Metagenomics and In Vitro Growth-Promoting Experiments Revealed the Potential Roles of Mycorrhizal Fungus Humicolopsis cephalosporioides and Helper Bacteria in Cheilotheca humilis Growth
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Morphological Observation
2.2. Sampling of the Rhizosphere Soil Samples
2.3. DNA Extraction and Metagenomic Sequencing
2.4. Metagenomic Assembly and Gene Function Annotation
2.5. Metagenome Binning, Taxonomy Assignment, and Phylogenetic Analysis
2.6. Statistical Analysis and Visualization
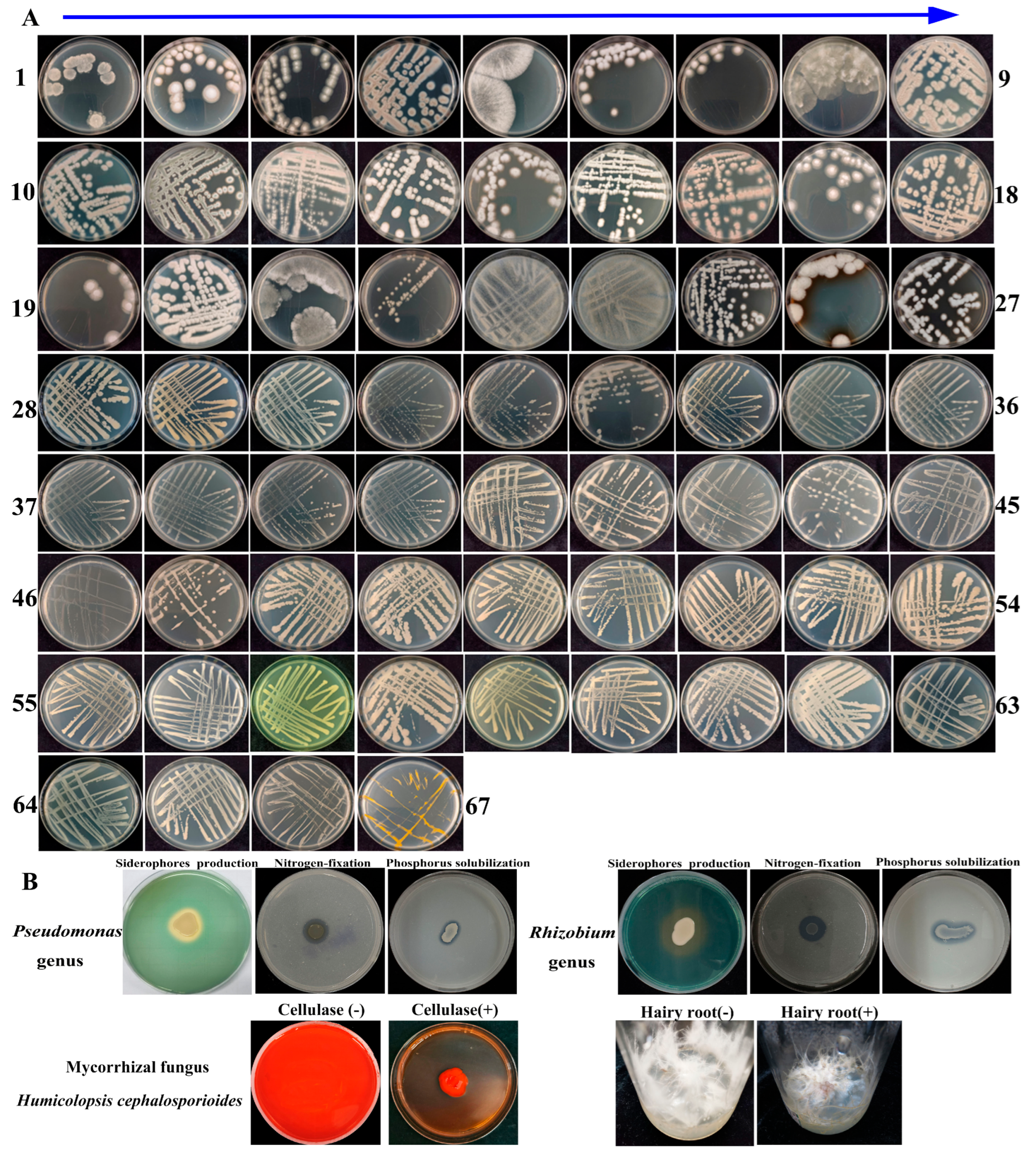
2.7. Isolation and Identification of Microorganisms
2.8. Assay for In Vitro Growth-Promoting Characteristics of Isolated Microorganisms
3. Results
3.1. Morphological Observation of Myco-Heterotrophic Plants
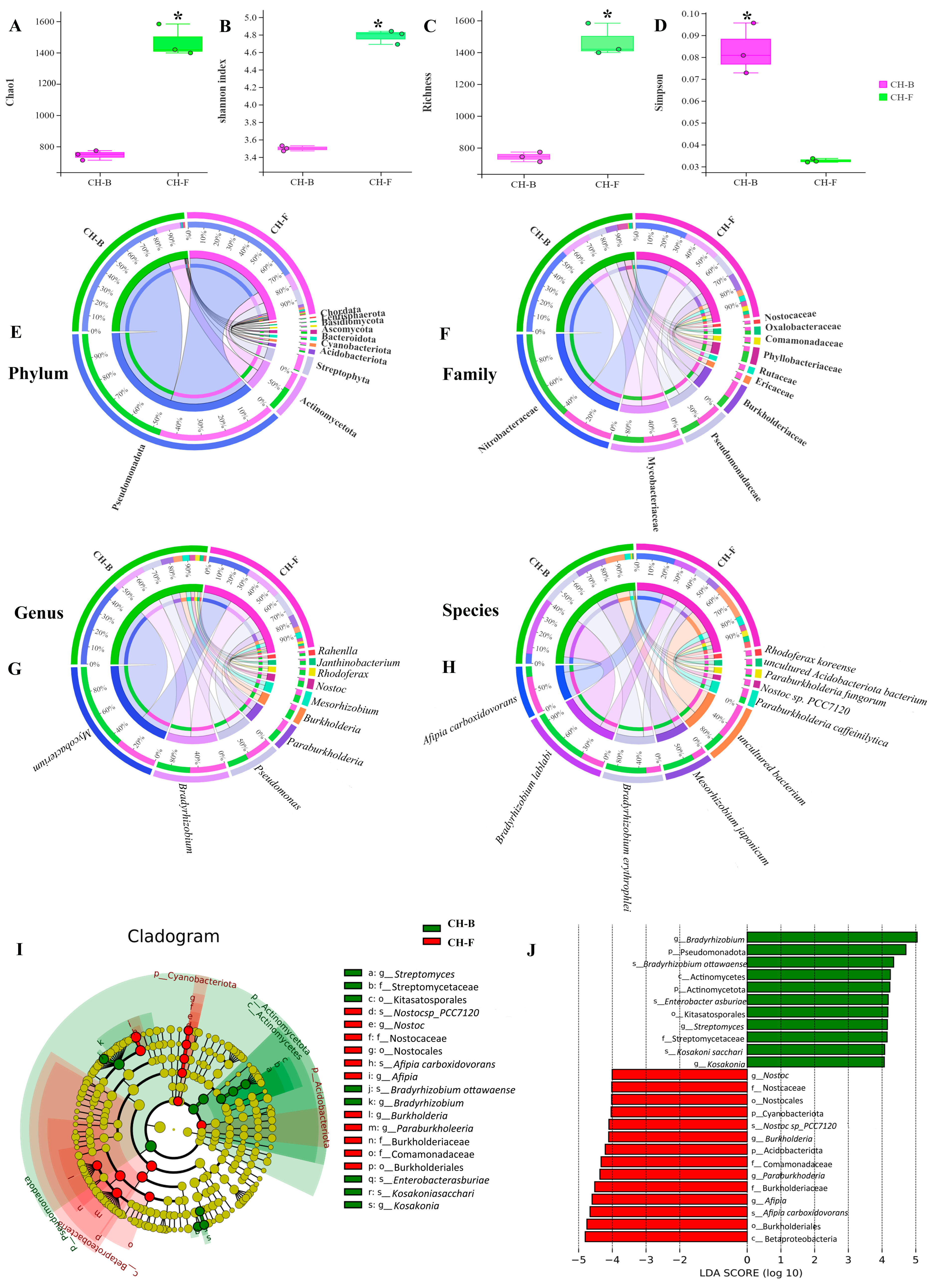
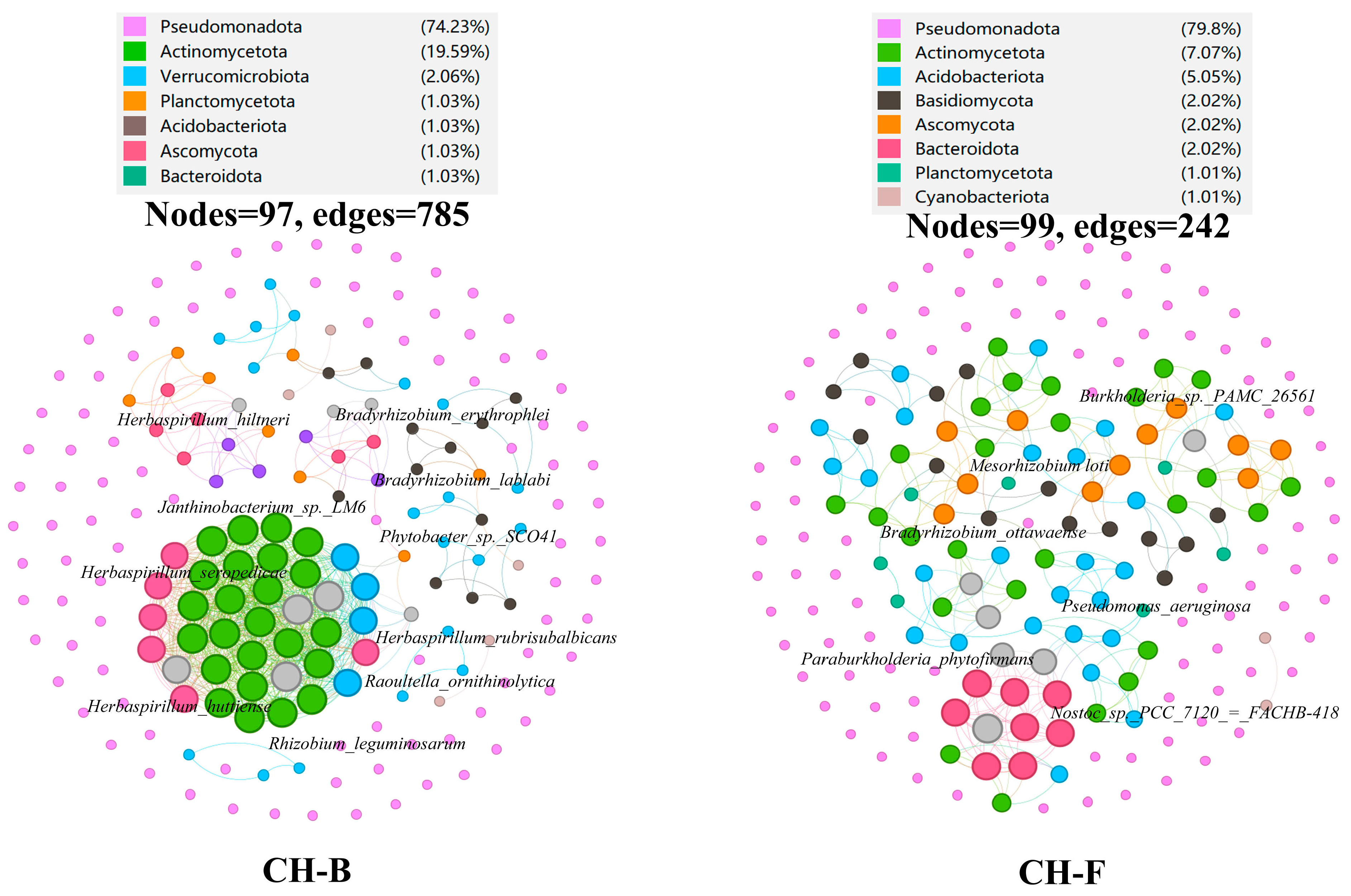
3.2. The Diversity, Composition, and Structure of Rhizosphere Microorganisms in the Cheilotheca humilis
3.3. Predictive Core Taxa of Rhizosphere Microbial Community in the Mycoheterotrophic Plant Cheilotheca humilis
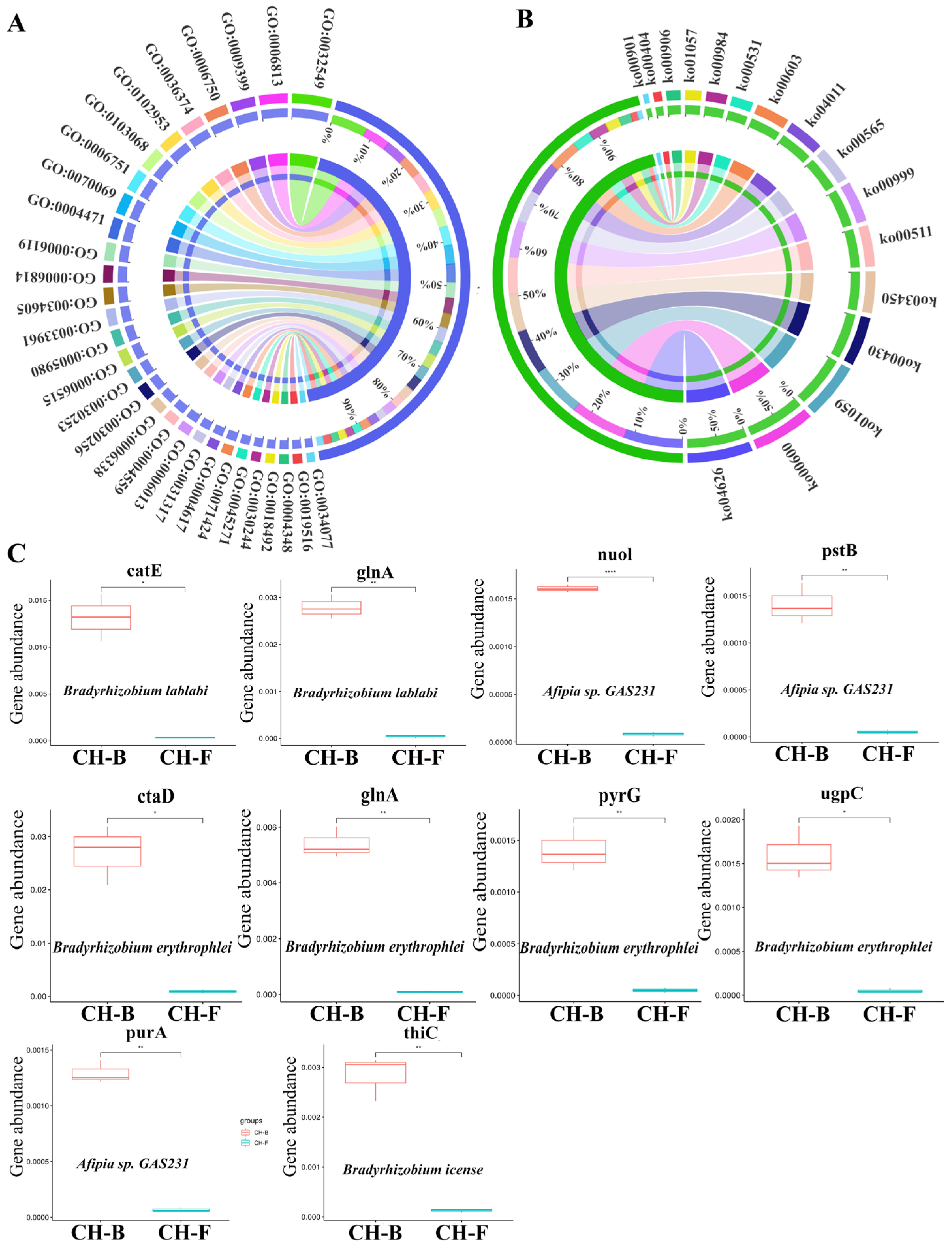
3.4. Functional Characteristics Analysis of Genes Associated with Rhizosphere Microbes in the Myco-Heterotrophic Plant Cheilotheca humilis
3.5. Metagenome-Assembled Genomes (MAGs) in the Rhizosphere of Myco-Heterotrophic Plant Cheilotheca humilis
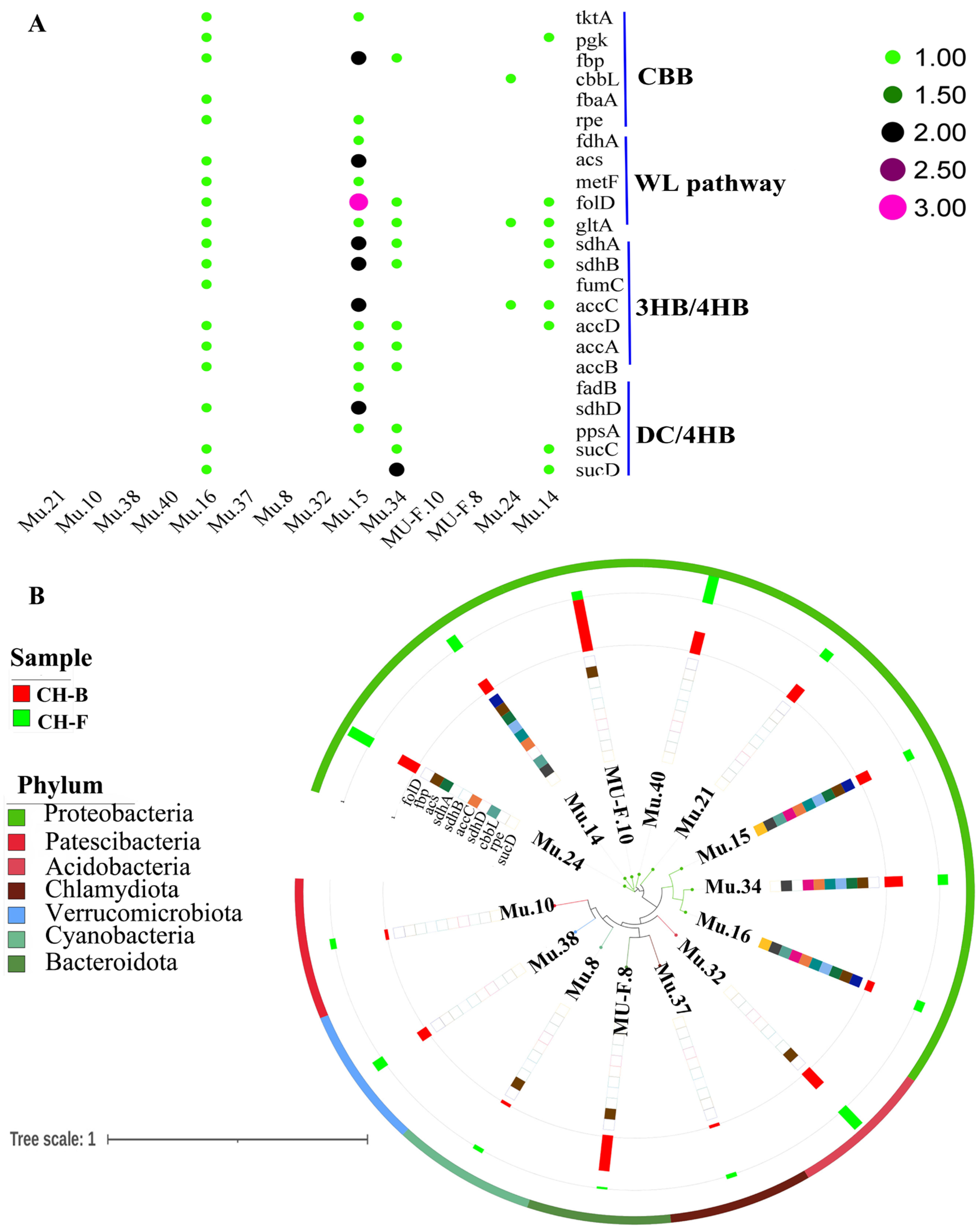
3.6. Carbon Fixation Pathways in the Rhizosphere Metagenome of Myco-Heterotrophic Plant Cheilotheca humilis
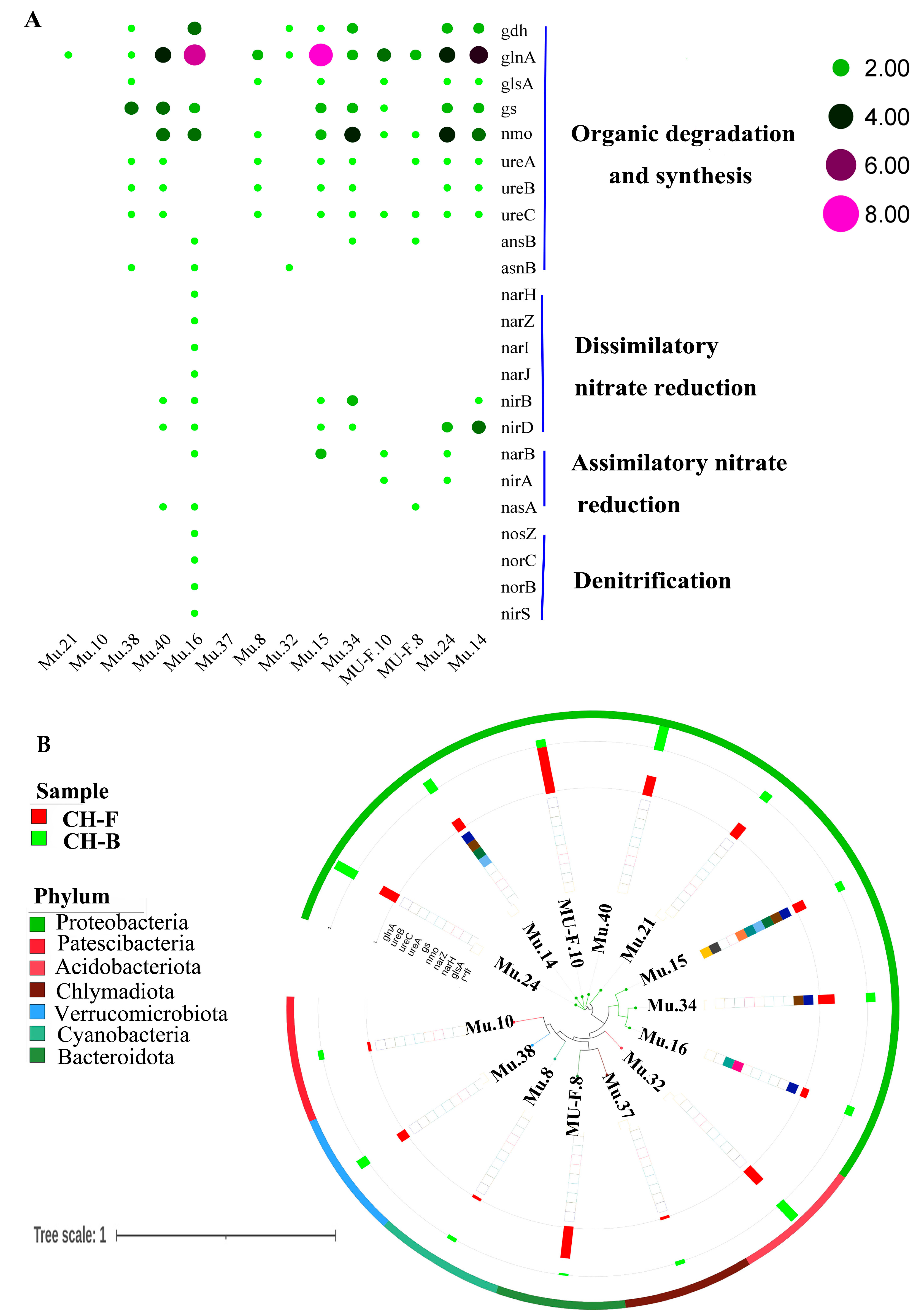
3.7. Nitrogen-Transforming Network in the Rhizosphere Metagenome of Myco-Heterotrophic Plant Cheilotheca humilis
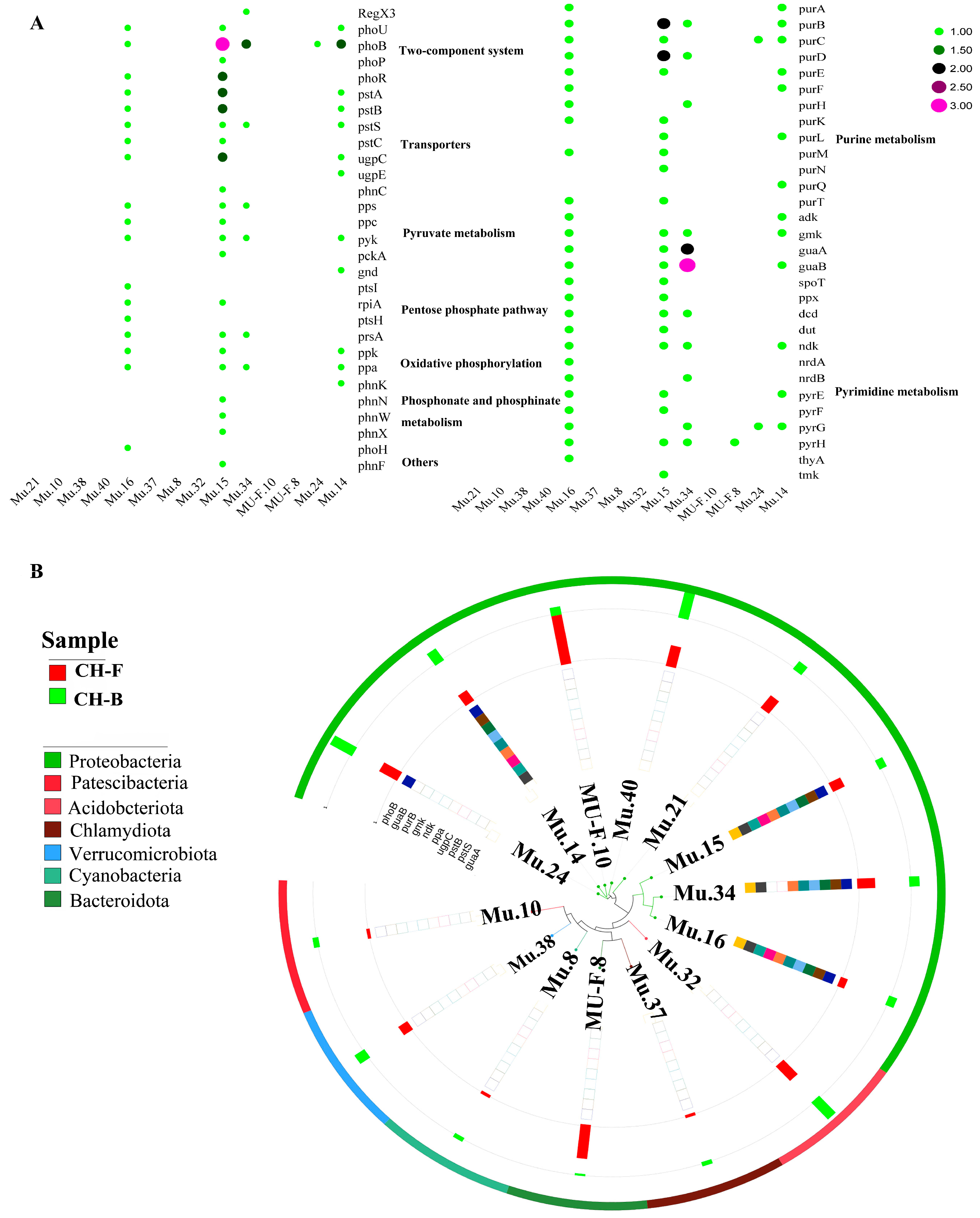
3.8. Phosphorus Metabolism in the Rhizosphere Metagenome of Myco-Heterotrophic Plant Cheilotheca humilis
3.9. Evaluation of the In Vitro Growth-Promoting Ability of Isolated Microorganisms
4. Discussion
4.1. Metagenomics and Microbial Cultivation Demonstrated the Community Diversity and Key Roles of Mycorrhizal Fungus H. cephalosporioides
4.2. Mycorrhizal Helper Bacteria Might Involve in the Growth of Cheilotheca humilis by Carbon Fixation Pathway
4.3. Mycorrhizal Helper Bacteria Might Be Involved in the Growth of Cheilotheca humilis by Nitrogen Transformation Pathway
4.4. Mycorrhizal Helper Bacteria Might Be Involved in the Growth of Cheilotheca humilis by Phosphorus Metabolism Pathway
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wallace, G.D. Studies of the Monotropoideae (Ericaceae): Taxonomy and distribution. Wasmann J. Biol. 1975, 33, 1–88. [Google Scholar]
- Logacheva, M.D.; Schelkunov, M.I.; Shtratnikova, V.Y.; Matveeva, M.V.; Penin, A.A. Comparative analysis of plastid genomes of non-photosynthetic Ericaceae and their photosynthetic relatives. Sci. Rep. 2016, 6, 30042. [Google Scholar] [CrossRef]
- Liu, X.; Liao, X.; Chen, D.; Zheng, Y.; Yu, X.; Xu, X.; Liu, Z.; Lan, S. The complete chloroplast genome sequence of Monotropa uniflora (Ericaceae). Mitochondrial DNA Part B 2020, 5, 3168–3169. [Google Scholar] [CrossRef]
- Leake, J.R. The biology of myco-heterotrophic (‘saprophytic’) plants. New Phytol. 1994, 127, 171–216. [Google Scholar] [CrossRef]
- Hsu, T.-W.; Kuoh, C.-S.; Hsieh, C.-F. Cheilotheca. In Editorial Committee of the Flora of Taiwan (ed) Flora of Taiwan, 2nd ed.; Department of Botany, National Taiwan University: Taipei, Taiwan, 1998; pp. 5–6. [Google Scholar]
- Massicotte, H.B.; Melville, L.H.; Peterson, R.L. Structural features of mycorrhizal associations in two members of the Monotropoideae, Monotropa uniflora and Pterospora andromedea. Mycorrhiza 2005, 15, 101–110. [Google Scholar] [CrossRef]
- Matsuda, Y.; Okochi, S.; Katayama, T.; Yamada, A.; Ito, S.I. Mycorrhizal fungi associated with Monotropastrum humile (Ericaceae) in central Japan. Mycorrhiza 2011, 21, 569–576. [Google Scholar] [CrossRef]
- Johansson, V.A.; Bahram, M.; Tedersoo, L.; Kõljalg, U.; Eriksson, O. Specificity of fungal associations of Pyroleae and Monotropa hypopitys during germination and seedling development. Mol. Ecol. 2017, 26, 2591–2604. [Google Scholar] [CrossRef]
- Ogura-Tsujita, Y.; Gebauer, G.; Hashimoto, T.; Umata, H.; Yukawa, T. Evidence for novel and specialized mycorrhizal parasitism: The orchid Gastrodia confusa gains carbon from saprotrophic Mycena. Proc. R. Soc. B-Biol. Sci. 2017, 276, 761–767. [Google Scholar] [CrossRef]
- Li, T.; Cui, R.; Geng, G.; Dong, Y.Z.; Xu, Y.; Sun, Y.C.; Stevanato, P.; Yu, L.H.; Liu, J.H.; Nurminsky, V.N.; et al. Sugar accumulation stage in sugar beets is a key stage in response to continuous cropping soil microbial community assembly. Plant Soil. 2024, 504, 457–473. [Google Scholar] [CrossRef]
- Ali, A.; Ghani, M.I.; Ding, H.; Iqbal, M.; Cheng, Z.; Cai, Z. Garlic substrate induces cucumber growth development and decreases fusarium wilt through regulation of soil microbial community structure and diversity in replanted disturbed soil. Int. J. Mol. Sci. 2020, 21, 6008. [Google Scholar] [CrossRef]
- Jia, T.; Cao, M.; Wang, R. Effects of restoration time on microbial diversity in rhizosphere and non-rhizosphere soil of Bothriochloa ischaemum. Int. J. Environ. Res. Public Health 2018, 15, 2155. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.W.; Shen, H.Y.; Liu, H.M.; Song, F.; Li, P.; Peng, N.; Zhao, S.M. Unraveling the microbial community and succession during zha-chili fermentation and their relationships with flavor formation. Food Res. Int. 2022, 157, 111239. [Google Scholar] [CrossRef] [PubMed]
- Lv, N.N.; Liu, Y.; Guo, T.F.; Liang, P.Z.; Li, R.; Liang, P.; Gao, X.W. The influence of Bt cotton cultivation on the structure and functions of the soil bacterial community by soil metagenomics. Ecotoxicol. Environ. Saf. 2022, 236, 113452. [Google Scholar] [CrossRef]
- Chang, W.N.; Chen, W.J.; Hu, Y.L.; Wang, Z.G. Bacillus altitudinis LZP02 improves rice growth by reshaping the rhizosphere microbiome. Plant Soil 2023, 498, 279–294. [Google Scholar] [CrossRef]
- Li, D.; Chen, W.; Luo, W.; Zhang, H.; Liu, Y.; Shu, D.; Wei, G. Seed microbiomes promote Astragalus mongholicus seed germination through pathogen suppression and cellulose degradation. Microbiome 2025, 13, 23. [Google Scholar] [CrossRef]
- Wang, J.; Tang, X.; Mo, Z.; Mao, Y. Metagenome-assembled genomes from Pyropia haitanensis microbiome provide insights into the potential metabolic functions to the seaweed. Front. Microbiol. 2022, 13, 857901. [Google Scholar] [CrossRef]
- Xu, L.; Dong, Z.B.; Chiniquy, D.; Pierroz, G.; Deng, S.; Gao, C.; ColemanDerr, D. Genome-resolved metagenomics reveals role of iron metabolism in drought-induced rhizosphere microbiome dynamics. Nat. Commun. 2021, 12, 3209. [Google Scholar] [CrossRef]
- Sangwan, S.; Prasanna, R. Mycorrhizae helper bacteria: Unlocking their potential as bioenhancers of plant-arbuscular mycorrhizal fungal associations. Microb. Ecol. 2022, 84, 1–10. [Google Scholar] [CrossRef]
- Xiang, T.H.; Li, J.S.; Bao, S.Y.; Xu, Z.X.; Wang, L.Z.; Long, F.Z.; He, C.J. Digital RNA-seq transcriptome plus tissue anatomy analyses reveal the developmental mechanism of the calabash-shaped root in Tetrastigma hemsleyanum. Tree Physiol. 2021, 41, 1729–1748. [Google Scholar] [CrossRef]
- Kuang, L.; Li, T.; Wang, B.; Peng, J.; Li, J.; Li, P.; Jiang, J. Diseased-induced multifaceted variations in community assembly and functions of plant-associated microbiomes. Front. Microbiol. 2023, 14, 1141585. [Google Scholar] [CrossRef]
- Kui, L.; Chen, B.; Chen, J.; Sharifi, R.; Dong, Y.; Zhang, Z.; Miao, J. A comparative analysis on the structure and function of the Panax notoginseng rhizosphere microbiome. Front. Microbiol. 2021, 12, 673512. [Google Scholar] [CrossRef]
- Wu, X.; Bei, S.; Zhou, X.; Luo, Y.; He, Z.; Song, C.; Yuan, H.; Pivato, B.; Liesack, W.; Peng, J. Metagenomic insights into genetic factors driving bacterial niche differentiation between bulk and rhizosphere soils. Sci. Total Environ. 2023, 891, 164221. [Google Scholar] [CrossRef]
- Huang, Y.; Hu, H.; Yue, E.; Ying, W.; Niu, T.; Yan, J.; Lu, Q.; Ruan, S. Role of plant metabolites in the formation of bacterial communities in the rhizosphere of Tetrastigma hemsleyanum. Front. Microbiol. 2023, 14, 1292896. [Google Scholar] [CrossRef]
- Li, Y.; Shi, X.; Tan, W.; Ling, Q.; Pei, F.; Luo, S.; Qin, P.; Yuan, H.; Huang, L.; Yu, F. Metagenomics combined with metabolomics reveals the effect of Enterobacter sp. inoculation on the rhizosphere microenvironment of Bidens pilosa L. in heavy metal contaminated soil. J. Hazard. Mater. 2023, 458, 132033. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.B.; Mao, X.Z.; Yang, J.C.; Chen, X.; Mao, F.L.; Xu, Y. dbCAN: A web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012, 40, W445–W451. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.X.; Jiang, H.D.; Huang, Y.; Zhong, L.S.; Xu, Q.; Yang, Q.G.; Chai, S.F. Combined analysis of metagenome and transcriptome revealed the adaptive mechanism of different golden camellia species in karst regions. Front. Plant Sci. 2023, 14, 1180472. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.D.; Li, F.; Kirton, E.; Thomas, A.; Egan, R.; An, H.; Wang, Z. MetaBAT 2: An adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. Peer J 2019, 7, e7359. [Google Scholar] [CrossRef]
- Wu, Y.W.; Simmons, B.A.; Singer, S.W. MaxBin 2.0: An automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 2016, 32, 605–607. [Google Scholar] [CrossRef]
- Alneberg, J.; Bjarnason, B.S.; de Bruijn, I.; Schirmer, M.; Quick, J.; Ijaz, U.Z.; Lahti, L.; Loman, N.J.; Andersson, A.F.; Quince, C. Binning metagenomic contigs by coverage and composition. Nat. Methods 2014, 1, 1144–1146. [Google Scholar] [CrossRef]
- Uritskiy, G.V.; DiRuggiero, J.; Taylor, J. MetaWRAP-a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome 2018, 6, 158. [Google Scholar] [CrossRef]
- Bowers, R.M.; Kyrpides, N.C.; Stepanauskas, R.; Harmon-Smith, M.; Doud, D.; Reddy, T.B.K.; Schulz, F.; Jarett, J.; Rivers, A.R.; Eloe-Fadrosh, E.A.; et al. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat. Biotechnol. 2017, 35, 725–731, Erratum in Nat. Biotechnol. 2018, 36, 196. https://doi.org/10.1038/nbt0218-196a. [Google Scholar] [CrossRef]
- Chaumeil, P.A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk v2: Memory friendly classification with the genome taxonomy database. Bioinformatics 2022, 38, 5315–5316. [Google Scholar] [CrossRef]
- Parks, D.H.; Chuvochina, M.; Rinke, C.; Mussig, A.J.; Chaumeil, P.A.; Hugenholtz, P. GTDB: An ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res. 2022, 50, D785–D794. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Wagner, H. Vegan: Community Ecology Package; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer International Publishing: Cham, Germany, 2016; pp. 189–201. [Google Scholar] [CrossRef]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An open-source software for exploring and manipulating networks. In Proceedings of the International AAAI Conference on Web and Social Media, San Jose, CA, USA, 17–20 May 2009; Volume 3, pp. 361–362. [Google Scholar]
- Mendes, R.; Kruijt, M.; de Bruijn, I.; Dekkers, E.; van der Voort, M.; Schneider, J.H.; Piceno, Y.M.; DeSantis, T.Z.; Andersen, G.L.; Bakker, P.A.; et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 2011, 332, 1097–1100. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, D.J.; David, A.S.; Menges, E.S.; Searcy, C.A.; Afkhami, M.E. Environmental stress destabilizes microbial networks. ISME J. 2021, 15, 1722–1734. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Wang, J.; Xie, X.; Li, B.; Yang, L.; Song, F.; Zhou, Y.; Jiang, M. Complete genome analysis and antimicrobial mechanism of Bacillus velezensis GX0002980 reveals its biocontrol potential against mango anthracnose disease. Microbiol. Spectr. 2025, 13, e02685-24. [Google Scholar] [CrossRef] [PubMed]
- Arora, N.K.; Verma, M. Modified microplate method for rapid and efficient estimation of siderophore produced by bacteria. 3 Biotech 2017, 7, 381. [Google Scholar] [CrossRef] [PubMed]
- Schwyn, B.; Neilands, J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Gaete, A.; Andreani-Gerard, C.; Maldonado, J.E.; Muñoz-Torres, P.A.; SepúlvedaChavera, G.F.; González, M. Bioprospecting of plant growth-promoting traits of Pseudomonas sp. strain C3 isolated from the atacama desert: Molecular and culture-based analysis. Diversity 2022, 14, 388. [Google Scholar] [CrossRef]
- Nacoon, S.; Seemakram, W.; Ekprasert, J.; Jogloy, S.; Kuyper, T.W.; Mongkolthanaruk, W.; Riddech, N.; Somdee, T.; Boonlue, S. Promoting growth and production of sunchoke (Helianthus tuberosus) by co-inoculation with phosphate solubilizing bacteria and arbuscular mycorrhizal fungi under drought. Front. Plant Sci. 2022, 13, 1022319. [Google Scholar] [CrossRef]
- Stephan, C.; Al Assaad, M.; Levine, M.F.; Deshpande, A.; Sigouros, M.; Manohar, J.; Sboner, A.; Elemento, O.; Pavlick, A.C.; Mosquera, J.M. Whole genome sequencing elucidates etiological differences in MCPyV-negative Merkel cell carcinoma. Pathol. Res. Pract. 2024, 263, 155668. [Google Scholar] [CrossRef]
- Zhao, X.L. A new variety of Monotropa (Pyrolaceae)-Monotropa uniflora L. var. huoshanensis. Subtrop. Plant Sci. 2016, 45, 153–155. (In Chinese) [Google Scholar]
- Trudell, S.A.; Rygiewicz, P.T.; Edmonds, R.L. Nitrogen and carbon stable isotope abundances support the myco-heterotrophic nature and host-specificity of certain achlorophyllous plants. New Phytol. 2003, 160, 391–401. [Google Scholar] [CrossRef]
- Hynson, N.A.; Preiss, K.; Gebauer, G.; Bruns, T.D. Isotopic evidence of full and partial myco-heterotrophy in the plant tribe Pyroleae (Ericaceae). New Phytol. 2009, 182, 719–726. [Google Scholar] [CrossRef]
- Graham, S.W.; Lam, V.K.; Merckx, V.S. Plastomes on the edge: The evolutionary breakdown of mycoheterotroph plastid genomes. New Phytol. 2017, 214, 48–55. [Google Scholar] [CrossRef]
- Barrett, C.F.; Kennedy, A.H. Plastid genome degradation in the endangered, mycoheterotrophic, North American orchid Hexalectris warnockii. Genome Biol. Evol. 2018, 10, 1657–1662. [Google Scholar] [CrossRef]
- Givnish, T.J.; Zuluaga, A.; Spalink, D.; Soto Gomez, M.; Lam, V.K.; Saarela, J.M.; Sass, C.; Iles, W.J.; de Sousa, D.J.; Leebens-Mack, J.; et al. Monocot plastid phylogenomics, timeline, net rates of species diversification, the power of multi-gene analyses, and a functional model for the origin of monocots. Am. J. Bot. 2018, 105, 1888–1910. [Google Scholar] [CrossRef] [PubMed]
- Li, H.F.; Song, F.; Wu, X.X.; Deng, C.L.; Xu, Q.; Peng, S.A.; Pan, Z.Y. Microbiome and metagenome analysis reveals Huanglongbing affects the abundance of citrus rhizosphere bacteria associated with resistance and energy metabolism. Horticulturae 2021, 7, 151. [Google Scholar] [CrossRef]
- Zheng, Y.; Cao, X.; Zhou, Y.; Ma, S.; Wang, Y.; Li, Z.; Zhao, D.; Yang, Y.; Zhang, H.; Meng, C.; et al. Purines enrich root-associated Pseudomonas and improve wild soybean growth under salt stress. Nat. Commun. 2024, 15, 3520. [Google Scholar] [CrossRef] [PubMed]
- Strullu-Derrien, C.; Selosse, M.A.; Kenrick, P.; Martin, F.M. The origin and evolution of mycorrhizal symbioses: From palaeomycology to phylogenomics. New Phytol. 2018, 220, 1012–1030. [Google Scholar] [CrossRef]
- Tedersoo, L.; Bahram, M.; Zobel, M. How mycorrhizal associations drive plant population and community biology. Science 2020, 367, eaba1223. [Google Scholar] [CrossRef]
- Shi, J.; Wang, X.; Wang, E. Mycorrhizal symbiosis in plant growth and stress adaptation: From genes to ecosystems. Annu. Rev. Plant Biol. 2022, 74, 569–607. [Google Scholar] [CrossRef]
- Pellitier, P.T.; Zak, D.R. Ectomycorrhizal fungi and the enzymatic liberation of nitrogen from soil organic matter: Why evolutionary history matters. New Phytol. 2018, 217, 68–73. [Google Scholar] [CrossRef]
- Gupta, S.K.; Chakraborty, A.P. Mycorrhiza helper bacteria: Future prospects. Int. J. Res. Rev. 2020, 7, 387–391. [Google Scholar]
- Zhang, L.; Zhou, J.; George, T.S.; Limpens, E.; Feng, G. Arbuscular mycorrhizal fungi conducting the hyphosphere bacterial orchestra. Trends Plant Sci. 2022, 27, 402–411. [Google Scholar] [CrossRef]
- Zhang, C.; van der Heijden, M.G.A.; Dodds, B.K.; Nguyen, T.B.; Spooren, J.; Valzano-Held, A.; Cosme, M.; Berendsen, R.L. A tripartite bacterial-fungal-plant symbiosis in the mycorrhiza-shaped microbiome drives plant growth and mycorrhization. Microbiome 2024, 12, 13, Erratum in Microbiome 2024, 12, 30. https://doi.org/10.1186/s40168-024-01776-2. [Google Scholar] [CrossRef]
- Wilcoxen, J.; Snider, S.; Hille, R. Substitution of silver for copper in the binuclear Mo/Cu center of carbon monoxide dehydrogenase from Oligotropha carboxidovorans. J. Am. Chem. Soc. 2011, 133, 12934–12936. [Google Scholar] [CrossRef]
- Kirti, A.; Prashar, V.; Kumar, A.; Pandey, S.; Rajaram, H. Thymidylate kinase (TMK) of the photosynthetic, nitrogen-fixing cyanobacterium Nostoc sp. strain PCC7120: Biophysical, biochemical and physiological characterisation. Plant Physiol. Biochem. 2021, 166, 416–426. [Google Scholar] [CrossRef]
- Srisawat, P.; Higuchi, T.M.; Numata, K. Microbial autotrophic biorefineries: Perspectives for biopolymer production. Polym. J. 2022, 54, 1139–1151. [Google Scholar] [CrossRef]
- Shen, H.; Wang, T.; Dong, W.; Sun, G.; Liu, J.; Peng, N.; Zhao, S. Metagenome-assembled genome reveals species and functional composition of Jianghan chicken gut microbiota and isolation of Pediococcus acidilactic with probiotic properties. Microbiome 2024, 12, 25. [Google Scholar] [CrossRef]
- van der Heijden, M.G.A.; Martin, F.M.; Selosse, M.A.; Sanders, I.R. Mycorrhizal ecology and evolution: The past, the present and the future. New Phytol. 2015, 205, 1406–1423. [Google Scholar] [CrossRef]
- Jin, H.; Pfeffer, P.E.; Douds, D.D.; Piotrowski, E.; Lammers, P.J.; Shachar-Hill, Y. The uptake, metabolism, transport and transfer of nitrogen in an arbuscular mycorrhizal symbiosis. New Phytol. 2005, 168, 687–696. [Google Scholar] [CrossRef]
- Mendoza-Suárez, M.; Akyol, T.Y.; Nadzieja, M.; Andersen, S.U. Increased diversity of beneficial rhizobia enhances faba bean growth. Nat. Commun. 2024, 15, 10673. [Google Scholar] [CrossRef]
- Schulte, C.C.M.; Borah, K.; Wheatley, R.M.; Terpolilli, J.J.; Saalbach, G.; Crang, N.; de Groot, D.H.; Ratcliffe, R.G.; Kruger, N.J.; Papachristodoulou, A.; et al. Metabolic control of nitrogen fixation in rhizobium-legume symbioses. Sci. Adv. 2021, 7, eabh2433. [Google Scholar] [CrossRef] [PubMed]
- Wasai-Hara, S.; Itakura, M.; Fernandes Siqueira, A.; Takemoto, D.; Sugawara, M.; Mitsui, H.; Sato, S.; Inagaki, N.; Yamazaki, T.; Imaizumi-Anraku, H.; et al. Bradyrhizobium ottawaense efficiently reduces nitrous oxide through high nosZ gene expression. Sci. Rep. 2023, 13, 18862. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.G.; Peng, J.; Chen, C.; Xiong, C.; Li, S.; Ge, A.; Wang, E.; Liesack, W. Harnessing biological nitrogen fixation in plant leaves. Trends Plant Sci. 2023, 28, 1391–1405. [Google Scholar] [CrossRef]
- Zhang, W.; Gao, M.; Hu, R.; Shang, Y.; Liu, M.; Lan, P.; Jiao, S.; Wei, G.; Chen, S. Nitrogen-fixing Paenibacillus haidiansis and Paenibacillus sanfengchensis: Two novel species from plant rhizospheres. Microorganisms 2024, 12, 2561. [Google Scholar] [CrossRef]
- Trovero, M.F.; Scavone, P.; Platero, R.; de Souza, E.M.; Fabiano, E.; Rosconi, F. Herbaspirillum seropedicae differentially expressed genes in response to iron availability. Front. Microbiol. 2018, 9, 1430. [Google Scholar] [CrossRef]
- Kuang, W.; Sanow, S.; Kelm, J.M.; Müller Linow, M.; Andeer, P.; Kohlheyer, D.; Northen, T.; Vogel, J.P.; Watt, M.; Arsova, B. N-dependent dynamics of root growth and nitrate and ammonium uptake are altered by the bacterium Herbaspirillum seropedicae in the cereal model Brachypodium distachyon. J. Exp. Bot. 2022, 73, 5306–5321. [Google Scholar] [CrossRef]
- Wang, W.; Shi, J.; Xie, Q.; Jiang, Y.; Yu, N.; Wang, E. Nutrient exchange and regulation in arbuscular mycorrhizal symbiosis. Mol. Plant 2017, 10, 1147–1158. [Google Scholar] [CrossRef] [PubMed]
- Abraham, J.; Silambarasan, S. Plant growth promoting bacteria Enterobacter asburiae JAS5 and Enterobacter cloacae JAS7 in mineralization of endosulfan. Appl. Biochem. Biotech. 2015, 175, 3336–3348. [Google Scholar] [CrossRef] [PubMed]
- Herpell, J.B.; Alickovic, A.; Diallo, B.; Schindler, F.; Weckwerth, W. Phyllosphere symbiont promotes plant growth through ACC deaminase production. ISME J. 2023, 17, 1267–1277. [Google Scholar] [CrossRef] [PubMed]

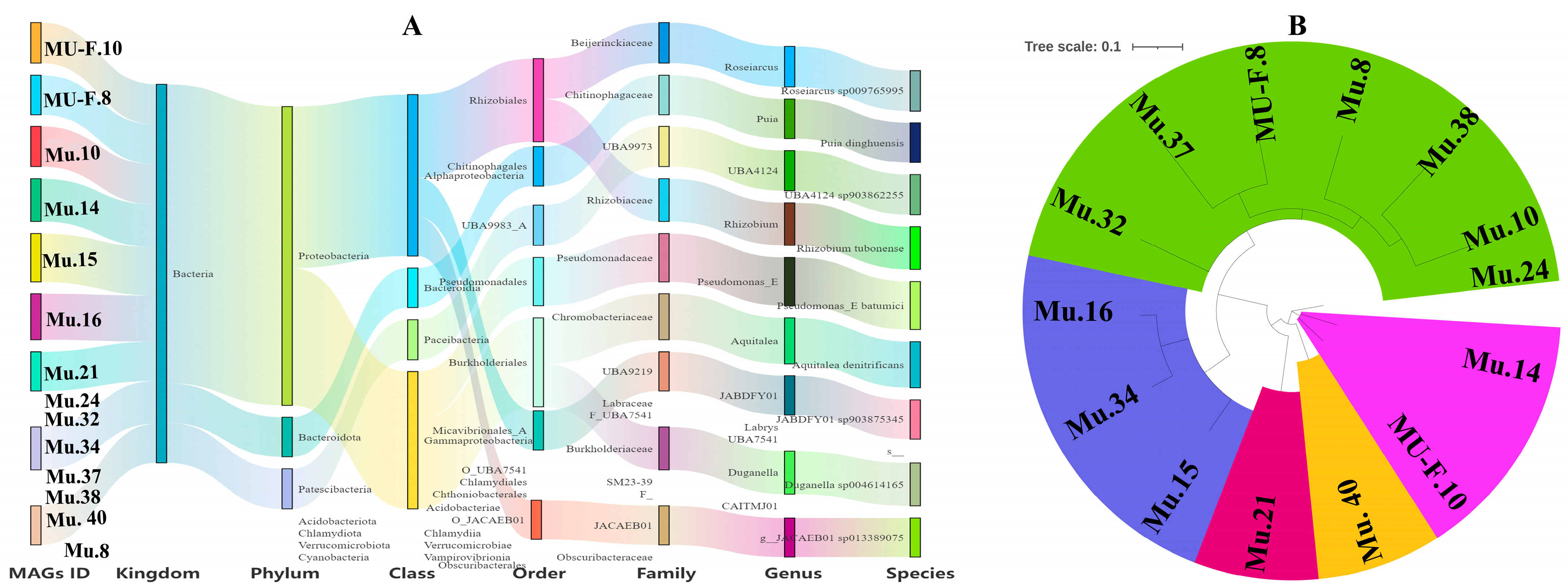
| MAG ID | Species Taxonomy | Closest Placement ANI | Completeness | Contamination | Contigs | Genome | N50 | GC |
|---|---|---|---|---|---|---|---|---|
| (%) | (%) | Num | Size | (%) | ||||
| Mu.10 | UBA4124 sp. | 79.31 | 81.24 | 0 | 29 | 804,524 | 44,130 | 41.84 |
| Mu.21 | JABDFY01 sp. | 77.59 | 96.4 | 2.94 | 136 | 2,157,904 | 23,608 | 57.59 |
| Mu.16 | Aquitalea denitrificans | 90.99 | 97.86 | 0.43 | 63 | 4,193,559 | 106,226 | 58.73 |
| Mu.37 | N/A | N/A | 77.53 | 3.04 | 216 | 1,326,563 | 7456 | 50.63 |
| Mu.32 | N/A | N/A | 96.15 | 5.22 | 71 | 4,659,288 | 115,614 | 59.11 |
| Mu.40 | JACAEB01 sp. | 77.13 | 99.13 | 1.16 | 80 | 3,422,884 | 80,120 | 63.76 |
| Mu.24 | N/A | N/A | 96.09 | 2.69 | 630 | 8,226,318 | 31,297 | 60.93 |
| Mu.38 | N/A | N/A | 93.88 | 7.6 | 519 | 4,456,784 | 12,606 | 54.57 |
| Mu.14 | Rhizobium tubonense | 84.43 | 89.81 | 3.27 | 455 | 5,778,709 | 23,353 | 59.38 |
| Mu.8 | N/A | N/A | 91.88 | 2.78 | 606 | 5,927,924 | 13,162 | 46.75 |
| Mu.15 | Pseudomonas_E batumici | 95.48 | 75.44 | 8.77 | 1698 | 6,777,919 | 4957 | 62.31 |
| Mu.34 | Duganella sp. | 84.75 | 78.06 | 7.64 | 1233 | 5,166,334 | 4986 | 62.06 |
| MU-F.10 | Roseiarcus | 79.98 | 74.25 | 6.28 | 1029 | 4,617,664 | 5534 | 62.82 |
| MU-F.8 | Puia dinghuensis | 78.15 | 91.01 | 7.44 | 1585 | 6,948,697 | 5212 | 53.16 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Shang, Y.; Wang, X.; Li, X.; Yu, Z.; Zeng, Z.; Chen, Z.; Wang, L.; Xiang, T.; Huang, X. Metagenomics and In Vitro Growth-Promoting Experiments Revealed the Potential Roles of Mycorrhizal Fungus Humicolopsis cephalosporioides and Helper Bacteria in Cheilotheca humilis Growth. Microorganisms 2025, 13, 2387. https://doi.org/10.3390/microorganisms13102387
Liu Y, Shang Y, Wang X, Li X, Yu Z, Zeng Z, Chen Z, Wang L, Xiang T, Huang X. Metagenomics and In Vitro Growth-Promoting Experiments Revealed the Potential Roles of Mycorrhizal Fungus Humicolopsis cephalosporioides and Helper Bacteria in Cheilotheca humilis Growth. Microorganisms. 2025; 13(10):2387. https://doi.org/10.3390/microorganisms13102387
Chicago/Turabian StyleLiu, Yawei, Yuhao Shang, Xin Wang, Xiao Li, Zhiming Yu, Zhanghui Zeng, Zhehao Chen, Lilin Wang, Taihe Xiang, and Xiaoping Huang. 2025. "Metagenomics and In Vitro Growth-Promoting Experiments Revealed the Potential Roles of Mycorrhizal Fungus Humicolopsis cephalosporioides and Helper Bacteria in Cheilotheca humilis Growth" Microorganisms 13, no. 10: 2387. https://doi.org/10.3390/microorganisms13102387
APA StyleLiu, Y., Shang, Y., Wang, X., Li, X., Yu, Z., Zeng, Z., Chen, Z., Wang, L., Xiang, T., & Huang, X. (2025). Metagenomics and In Vitro Growth-Promoting Experiments Revealed the Potential Roles of Mycorrhizal Fungus Humicolopsis cephalosporioides and Helper Bacteria in Cheilotheca humilis Growth. Microorganisms, 13(10), 2387. https://doi.org/10.3390/microorganisms13102387






