Genome Analysis of Alginate-Degrading Bacterium Vibrio sp. 32415 and Optimization of Alginate Lyase Production
Abstract
1. Introduction
2. Materials and Methods
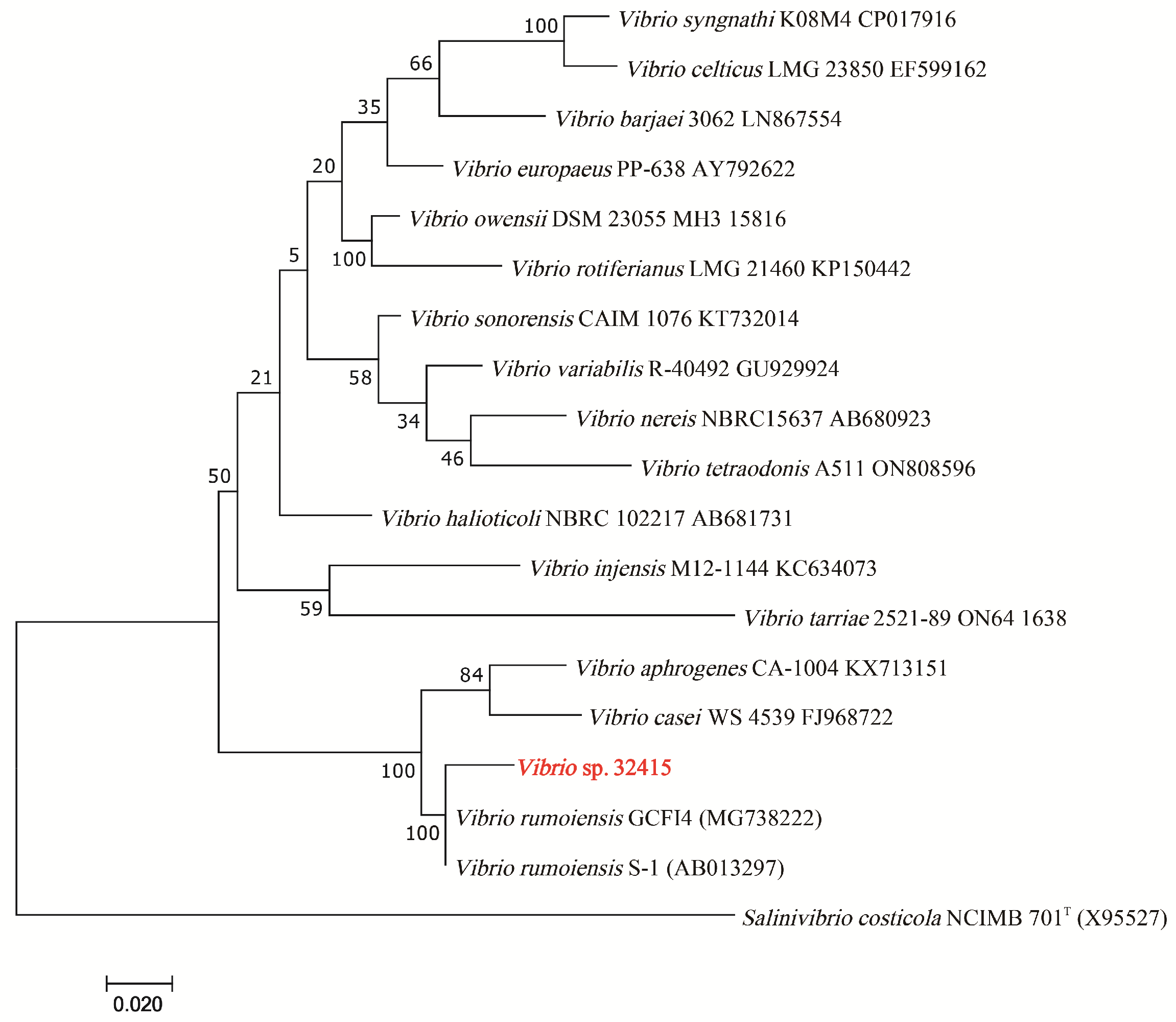
2.1. Screening and Identification of V. sp. 32415
2.2. Genome Sequencing and Annotation
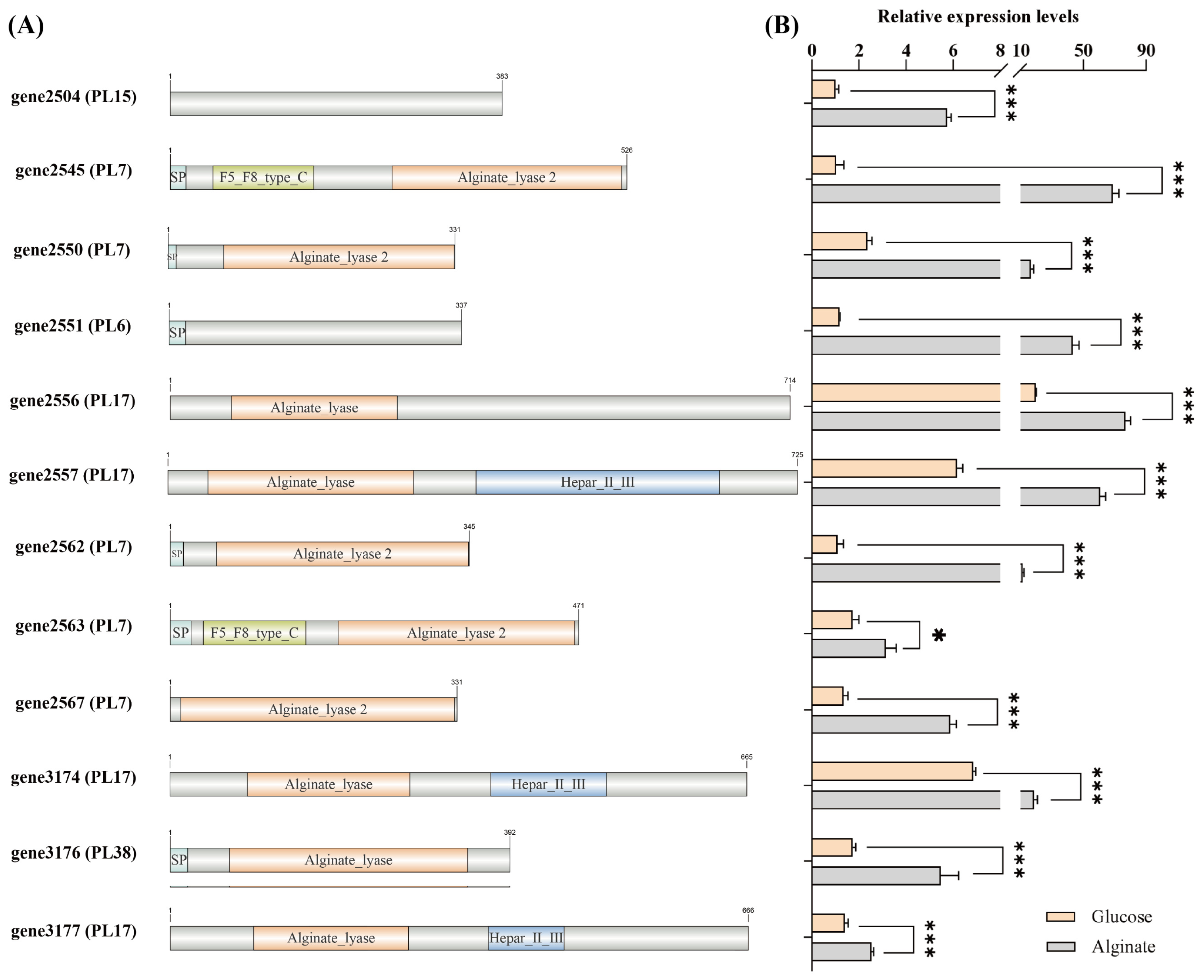
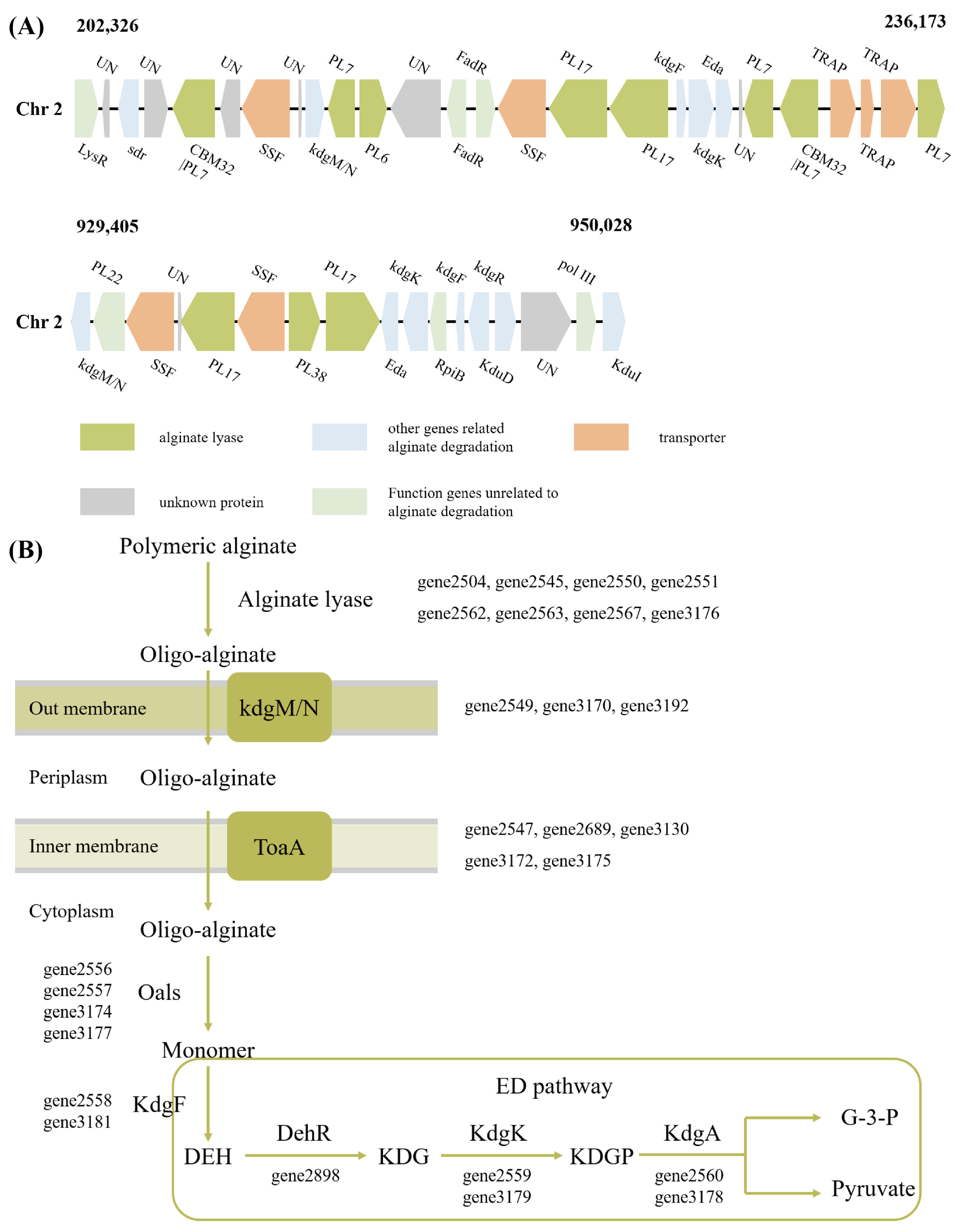
2.3. Carbohydrate Utilization Capacity and Alginate Metabolism
2.4. Optimization of Enzyme Production
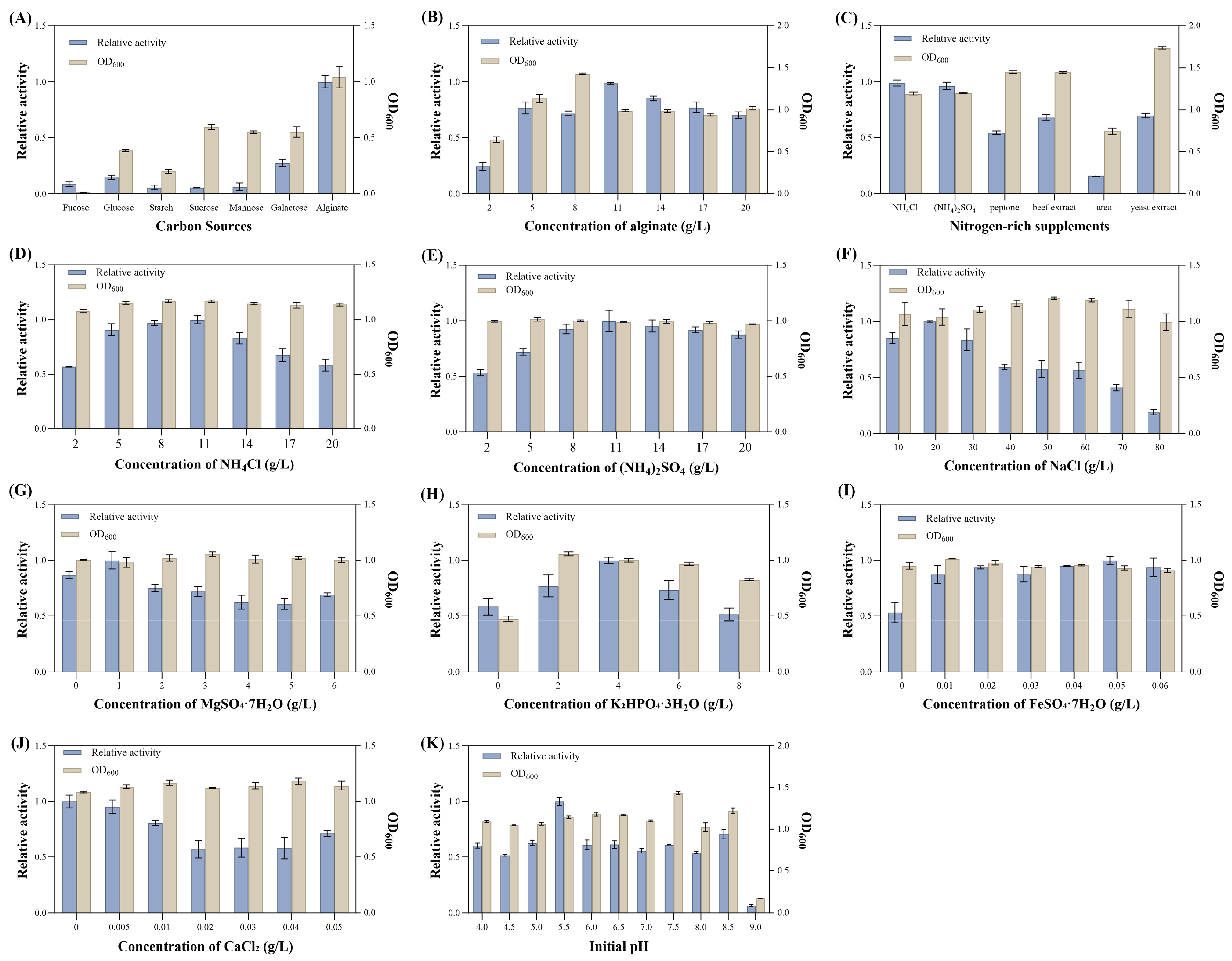
2.4.1. Optimization of Medium Components
One-Factor-at-a-Time Experiment
Plackett–Burman Experiment
Box–Behnken Design
2.4.2. Optimization of Culture Conditions
2.5. Detection of Alginate Lyase Activity and Enzymatic Properties
3. Results and Discussion
3.1. Separation, Identification and Characterization of V. sp. 32415
3.2. Genome Specifics
3.3. Alginate Metabolism and Utilization
3.4. Optimization of Enzyme Production
3.4.1. One-Factor-at-a-Time Fermentation Condition Optimization
3.4.2. Screening the Significant Factors Affecting the Enzyme Production of the Strain Through Plackett–Burman Experiment
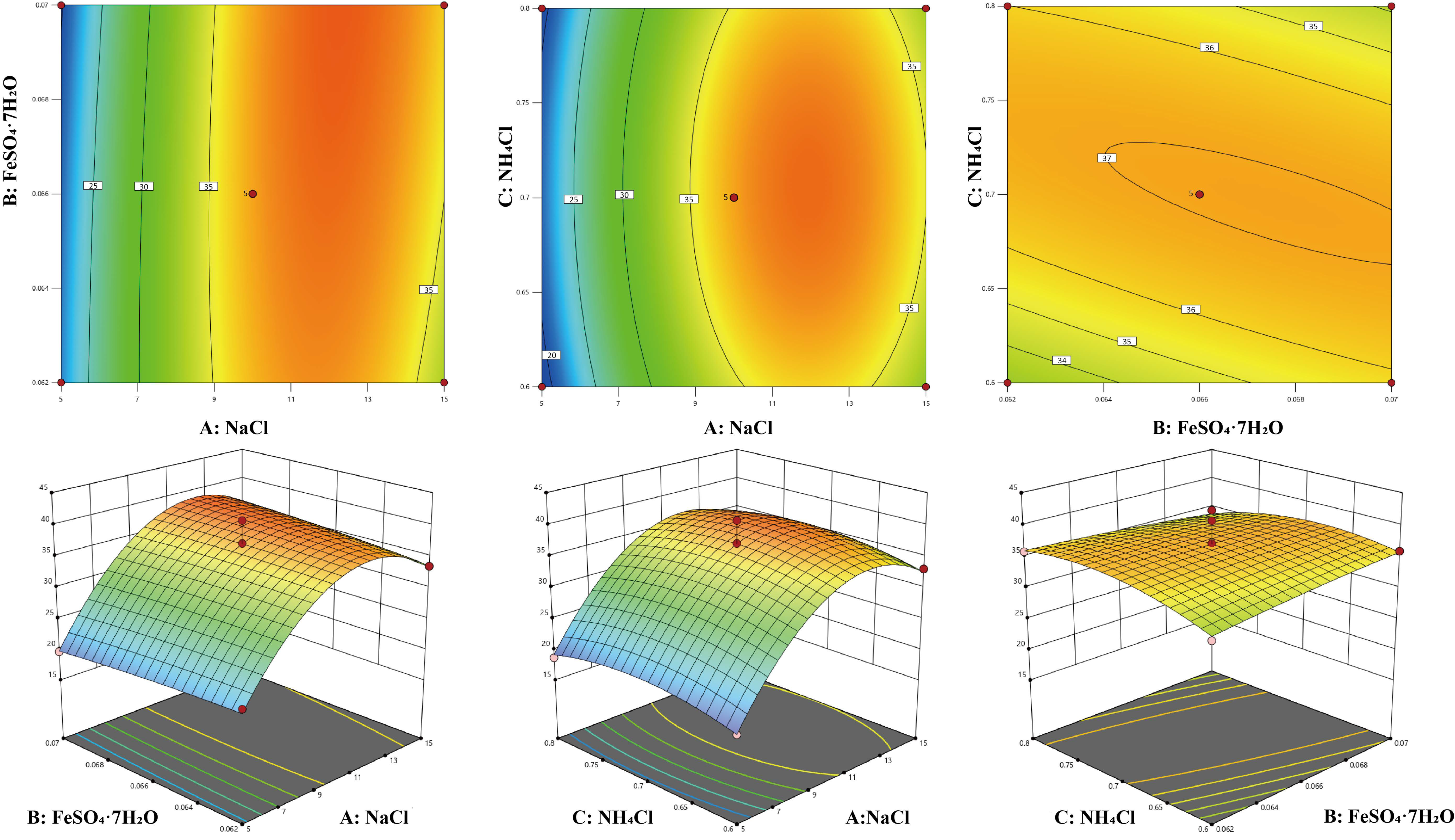
3.4.3. Box–Behnken Design Optimization Factor Parameters
3.4.4. Verification of Optimization Results
3.4.5. Fermentation Condition Optimization
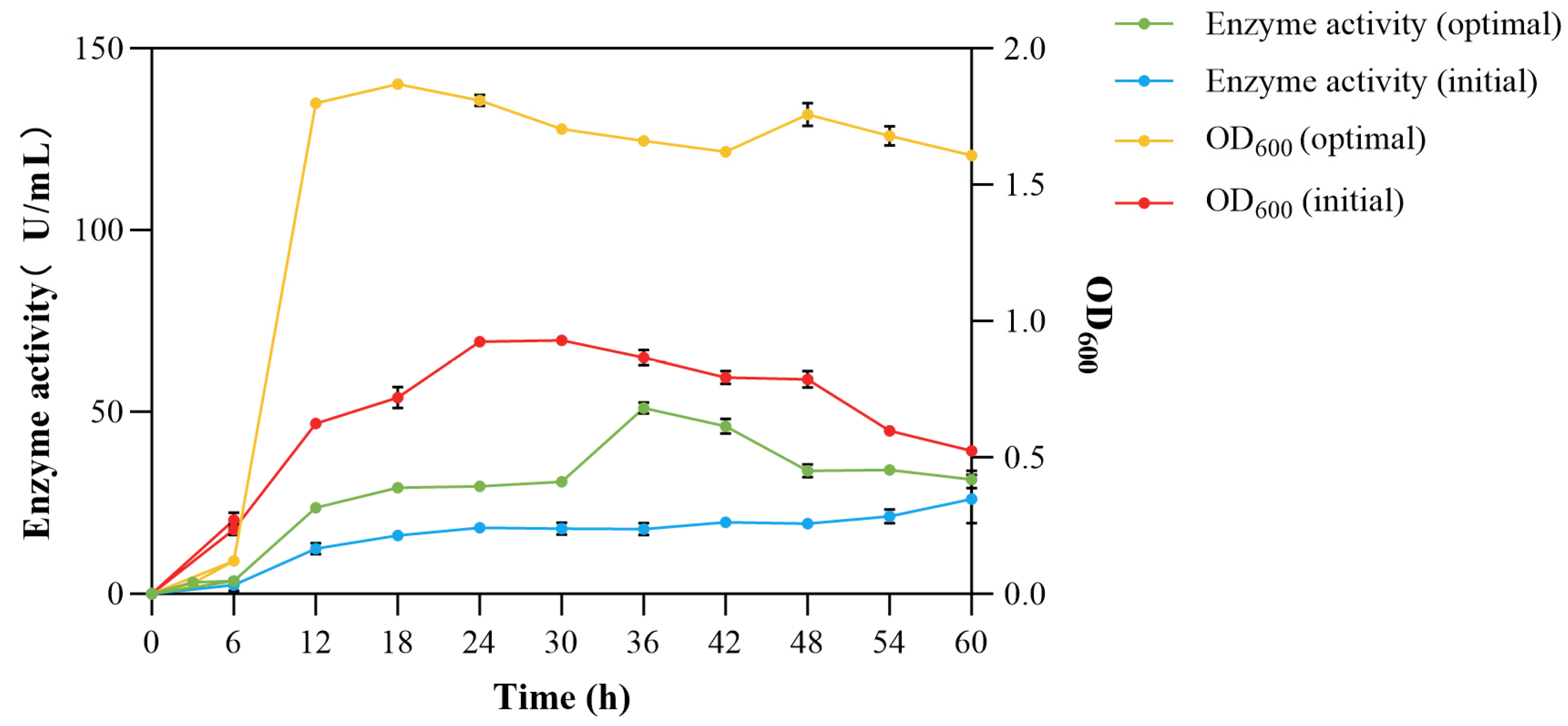
3.4.6. Growth and Enzyme Production Curves Before and After Optimization
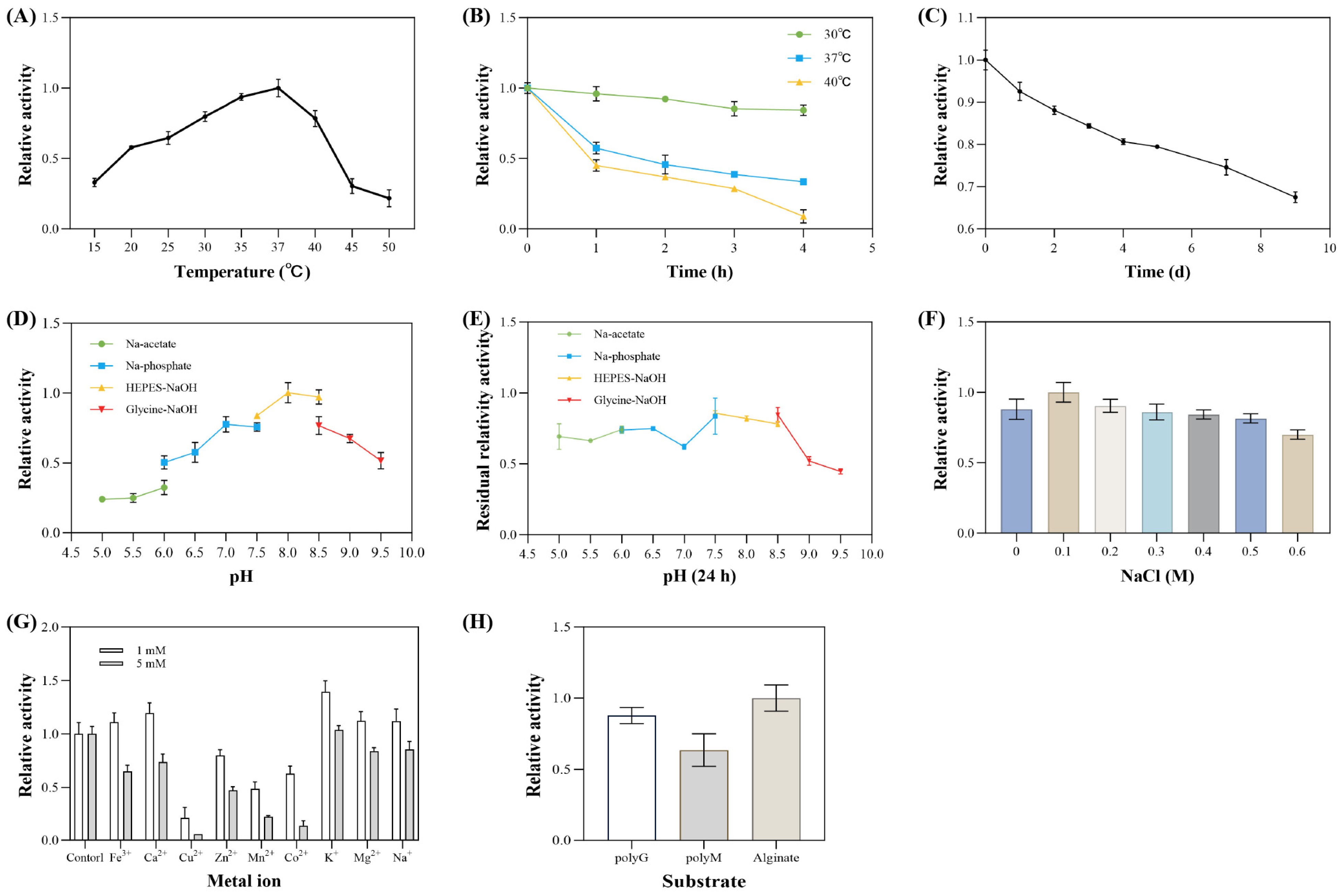
3.5. Enzymatic Properties of Extracellular Alginate Lyase
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AOS | Alginate oligosaccharides |
| AUL | Alginate Utilization Locus |
| polyM | Poly mannuronic acid |
| polyG | Poly guluronic acid |
| PUL | Polysaccharide Utilization Locus |
References
- Mazéas, L.; Yonamine, R.; Barbeyron, T.; Henrissat, B.; Drula, E.; Terrapon, N.; Nagasato, C.; Hervé, C. Assembly and synthesis of the extracellular matrix in brown algae. Semin. Cell Dev. Biol. 2023, 134, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Beuder, S.; Braybrook, S.A. Brown algal cell walls and development. Semin. Cell Dev. Biol. 2023, 134, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Guo, Y.F.; Wang, Y.L.; Wang, C.B.; Liu, J.H.; Meng, G.Q.; Wang, Z.P. Preparation of alginate oligosaccharides from Laminaria japonica biomass by a novel biofunctional alginate lyase with pH and salt tolerance. Processes 2023, 11, 1495. [Google Scholar] [CrossRef]
- Li, L.; Jiang, J.J.; Yao, Z.; Zhu, B.W. Recent advances in the production, properties and applications of alginate oligosaccharides—A mini review. World J. Microbiol. Biotechnol. 2023, 39, 207. [Google Scholar] [CrossRef]
- Bi, D.C.; Yang, X.; Lu, J.; Xu, X. Preparation and potential applications of alginate oligosaccharides. Crit. Rev. Food Sci. Nutr. 2023, 63, 10130–10147. [Google Scholar] [CrossRef]
- Li, L.; Zhu, B.W.; Yao, Z.; Jiang, J.J. Directed preparation, structure-activity relationship and applications of alginate oligosaccharides with specific structures: A systematic review. Food Res. Int. 2023, 170, 112990. [Google Scholar] [CrossRef]
- Zhang, C.H.; Li, M.X.; Rauf, A.; Khalil, A.A.; Shan, Z.G.; Chen, C.Y.; Rengasamy, K.R.R.; Wan, C.P. Process and applications of alginate oligosaccharides with emphasis on health beneficial perspectives. Crit. Rev. Food Sci. Nutr. 2023, 63, 303–329. [Google Scholar] [CrossRef]
- Yang, J.; Cui, D.D.; Ma, S.; Chen, W.K.; Chen, D.W.; Shen, H. Characterization of a novel PL17 family alginate lyase with exolytic and endolytic cleavage activity from marine bacterium Microbulbifer sp. SH-1. Int. J. Biol. Macromol. 2021, 169, 551–563. [Google Scholar] [CrossRef]
- Xiao, Z.B.; Li, K.K.; Li, T.; Zhang, F.X.; Xue, J.Y.; Zhao, M.; Yin, H. Characterization and mechanism study of a novel PL7 family exolytic alginate lyase from marine bacteria Vibrio sp. W13. Appl. Biochem. Biotechnol. 2024, 196, 68–84. [Google Scholar] [CrossRef]
- Li, B.; Liu, J.W.; Zhou, S.; Fu, L.; Yao, P.; Chen, L.; Yang, Z.S.; Wang, X.L.; Zhang, X.H. Vertical variation in Vibrio community composition in Sansha Yongle Blue Hole and its ability to degrade macromolecules. Mar. Life Sci. Technol. 2020, 2, 60–72. [Google Scholar] [CrossRef]
- Zhu, B.W.; Li, K.K.; Wang, W.X.; Ning, L.M.; Tan, H.D.; Zhao, X.M.; Yin, H. Preparation of trisaccharides from alginate by a novel alginate lyase Alg7A from marine bacterium Vibrio sp. W13. Int. J. Biol. Macromol. 2019, 139, 879–885. [Google Scholar] [CrossRef] [PubMed]
- Lyu, Q.Q.; Zhang, K.K.; Shi, Y.H.; Li, W.H.; Diao, X.T.; Liu, W.Z. Structural insights into a novel Ca2+-independent PL-6 alginate lyase from Vibrio OU02 identify the possible subsites responsible for product distribution. Biochim. Biophys. Acta Gen. Subj. 2019, 1863, 1167–1176. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.Y.; Wang, Y.; Gao, S.; Wu, H.; Wang, D.N.; Yu, W.G.; Han, F. Biochemical characteristics and molecular mechanism of an exo-type alginate lyase VxAly7D and its use for the preparation of unsaturated monosaccharides. Biotechnol. Biofuels 2020, 13, 99. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Y.; Tang, Y.Q.; Gao, F.; Xu, X.D.; Chen, G.J.; Li, Y.J.; Wang, L.S. Low-cost and efficient strategy for brown algal hydrolysis: Combination of alginate lyase and cellulase. Bioresour. Technol. 2024, 397, 130481. [Google Scholar] [CrossRef]
- Bao, K.; Yang, M.; Sun, Q.H.; Zhang, K.S.; Huang, H.Q. Genome analysis of a potential novel Vibrio species secreting pH- and thermo-stable alginate lyase and its application in producing alginate oligosaccharides. Mar. Drugs 2024, 22, 414. [Google Scholar] [CrossRef]
- Wang, H.Y.; Chen, Z.F.; Zheng, Z.H.; Lei, H.W.; Cong, H.H.; Zhou, H.X. A Novel Cold-Adapted and High-Alkaline Alginate Lyase with Potential for Alginate Oligosaccharides Preparation. Molecules 2023, 28, 6190. [Google Scholar] [CrossRef]
- Wang, X.H.; Zhang, Y.Q.; Zhang, X.R.; Zhang, X.D.; Sun, X.M.; Wang, X.F.; Sun, X.H.; Song, X.Y.; Zhang, Y.Z.; Wang, N.; et al. High-level extracellular production of a trisaccharide-producing alginate lyase AlyC7 in Escherichia coli and its agricultural application. Mar. Drugs 2024, 22, 230. [Google Scholar] [CrossRef]
- Huang, Z.; Liang, S.; Jiang, W.L.; Wang, L.; Wang, Y.; Wang, H.; Wang, L.S.; Cong, Y.T.; Lu, Y.N.; Yang, G.J. Multi-functional alginate lyase AlgVR7 from Vibrio rumoiensis: Structural insights and catalytic mechanisms. Mar. Drugs 2025, 23, 124. [Google Scholar] [CrossRef]
- Huang, H.Q.; Zheng, Z.G.; Zou, X.X.; Wang, Z.X.; Gao, R.; Zhu, J.; Hu, Y.H.; Bao, S.X. Genome analysis of a novel polysaccharide-degrading bacterium Paenibacillus algicola and determination of alginate lyases. Mar. Drugs 2022, 20, 388. [Google Scholar] [CrossRef]
- Cheng, W.W.; Yan, X.Y.; Xiao, J.L.; Chen, Y.Y.; Chen, M.H.; Jin, J.Y.; Bai, Y.; Wang, Q.; Liao, Z.Y.; Chen, Q.Z. Isolation, identification, and whole genome sequence analysis of the alginate-degrading bacterium Cobetia sp. cqz5-12. Sci. Rep. 2020, 10, 10920. [Google Scholar] [CrossRef]
- Zhou, J.S.; Cai, M.H.; Jiang, T.; Zhou, W.Q.; Shen, W.; Zhou, X.S.; Zhang, Y.X. Mixed carbon source control strategy for enhancing alginate lyase production by marine Vibrio sp. QY102. Bioprocess Biosyst. Eng. 2014, 37, 575–584. [Google Scholar] [CrossRef]
- Butt, M.; Jung, J.; Kim, J.M.; Bayburt, H.; Han, D.M.; Jeon, C.O. Shewanella phaeophyticola sp. nov. and Vibrio algarum sp. nov.; isolated from marine brown algae. Int. J. Syst. Evol. Microbiol. 2024, 74, 006378. [Google Scholar]
- Kurilenko, V.; Bystritskaya, E.; Otstavnykh, N.; Velansky, P.; Lichmanuk, D.; Savicheva, Y.; Romanenko, L.; Isaeva, M. Description and Genome-Based Analysis of Vibrio chaetopteri sp. nov.; a New Species of the Mediterranei Clade Isolated from a Marine Polychaete. Microorganisms 2025, 13, 638. [Google Scholar] [PubMed]
- Yoon, S.H.; Ha, S.M.; Lim, J.; Kwon, S.; Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Carbasse, J.S.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 2022, 50, D801–D807. [Google Scholar] [CrossRef]
- Liu, X.; Zhao, W.T.; Li, Y.; Sun, Z.L.; Lu, C.; Sun, L.Q. Genome analysis of a polysaccharide-degrading bacterium Microbulbifer sp. HZ11 and degradation of alginate. Mar. Drugs 2024, 22, 569. [Google Scholar] [CrossRef]
- Sha, L.; Huang, M.H.; Huang, X.N.; Huang, Y.T.; Shao, E.S.; Guan, X.; Huang, Z.P. Cloning and characterization of a novel endo-type metal-independent alginate lyase from the marine bacteria Vibrio sp. Ni1. Mar. Drugs 2022, 20, 479. [Google Scholar] [CrossRef]
- Meng, Q.; Zhou, L.C.; Hassanin, H.A.M.; Jiang, B.; Liu, Y.C.; Chen, J.J.; Zhang, T. A new role of family 32 carbohydrate binding module in alginate lyase from Vibrio natriegens SK42.001 in altering its catalytic activity, thermostability and product distribution. Food Biosci. 2021, 42, 101112. [Google Scholar]
- Brenner, D.J.; Krieg, N.R.; Staley, J.T. Bergey’s Manual of Systematic Bacteriology, 2nd ed.; Springer: New York, NY, USA, 2005; Volume 2, pp. 491–545. [Google Scholar]
- Muhammad, N.; Avila, F.; Nedashkovskaya, O.I.; Kim, S.G. Three novel marine species of the genus Reichenbachiella exhibiting degradation of complex polysaccharides. Front. Microbiol. 2023, 14, 1265676. [Google Scholar] [CrossRef]
- Tanaka, M.; Kumakura, D.; Mino, S.; Doi, H.; Ogura, Y.; Hayashi, T.; Yumoto, I.; Cai, M.; Zhou, Y.G.; Gomez-Gil, B.; et al. Genomic characterization of closely related species in the Rumoiensis clade infers ecogenomic signatures to non-marine environments. Environ. Microbiol. 2020, 22, 3205–3217. [Google Scholar] [CrossRef]
- Chernysheva, N.; Bystritskaya, E.; Likhatskaya, G.; Nedashkovskaya, O.; Isaeva, M. Genome-wide analysis of PL7 alginate lyases in the genus Zobellia. Molecules 2021, 26, 2387. [Google Scholar] [CrossRef] [PubMed]
- Li, J.J.; Pei, X.J.; Xue, C.H.; Chang, Y.G.; Shen, J.J.; Zhang, Y.Y. A repertoire of alginate lyases in the alginate polysaccharide utilization loci of marine bacterium Wenyingzhuangia fucanilytica: Biochemical properties and action pattern. J. Sci. Food Agric. 2024, 104, 134–140. [Google Scholar] [CrossRef] [PubMed]
- He, X.X.; Zhang, Y.H.; Wang, X.L.; Zhu, X.Y.; Chen, L.R.; Liu, W.Z.; Lyu, Q.Q.; Ran, L.M.; Cheng, H.J.; Zhang, X.H. Characterization of multiple alginate lyases in a highly efficient alginate-degrading Vibrio strain and its degradation strategy. Appl. Environ. Microbiol. 2022, 88, e0138922. [Google Scholar] [CrossRef]
- BrenZhang, L.Z.; Li, X.; Zhang, X.Y.; Li, Y.J.; Wang, L.S. Bacterial alginate metabolism: An important pathway for bioconversion of brown algae. Biotechnol. Biofuels 2021, 14, 158. [Google Scholar] [CrossRef]
- Sun, X.K.; Gong, Y.; Shang, D.D.; Liu, B.T.; Du, Z.J.; Chen, G.J. Degradation of alginate by a newly isolated marine bacterium Agarivorans sp. B2Z047. Mar. Drugs 2022, 20, 254. [Google Scholar] [CrossRef]
- Fang, X.R.; Li, S.; Kang, W.X.; Lin, C.Y.; Wang, J.M.; Ke, Q.; Wang, C.H.; Wang, Q.; Chen, Q.Z. Enhanced algin oligosaccharide production through selective breeding and optimization of growth and degradation conditions in Cobetia sp. cqz5-12-M1. Sci. Rep. 2024, 14, 19550. [Google Scholar] [CrossRef]
- Ramya, P.; Selvaraj, K.; Suthendran, K.; Sundar, K.; Vanavil, B. Optimization of alginate lyase production using Enterobacter tabaci RAU2C isolated from marine environment by RSM and ANFIS modelling. Aquac. Int. 2023, 31, 3207–3237. [Google Scholar] [CrossRef]
- Ali, N.L.; Foo, H.L.; Ramli, N.; Halim, M.; Thalij, K.M. Efficient Assessment and Optimisation of Medium Components Influencing Extracellular Xylanase Production by Pediococcus pentosaceus G4 Using Statistical Approaches. Int. J. Mol. Sci. 2025, 26, 7219. [Google Scholar] [CrossRef]
- Neethu, K.P.; Sobhana, K.S.; Keerthi, R.B.; Varghese, E.; Ranjith, L.; Jasmine, S.; Joshi, K.K.; George, G. Optimizing α-amylase production by an extremely halophilic archaeon Haloferax mucosum MS1.4, using response surface methodology. J. Microbiol. Methods 2025, 232–234, 107136. [Google Scholar] [CrossRef]
- Jeyabalan, J.; Veluchamy, A.; Narayanasamy, S. Production optimization, characterization, and application of a novel thermo- and pH-stable laccase from Bacillus drentensis 2E for bioremediation of industrial dyes. Int. J. Biol. Macromol. 2025, 308, 142557. [Google Scholar] [CrossRef]
- Kanwar, K.; Sharma, D.; Singh, H.; Pal, M.; Bandhu, R.; Azmi, W. In vitro effects of alginate lyase SG4 + produced by Paenibacillus lautus alone and combined with antibiotics on biofilm formation by mucoid Pseudomonas aeruginosa. Braz. J. Microbiol. 2024, 55, 1189–1203. [Google Scholar] [CrossRef]
- Zhou, L.C.; Meng, Q.; Zhang, R.; Jiang, B.; Liu, X.Y.; Chen, J.J.; Zhang, T. Characterization of a novel polysaccharide lyase family 5 alginate lyase with polyM substrate specificity. Foods 2022, 11, 3527. [Google Scholar] [CrossRef]
- Li, H.B.; Huang, X.Y.; Yao, S.X.; Zhang, C.H.; Hong, X.; Wu, T.; Jiang, Z.D.; Ni, H.; Zhu, Y.B. Characterization of a bifunctional and endolytic alginate lyase from Microbulbifer sp. ALW1 and its application in alginate oligosaccharides production from Laminaria japonica. Protein Expr. Purif. 2022, 200, 106171. [Google Scholar] [CrossRef]
- Dobruchowska, J.M.; Bjornsdottir, B.; Fridjonsson, O.H.; Altenbuchner, J.; Watzlawick, H.; Gerwig, G.J.; Dijkhuizen, L.; Kamerling, J.P.; Hreggvidsson, G.O. Enzymatic depolymerization of alginate by two novel thermostable alginate lyases from Rhodothermus marinus. Front. Plant Sci. 2022, 13, 981602. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Cao, S.S.; Zhu, B.W.; Jiang, L.; Yao, Z. Biochemical characterization and elucidation the degradation pattern of a new cold-adapted and Ca2+ activated alginate lyase for efficient preparation of alginate oligosaccharides. Enzym. Microb. Technol. 2023, 162, 110146. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.J.; Chen, P.; Zeng, Y.; Men, Y.; Mu, S.C.; Zhu, Y.M.; Chen, Y.F.; Sun, Y.X. The characterization and modification of a novel bifunctional and robust alginate lyase derived from Marinimicrobium sp. H1. Mar. Drugs 2019, 17, 545. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Hu, F.; Wang, M.Y.; Zhu, B.W.; Ni, F.; Yao, Z. Elucidation of degradation pattern and immobilization of a novel alginate lyase for preparation of alginate oligosaccharides. Int. J. Biol. Macromol. 2020, 146, 579–587. [Google Scholar] [CrossRef]
- Saeed, S.; Shakir, H.A.; Qazi, J.I. Statistical optimization of amylase production from Bacillus amyloliquefaciens using agro-industrial waste (mango peels) under submerged fermentation by response surface methodology. J. Microbiol. Methods 2025, 237, 107208. [Google Scholar] [CrossRef]




| Number | Level | Enzyme Activity (U/mL) | ||||||
|---|---|---|---|---|---|---|---|---|
| Alginate (g/L) | NH4Cl (g/L) | NaCl (g/L) | K2HPO4·3H2O (g/L) | MgSO4·7H2O (g/L) | FeSO4·7H2O (g/L) | G: pH | ||
| 1 | 14 | 14 | 10 | 2 | 0 | 0.06 | 5 | 36.055 |
| 2 | 14 | 8 | 30 | 6 | 2 | 0.04 | 5 | 25.295 |
| 3 | 14 | 8 | 10 | 2 | 2 | 0.04 | 6 | 34.879 |
| 4 | 14 | 14 | 10 | 6 | 2 | 0.06 | 5 | 34.275 |
| 5 | 14 | 14 | 30 | 2 | 0 | 0.04 | 6 | 17.828 |
| 6 | 14 | 8 | 30 | 6 | 0 | 0.06 | 6 | 25.233 |
| 7 | 8 | 8 | 10 | 2 | 0 | 0.04 | 5 | 28.921 |
| 8 | 8 | 8 | 30 | 2 | 2 | 0.06 | 5 | 26.543 |
| 9 | 8 | 14 | 10 | 6 | 2 | 0.04 | 6 | 30.941 |
| 10 | 8 | 8 | 10 | 6 | 0 | 0.06 | 6 | 35.579 |
| 11 | 8 | 14 | 30 | 2 | 2 | 0.06 | 6 | 21.643 |
| 12 | 8 | 14 | 30 | 6 | 0 | 0.04 | 5 | 13.348 |
| Number | Level | Enzyme Activity (U/mL) | ||
|---|---|---|---|---|
| NaCl (g/L) | FeSO4·7H2O (g/L) | NH4Cl (g/L) | ||
| 1 | 20 | 0.05 | 11 | 40.704 |
| 2 | 10 | 0.06 | 11 | 19.687 |
| 3 | 30 | 0.04 | 11 | 33.554 |
| 4 | 20 | 0.05 | 11 | 37.142 |
| 5 | 20 | 0.06 | 8 | 35.935 |
| 6 | 10 | 0.04 | 11 | 22.441 |
| 7 | 20 | 0.04 | 14 | 35.848 |
| 8 | 30 | 0.05 | 14 | 33.304 |
| 9 | 20 | 0.04 | 8 | 32.322 |
| 10 | 20 | 0.05 | 11 | 34.439 |
| 11 | 20 | 0.06 | 14 | 34.459 |
| 12 | 20 | 0.05 | 11 | 36.178 |
| 13 | 10 | 0.05 | 14 | 18.686 |
| 14 | 20 | 0.05 | 11 | 36.816 |
| 15 | 30 | 0.05 | 8 | 33.109 |
| 16 | 10 | 0.05 | 8 | 18.636 |
| 17 | 30 | 0.06 | 11 | 35.418 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, Y.; Xu, J.; Li, Z.; Wei, R.; Zhao, H.; Sun, L.; Lu, C. Genome Analysis of Alginate-Degrading Bacterium Vibrio sp. 32415 and Optimization of Alginate Lyase Production. Microorganisms 2025, 13, 2385. https://doi.org/10.3390/microorganisms13102385
Zeng Y, Xu J, Li Z, Wei R, Zhao H, Sun L, Lu C. Genome Analysis of Alginate-Degrading Bacterium Vibrio sp. 32415 and Optimization of Alginate Lyase Production. Microorganisms. 2025; 13(10):2385. https://doi.org/10.3390/microorganisms13102385
Chicago/Turabian StyleZeng, Yi, Jia Xu, Zhongran Li, Rujie Wei, Haiyang Zhao, Liqin Sun, and Chang Lu. 2025. "Genome Analysis of Alginate-Degrading Bacterium Vibrio sp. 32415 and Optimization of Alginate Lyase Production" Microorganisms 13, no. 10: 2385. https://doi.org/10.3390/microorganisms13102385
APA StyleZeng, Y., Xu, J., Li, Z., Wei, R., Zhao, H., Sun, L., & Lu, C. (2025). Genome Analysis of Alginate-Degrading Bacterium Vibrio sp. 32415 and Optimization of Alginate Lyase Production. Microorganisms, 13(10), 2385. https://doi.org/10.3390/microorganisms13102385




