
- Badri, D.V.; Weir, T.L.; van der Lelie, D.; Vivanco, J.M. Rhizosphere chemical dialogues: Plant-microbe interactions. Curr. Opin. Biotechnol. 2009, 20, 642–650. [Google Scholar] [CrossRef]
- Park, I.; Seo, Y.S.; Mannaa, M. Recruitment of the rhizo-microbiome army: Assembly determinants and engineering of the rhizosphere microbiome as a key to unlocking plant potential. Front. Microbiol. 2023, 14, 1163832. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, Y.; Fu, Y.; Shao, J.; Liu, Y.; Xuan, W.; Xu, G.; Zhang, R. Signal communication during microbial modulation of root system architecture. J. Exp. Bot. 2023, 75, 526–537. [Google Scholar] [CrossRef]
- Middleton, H.; Yergeau, É.; Monard, C.; Combier, J.-P.; El Amrani, A. Rhizospheric Plant-Microbe Interactions: miRNAs as a Key Mediator. Trends Plant Sci. 2021, 26, 132–141. [Google Scholar] [CrossRef]
- Haldar, S.; Sengupta, S. Plant-microbe Cross-talk in the Rhizosphere: Insight and Biotechnological Potential. Open Microbiol. J. 2015, 9, 1–7. [Google Scholar] [CrossRef]
- Hist, S.; Biol, P.; Sci, B.; Malley, M.A.O. ‘Everything is everywhere: But the environment selects’: Ubiquitous distribution and ecological determinism in microbial biogeography. Stud. Hist. Philos. Sci. Part C Stud. Hist. Philos. Biol. Biomed. Sci. 2008, 39, 314–325. [Google Scholar] [CrossRef]
- Ali, S.; Tyagi, A.; Park, S.; Mir, R.A.; Mushtaq, M.; Bhat, B.; Mahmoudi, H.; Bae, H. Deciphering the plant microbiome to improve drought tolerance: Mechanisms and perspectives. Environ. Exp. Bot. 2022, 201, 104933. [Google Scholar] [CrossRef]
- Naylor, D.; Coleman-derr, D. Drought Stress and Root-Associated Bacterial Communities. Front. Plant Sci. 2018, 8, 223. [Google Scholar] [CrossRef] [PubMed]
- Future, D.; Zhang, Q.; White, J.F. Bioprospecting Desert Plants for Endophytic and Biostimulant Microbes: A Strategy for Enhancing Agricultural Production in a Hotter, Drier Future. Biology 2021, 10, 961. [Google Scholar] [CrossRef] [PubMed]
- Glick, B.R. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 2014, 169, 30–39. [Google Scholar] [CrossRef]
- Khoury, C.K.; Brush, S.; Costich, D.E.; Curry, H.A.; de Haan, S.; Engels, J.M.M.; Guarino, L.; Hoban, S.; Mercer, K.L.; Miller, A.J.; et al. Crop genetic erosion: Understanding and responding to loss of crop diversity. New Phytol. 2022, 233, 84–118. [Google Scholar] [CrossRef]
- Orr, A.; Mwema, C.; Gierend, A.; Nedumaran, S. Sorghum and Millets in Eastern and Southern Africa Facts, Trends and Outlook; ICRISAT Working Paper No. 62; International Crops Research Institute for the Semi-Arid-Tropics (ICRISAT): Telangana, India, 2016; 76p. [Google Scholar]
- Xie, J.; Dawwam, G.E.; Sehim, A.E.; Li, X.; Wu, J.; Chen, S.; Zhang, D. Drought Stress Triggers Shifts in the Root Microbial Community and Alters Functional Categories in the Microbial Gene Pool. Front. Microbiol. 2021, 12, 744897. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Shi, G.; Wei, S.; Ma, J.; Zhang, X.; Wang, J.; Chen, L.; Liu, Y.; Zhao, X.; Lu, Z. Drought Sensitivity of Spring Wheat Cultivars Shapes Rhizosphere Microbial Community Patterns in Response to Drought. Plants 2023, 12, 3650. [Google Scholar] [CrossRef]
- Qi, M.; Berry, J.C.; Veley, K.M.; O’connor, L.; Finkel, O.M.; Salas-González, I.; Kuhs, M.; Jupe, J.; Holcomb, E.; del Rio, T.G.; et al. Identification of beneficial and detrimental bacteria impacting sorghum responses to drought using multi-scale and multi-system microbiome comparisons. ISME J. 2022, 16, 1957–1969, Correction in ISME J. 2023, 17, 1353. [Google Scholar] [CrossRef] [PubMed]
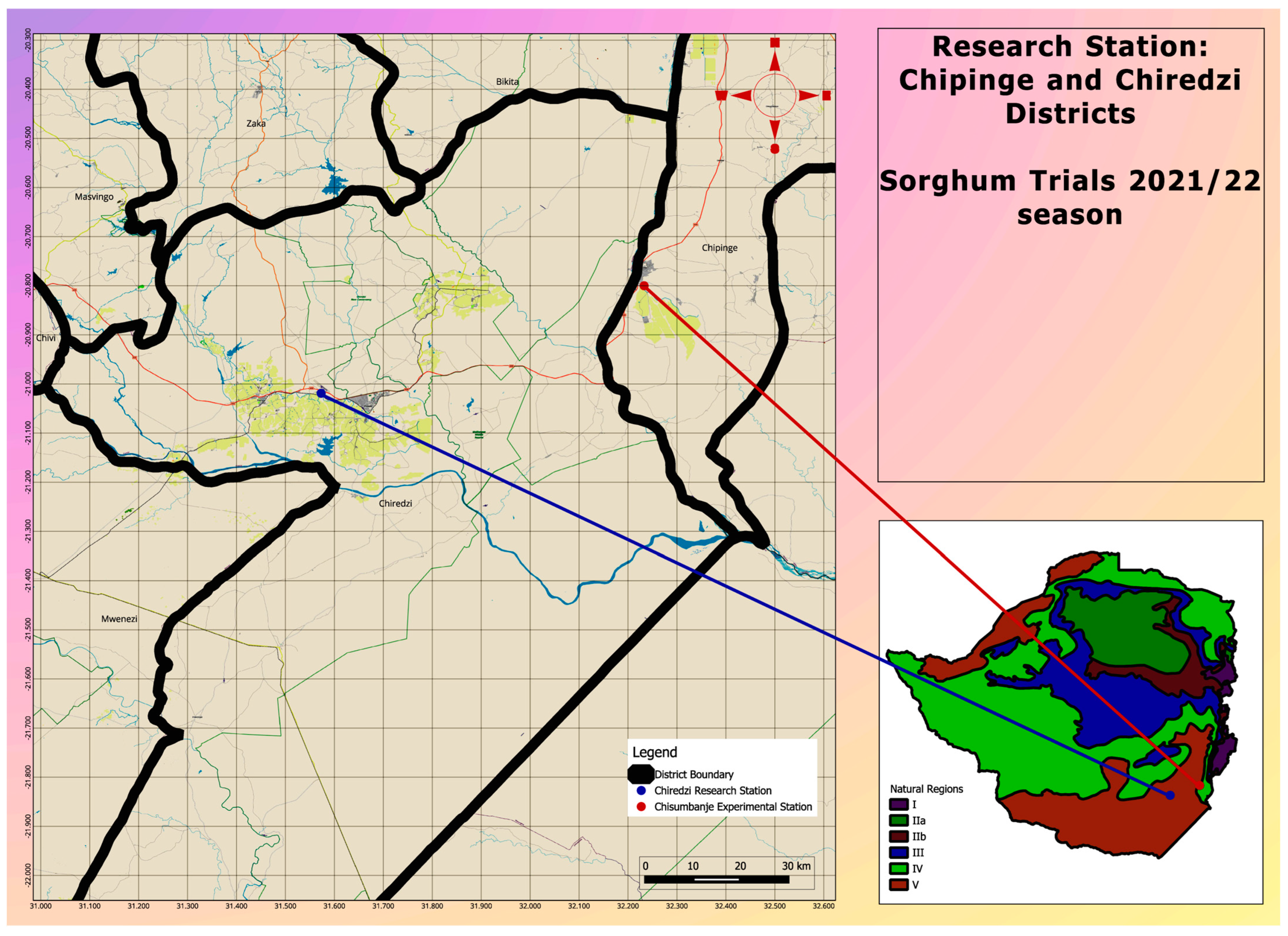







| Genotype Name | Origin | Status |
|---|---|---|
| SV4 | Crop Breeding Institute, Harare, Zimbabwe | Released grain commercial variety [check] |
| ICSV111IN | ICRISAT—Hyderabad, India | Advanced pre-release line [Experimental] |
| CHITICHI | Chiredzi Community Seed Bank Masvingo, Zimbabwe | Local landrace variety [check] |
| MACIA | Seed Company of Zimbabwe Harare, Zimbabwe | Released grain commercial variety [check] |
| IESV91070DL | ICRISAT—Hyderabad, India | Advanced pre-release line [Experimental] |
| ASAREACA12-3-1 | ICRISAT—Hyderabad, India | Advanced pre-release line [Experimental] |
| Source | DF | Seq SS | Contribution (%) | Adj SS | Adj MS | F-Value | p-Value |
|---|---|---|---|---|---|---|---|
| Regression | 2 | 0.54357 | 85.64 | 0.54357 | 0.27178 | 26.83 | 0.000 |
| Actinobacteriota Thermoleophilia | 1 | 0.40821 | 64.31 | 0.12504 | 0.12504 | 12.34 | 0.007 |
| Firmicutes Bacilli | 1 | 0.13536 | 21.33 | 0.13536 | 0.13536 | 13.36 | 0.005 |
| Error | 9 | 0.09116 | 14.36 | 0.09116 | 0.01013 | ||
| Total | 11 | 0.63473 | 100.00 |
| Source | DF | Seq SS | Contribution (%) | Adj SS | Adj MS | F-Value | p-Value |
|---|---|---|---|---|---|---|---|
| Regression | 2 | 6.395 | 52.23 | 6.395 | 3.1975 | 4.92 | 0.036 |
| Firmicutes Bacilli | 1 | 4.296 | 35.08 | 5.277 | 5.2769 | 8.12 | 0.019 |
| Actinobacteriota Actinobacteria | 1 | 2.099 | 17.14 | 2.099 | 2.0992 | 3.23 | 0.106 |
| Error | 9 | 5.850 | 47.77 | 5.850 | 0.6500 | ||
| Total | 11 | 12.245 | 100.00 |
| Study Site | Genotype | Genetic Worth Index (V1) | GY (t/ha) |
|---|---|---|---|
| Chisumbanje Research Station | IESV91070DL | 816.660 | 0.1910 c |
| ICSV111IN | 601.739 | 0.9050 a | |
| SV4 | 465.159 | 0.4150 bc | |
| CHITICHI | 463.827 | 0.6950 ab | |
| ASARECA12-3-1 | 382.508 | 0.5000 bc | |
| MACIA | 22.790 | 0.6000 ab | |
| Chiredzi Research Station | ASARECA12-3-1 | 361.162 | 1.320 bc |
| IESV91070DL | 271.417 | 1.335 bc | |
| CHITICHI | 251.939 | 0.480 c | |
| SV4 | 205.087 | 3.495 a | |
| MACIA | 174.311 | 1.520 b | |
| ICSV111IN | 123.343 | 2.695 a |
| Conventional | Conventional Integrated with Microbial Diversity Data | |||
|---|---|---|---|---|
| Genotype | Mean GY (t/ha) | Ranking | ∑[Mean (GY + VI)] | Ranking |
| SV4 | 1.955 | 1 | 337.078 | 5 |
| ICSV111IN | 1.8 | 2 | 364.341 | 3 |
| MACIA | 1.06 | 3 | 99.6105 | 6 |
| ASARECA12-3-1 | 0.91 | 4 | 372.745 | 2 |
| IESV91070DL | 0.763 | 5 | 544.8015 | 1 |
| CHITICHI | 0.5875 | 6 | 358.4705 | 4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Magaisa, A.; Ngadze, E.; Mamphogoro, T.P.; Moyo, M.P.; Kamutando, C.N. Exploring the Role of Rhizobacteria in Sorghum bicolor Adaptation to Combined Drought and Heat Stress. Microorganisms 2025, 13, 2454. https://doi.org/10.3390/microorganisms13112454
Magaisa A, Ngadze E, Mamphogoro TP, Moyo MP, Kamutando CN. Exploring the Role of Rhizobacteria in Sorghum bicolor Adaptation to Combined Drought and Heat Stress. Microorganisms. 2025; 13(11):2454. https://doi.org/10.3390/microorganisms13112454
Chicago/Turabian StyleMagaisa, Alec, Elizabeth Ngadze, Tshifhiwa Paris Mamphogoro, Martin Philani Moyo, and Casper Nyaradzai Kamutando. 2025. "Exploring the Role of Rhizobacteria in Sorghum bicolor Adaptation to Combined Drought and Heat Stress" Microorganisms 13, no. 11: 2454. https://doi.org/10.3390/microorganisms13112454
APA StyleMagaisa, A., Ngadze, E., Mamphogoro, T. P., Moyo, M. P., & Kamutando, C. N. (2025). Exploring the Role of Rhizobacteria in Sorghum bicolor Adaptation to Combined Drought and Heat Stress. Microorganisms, 13(11), 2454. https://doi.org/10.3390/microorganisms13112454







