Comparative Analysis of Meat Quality and Hindgut Microbiota of Cultured and Wild Bighead Carp (Hypophthalmichthys nobilis, Richardson 1845) from the Yangtze River Area
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples Collection
2.2. Muscle Body Composition Analysis
2.2.1. Proximate Composition Analysis
2.2.2. Amino Acid Composition Analysis
2.2.3. Fatty Acid Composition Analysis
2.3. Determination of Muscle Physicochemical Properties
2.3.1. Muscle Texture Determination
2.3.2. Determination of Muscle Water-Holding Capacity (WHC)
2.3.3. Determination of Muscle pH and Postmortem Glycolysis-Related Indices
2.3.4. Electronic Nose Analysis
2.4. Sequencing and Bioinformatics Analysis
2.5. Correlation Analysis
2.6. Statistical Analysis
3. Results
3.1. Muscle Proximate Composition and Physicochemical Properties of Cultured and Wild Bighead Carp from the Yangtze River Area
3.2. Amino Acid Composition of Cultured and Wild Bighead Carp from the Yangtze River Area
3.3. Fatty Acid Compositions of Cultured and Wild Bighead Carp from the Yangtze River Area
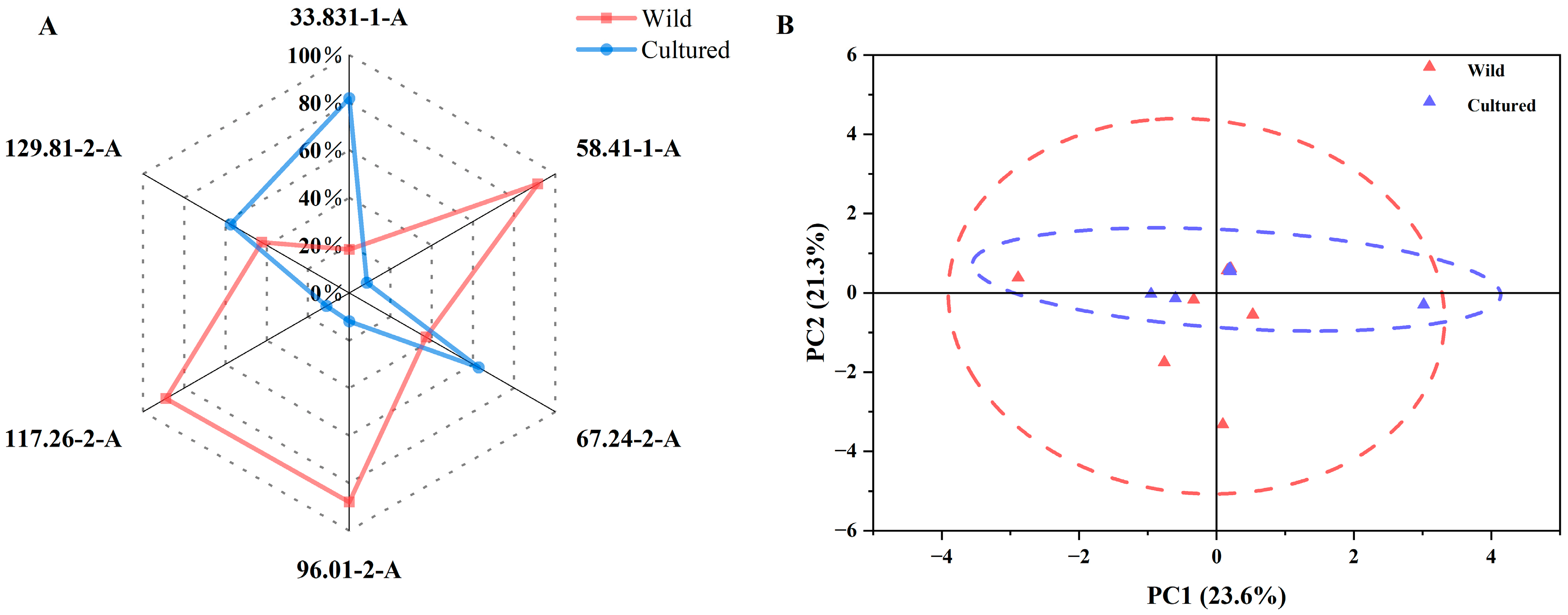
3.4. E-Nose Analysis of Cultured and Wild Bighead Carp from the Yangtze River Area
3.5. Gut Microbial Analysis
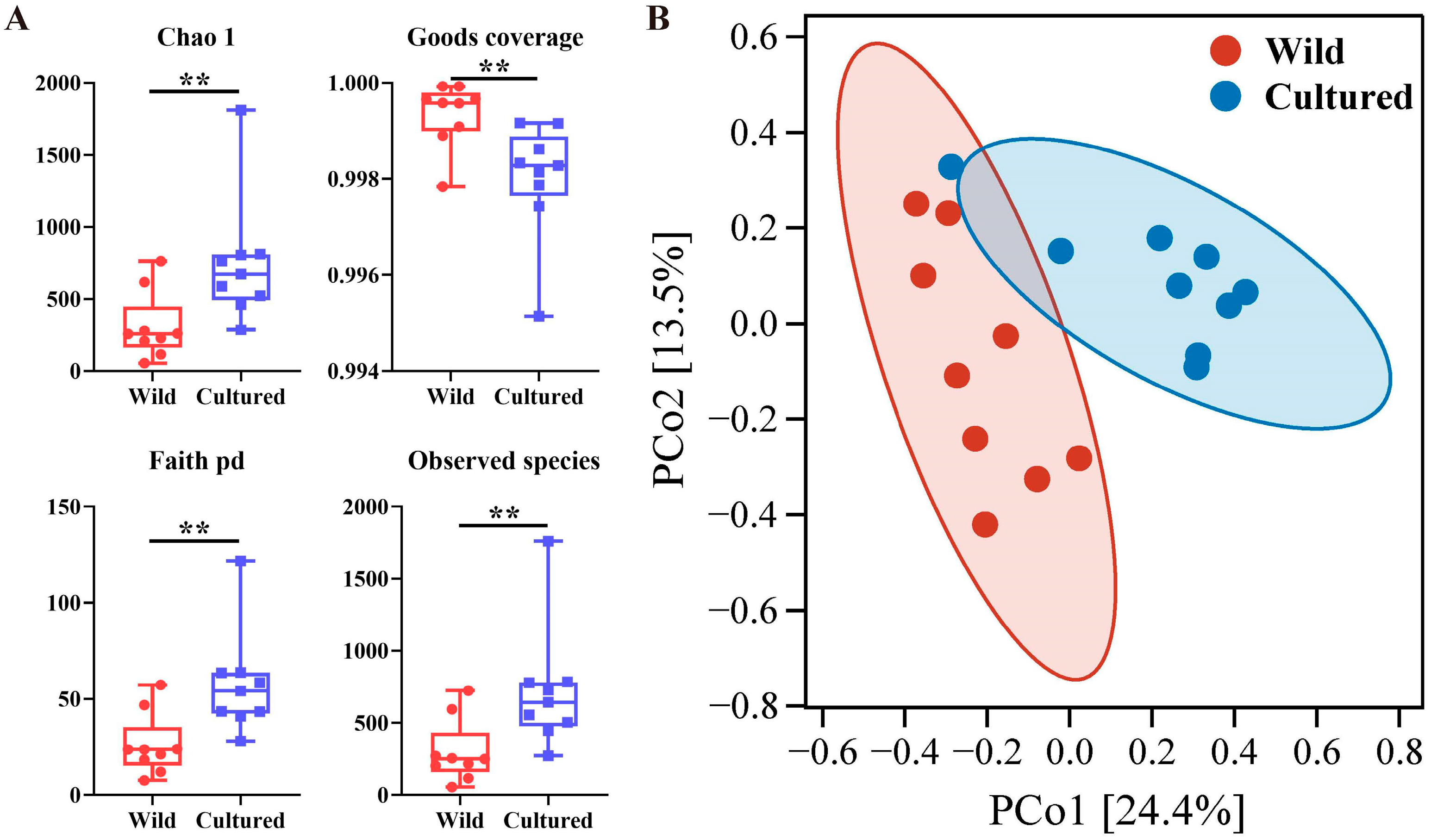
3.5.1. Gut Microbial Composition
3.5.2. Richness and Diversity Analysis
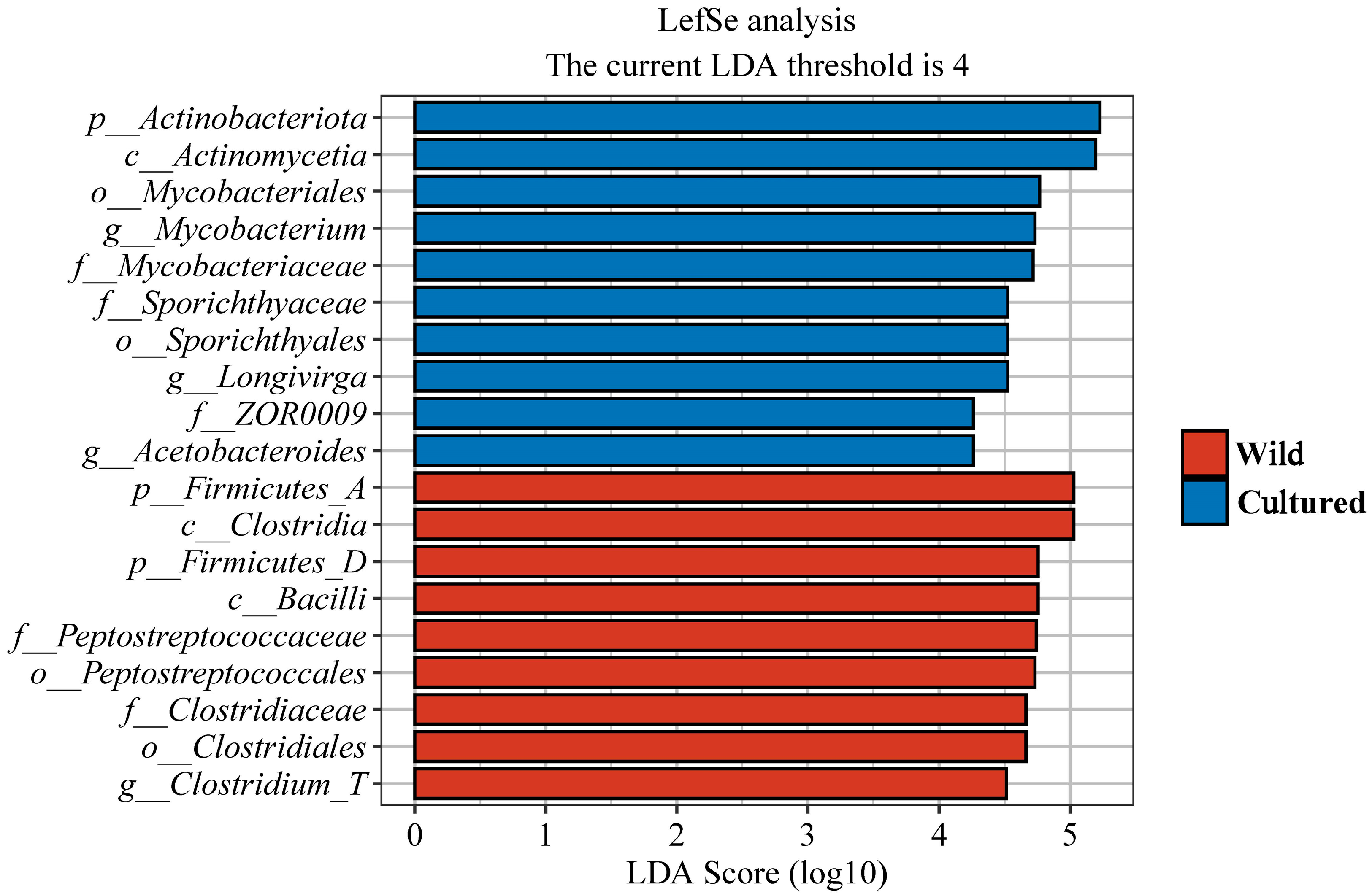
3.5.3. Bacterial Signatures in Wild and Cultured Bighead Carp
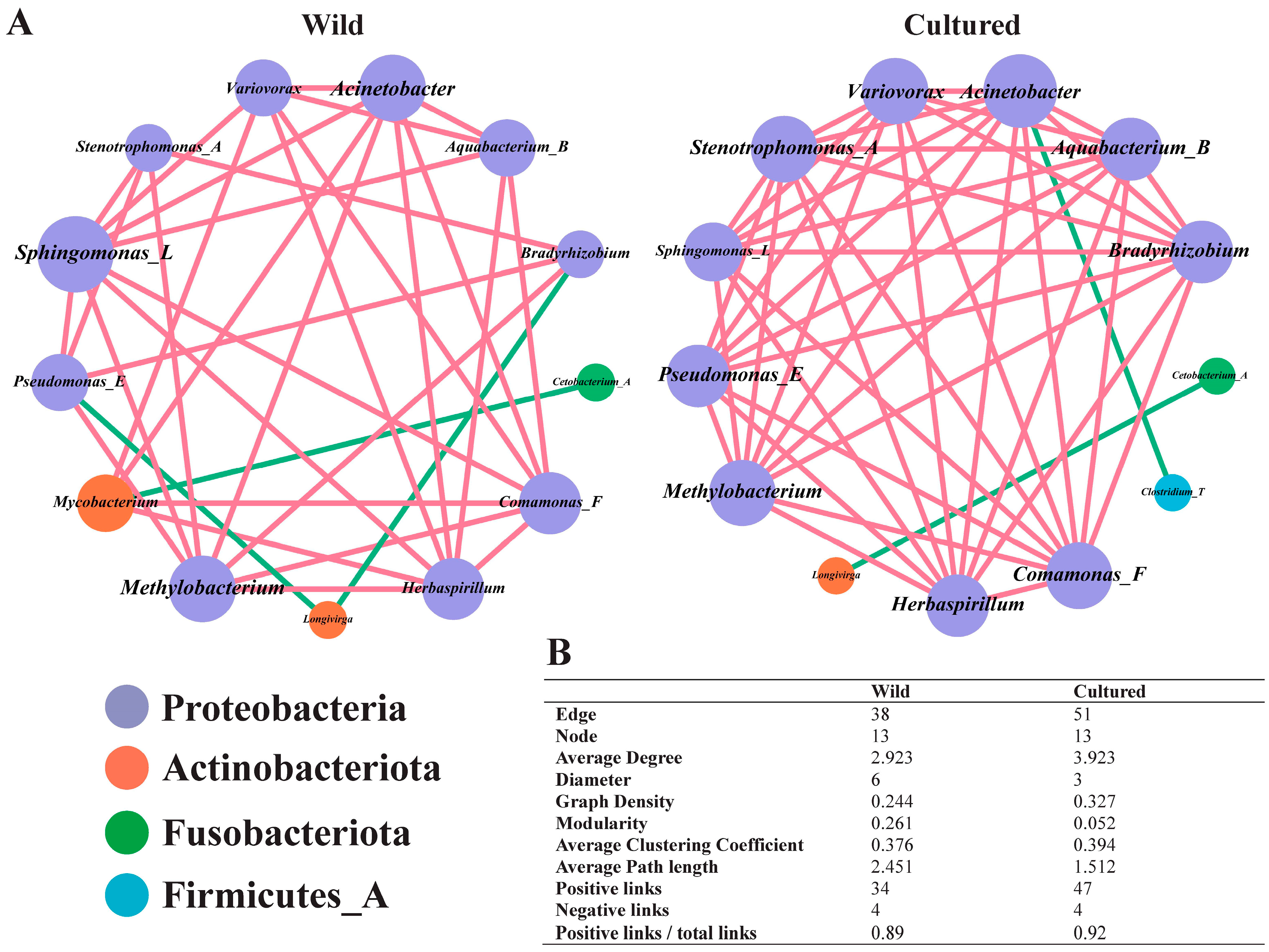
3.5.4. Functional Enrichment Analysis of Gut Microbiota Between Wild and Cultured Bighead Carp
3.6. Correlation Analysis
4. Discussion
4.1. Muscle Proximate Composition and Physicochemical Properties of Cultured and Wild Bighead Carp from the Yangtze River Area
4.2. Muscle Amino Acid, Fatty Acid Composition and E-Nose Flavor of Cultured and Wild Bighead Carp from the Yangtze River Area
4.3. Gut Microbiota of Cultured and Wild Bighead Carp from the Yangtze River Area
4.4. Correlation Analysis to Identify Gut Microbiota Effects on Muscle Quality and to Distinguish Wild and Cultured Bighead Carp
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Freitas, J.; Vaz-Pires, P.; Câmara, J.S. Quality Index Method for fish quality control: Understanding the applications, the appointed limits and the upcoming trends. Trends Food Sci. Technol. 2021, 111, 333–345. [Google Scholar] [CrossRef]
- Wang, L.; Xiong, J.; Xu, C.; Qin, C.; Zhang, Y.; Yang, L.; Zhi, S.; Feng, J.; Nie, G. Comparison of muscle nutritional composition, texture quality, carotenoid metabolites and transcriptome to underling muscle quality difference between wild-caught and pond-cultured Yellow River carp (Cyprinus carpio haematopterus). Aquaculture 2024, 581, 740392. [Google Scholar] [CrossRef]
- Thorslund, C.A.H.; Sandøe, P.; Aaslyng, M.D.; Lassen, J. A good taste in the meat, a good taste in the mouth—Animal welfare as an aspect of pork quality in three European countries. Livest. Sci. 2016, 193, 58–65. [Google Scholar] [CrossRef]
- Ma, R.; Meng, Y.; Zhang, W.; Mai, K. Comparative study on the organoleptic quality of wild and farmed large yellow croaker Larimichthys crocea. J. Oceanol. Limnol. 2020, 38, 260–274. [Google Scholar] [CrossRef]
- Saeed, R.; Feng, H.; Wang, X.; Xiaoshuan, Z.; Zetian, F. Fish quality evaluation by sensor and machine learning: A mechanistic review. Food Control 2022, 137, 108902. [Google Scholar] [CrossRef]
- Zhao, F.; Zhuang, P.; Zhang, L.; Shi, Z. Biochemical composition of juvenile cultured vs. Wild silver pomfret, Pampus argenteus: Determining the diet for cultured fish. Fish Physiol. Biochem. 2010, 36, 1105–1111. [Google Scholar] [CrossRef]
- Wang, L.; Jia, S.; Zhang, L.; Ma, F.; Zhang, M.; Yu, M.; Jiang, H.; Qiao, Z.; Li, X. Comparative study on nutritional quality and volatile flavor compounds of muscle in Cyprinus carpio haematopterus under wild, traditional pond and in-pond raceway system culture. Aquac. Rep. 2022, 25, 101194. [Google Scholar] [CrossRef]
- Raymond, J.K.; Onyango, A.N.; Onyango, C.A. Proximate Composition and Mineral Contents of Farmed and Wild Fish in Kenya. J. Food Res. 2020, 3, 53–62. [Google Scholar] [CrossRef]
- Feng, W.W.; Wu, S.G.; Hao, Y.T.; Li, W.X.; Li, M.; Zou, H.; Wang, G.T. Isolation and identification of anaerobes in the intestinal mucosa of grass carp Ctenopharyngodon idellus. Acta Hydrobiol. Sin. 2018, 42, 11–16. [Google Scholar] [CrossRef]
- Tran, N.T.; Li, Z.; Ma, H.; Zhang, Y.; Zheng, H.; Gong, Y.; Li, S. Clostridium butyricum: A promising pro-biotic confers positive health benefits in aquatic animals. Rev. Aquac. 2020, 12, 2573–2589. [Google Scholar] [CrossRef]
- Dougherty, M.W.; Kudin, O.; Mühlbauer, M.; Neu, J.; Ghariabeh, R.Z.; Jobin, C. Gut microbiota maturation during early human life induces enterocyte proliferation via microbial metabolites. BMC Microbiol. 2020, 20, 205. [Google Scholar] [CrossRef] [PubMed]
- Luan, Y.; Li, M.; Zhou, W.; Yao, Y.; Yang, Y.; Zhang, Z.; Ringø, E.; Erik Olsen, R.; Liu Clarke, J.; Xie, S.; et al. The Fish Microbiota: Research Progress and Potential Applications. Engineering 2023, 29, 137–146. [Google Scholar] [CrossRef]
- Bereded, N.K.; Abebe, G.B.; Fanta, S.W.; Curto, M.; Waidbacher, H.; Meimberg, H.; Doming, K.J. The gut bacterial microbiome of Nile tilapia (Oreochromis niloticus) from lakes across an altitudinal gradient. BMC Microbiol. 2022, 22, 87. [Google Scholar] [CrossRef] [PubMed]
- van Kuijk, S.J.A.; Han, Y.; Garcia-Ruiz, A.I.; Rodiles, A. Hydroxychloride trace minerals have a positive effect on growth performance, carcass quality and impact ileal and cecal microbiota in broiler chickens. J. Anim. Sci. Biotechnol. 2021, 12, 38. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Yan, Q.; Yu, Y.; Zhang, T. Factors influencing the grass carp gut microbiome and its effect on metabolism. FEMS Microbiol. Ecol. 2014, 87, 704–714. [Google Scholar] [CrossRef] [PubMed]
- Semova, I.; Carten, J.D.; Stombaugh, J.; Mackey, L.C.; Knight, R.; Farber, S.A.; Rawls, J.F. Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebra fish. Cell Host Microbe 2012, 12, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Xiang, Y.; Wang, X.; Dai, B.; Zhang, X.; Ma, L.; Yang, H.; Lyu, W. Exploring the possible link between the gut microbiome and fat deposition in pigs. Oxidative Med. Cell. Longev. 2022, 2022, 1098892. [Google Scholar] [CrossRef]
- Richards, J.D.; Gong, J.; De Lange, C.F.M. The gastrointestinal microbiota and its role in monogastric nutrition and health with an emphasis on pigs: Current understanding, possible modulations, and new technologies for ecological studies. Can. J. Anim. Sci. 2005, 85, 421–435. [Google Scholar] [CrossRef]
- Grosicki, G.J.; Fielding, R.A.; Lustgarten, M.S. Gut microbiota contribute to age-related changes in skeletal muscle size, composition, and function: Biological basis for a gut-muscle axis. Calcif. Tissue Int. 2018, 102, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Qi, R.; Sun, J.; Qiu, X.; Zhang, Y.; Wang, J.; Wang, Q.; Huang, J.; Ge, L.; Liu, Z. The intestinal microbiota contributes to the growth and physiological state of muscle tissue in piglets. Sci. Rep. 2021, 11, 11237. [Google Scholar] [CrossRef]
- Hu, B.; Zhou, J.; Qiu, H.; Lai, X.; Li, J.; Wu, D.; Shen, J. Comparison of nutritional quality and volatile flavor compounds among bighead carp from three aquaculture systems. Saudi J. Biol. Sci. 2021, 28, 4291–4299. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Luo, H.; Du, S.Y.; Xiong, Z.B.; Jin, T.S.; Zhang, J. Differences in meat quality of bighead carp between the three nutritional types of reservoirs and artificial culture pond. Prog. Fish. Sci. 2021, 42, 135–141, (in Chinese with English abstract). [Google Scholar] [CrossRef]
- Wang, Q.; Xiong, B.X.; Luo, G.Q.; Su, Y.Q.; Liu, H.Y. Gut microbiota of bighead carp (Aristichthys nobilis) and paddlefish (Polyodon spathula) affected by their diets. Acta Hydrobiol. Sin. 2016, 40, 814–822. [Google Scholar] [CrossRef]
- Zhu, W.; Fu, J.; Luo, M.; Wang, L.; Wang, P.; Liu, Q.; Dong, Z. Genetic diversity and population structure of bighead carp (Hypophthalmichthys nobilis) from the middle and lower reaches of the Yangtze River revealed using microsatellite markers. Aquac. Rep. 2022, 27, 101377. [Google Scholar] [CrossRef]
- Luo, M.; An, R.; Fu, J.; An, S.; Zhu, W.; Wang, L.; Dong, Z. Comparative analysis of the gut microbiota in bighead carp under different culture patterns. J. Appl. Microbiol. 2022, 132, 1357–1369. [Google Scholar] [CrossRef]
- AOAC. Official Methods of Analysis of AOAC International, 18th ed.; Association of Official Analytical Chemists: Washington DC, USA, 2003. [Google Scholar]
- Chen, D.W.; Zhang, M. Non-volatile taste active compounds in the meat of Chinese mitten crab (Eriocheir sinensis). Food Chem. 2007, 104, 1200–1205. [Google Scholar] [CrossRef]
- Gladyshev, M.I.; Makhrov, A.A.; Baydarov, I.V.; Safonova, S.S.; Golod, V.M.; Alekseyev, S.S.; Glushchenko, L.A.; Rudchenko, A.E.; Karpov, V.A.; Sushchik, N.N. Fatty Acid Composition and Contents of Fish of Genus Salvelinus from Natural Ecosystems and Aquaculture. Biomolecules 2022, 12, 144. [Google Scholar] [CrossRef] [PubMed]
- Caimi, C.; Biasato, I.; Chemello, G.; Oddon, S.B.; Lussiana, C.; Malfatto, V.M.; Capucchio, M.T.; Colombino, E.; Schiavone, A.; Gai, F.; et al. Dietary inclusion of a partially defatted black soldier fly (Hermetia illucens) larva meal in low fishmeal-based diets for rainbow trout (Oncorhynchus mykiss). J. Anim. Sci. Biotechnol. 2021, 12, 50. [Google Scholar] [CrossRef]
- Liu, H.; Guo, X.; Gooneratne, R.; Lai, R.; Zeng, C.; Zhan, F.; Wang, W. The gut microbiome and degradation enzyme activity of wild freshwater fishes influenced by their trophic levels. Sci. Rep. 2016, 6, 24340. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory, C.J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Zhai, H.; Peng, Z.; Yu, J.; Yan, L.; Wang, W.; Ren, T.; Han, Y. Comparison of nutritional quality, flesh quality, muscle cellularity, and expression of muscle growth-related genes between wild and recirculating aquaculture system (RAS)-farmed black rockfish (Sebastes schlegelii). Aquac. Int. 2023, 31, 2263–2280. [Google Scholar] [CrossRef]
- Wang, Z.; Qiao, F.; Zhang, W.B.; Parisi, G.; Du, Z.Y.; Zhang, M.L. The flesh texture of teleost fish: Characteristics and interventional strategies. Rev. Aquac. 2024, 16, 508–535. [Google Scholar] [CrossRef]
- Wang, C.L.; Wang, Z.Y.; Song, C.W.; Luo, S.; Yuan, X.; Huang, Y.; Desouky, H.E. A comparative study on growth, muscle cellularity and flesh quality of farmed and imitative ecological farming loach, Misgurnus anguillicaudatus. Aquaculture 2021, 543, 736933. [Google Scholar] [CrossRef]
- Yang, H.; Yuan, Q.; Rahman, M.M.; Lv, W.; Huang, W.; Hu, W.; Zhou, W. Biochemical, histological, and transcriptomic analyses reveal underlying differences in flesh quality between wild and farmed Ricefield Eel (Monopterus albus). Foods 2024, 13, 1751. [Google Scholar] [CrossRef]
- Pietsch, C.; Schulz, C.; Rovira, P.; Kloas, W.; Burkhardt-Holm, P. Organ damage and hepatic lipid accumulation in carp (Cyprinus carpio L.) after feed-borne exposure to the Mycotoxin, Deoxynivalenol (DON). Toxins 2014, 6, 756–778. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wu, P.; Feng, L.; Jiang, W.; Liu, Y.; Kuang, S.; Tang, L.; Zhou, X. Guanidinoacetic acid supplementation totally based on vegetable meal diet improved the growth performance, muscle flavor components and sensory characteristics of on-growing grass carp (Ctenopharygodon idella). Aquaculture 2021, 531, 735841. [Google Scholar] [CrossRef]
- Islam, S.; Bhowmik, S.; Majumdar, P.R.; Srzednicki, G.; Hossain, M.A. Nutritional profile of wild, pond-, gher- and cage-cultured tilapia in Bangladesh. Heliyon 2021, 7, e06968. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Koshio, S.; Nguyen, B.T.; Wang, W.; Cao, X. Comparative studies on lipid profiles and amino acid composition of wild and cultured Dojo loach Misgurnus anguillicaudatus obtained from southern Japan. Fish. Sci. 2012, 78, 1331–1336. [Google Scholar] [CrossRef]
- Chen, J.Y.; Tang, L.W.; Peng, F.Y.; Huang, Z.; Feng, X.L.; Liu, X.J.; Wang, Y.L.; Xiao, Y.M.; Liu, W.B. Study on biological characteristics and excellent characters of Baling Tieshan organic bighead carp. Reprod. Breed. 2023, 3, 219–228. [Google Scholar] [CrossRef]
- Amoussou, N.; Marengo, M.; Iko Afé, O.H.; Lejeune, P.; Durieux, E.D.H.; Douny, C.; Scippo, M.L.; Gobert, S. Comparison of fatty acid profiles of two cultivated and wild marine fish from Mediterranean Sea. Aquac. Int. 2022, 30, 1435–1452. [Google Scholar] [CrossRef]
- Dias, C.B.; Garg, R.; Wood, L.G.; Garg, M.L. Saturated fat consumption may not be the main cause of increased blood lipid levels. Med. Hypotheses 2014, 82, 187–195. [Google Scholar] [CrossRef]
- Gershuni, V.M. Saturated Fat: Part of a Healthy Diet. Curr. Nutr. Rep. 2018, 7, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, B.; Yang, L.; Jiang, S.; Lu, J.; Lin, L. Comparison of the nutritional qualities of the pond, rice-field and wild crayfish (Procambarus clarkii) meat. Food Chem. Adv. 2023, 2, 100272. [Google Scholar] [CrossRef]
- Fuentes, A.; Fern’andez-Segovia, I.; Serra, J.A.; Barat, J.M. Comparison of wild and cultured sea bass (Dicentrarchus labrax) quality. Food Chem. 2010, 119, 1514–1518. [Google Scholar] [CrossRef]
- Mozaffarian, D.; Wu, J.H. (n-3) fatty acids and cardiovascular health: Are effects of EPA and DHA shared or complementary? J. Nutr. 2012, 142, 614S–625S. [Google Scholar] [CrossRef]
- Jiang, H.F.; Cheng, X.F.; Geng, L.W.; Tang, S.Z.; Tong, G.X.; Xu, W. Comparative study of the nutritional composition and toxic elements of farmed and wild Chanodichthys mongolicus. Chin. J. Oceanol. Limnol. 2017, 35, 737–744. [Google Scholar] [CrossRef]
- Powell, M.S.; Hardy, R.W.; Huston, A.M.; Toya, L.A.; Tave, D. Comparison of Body Composition and Fatty Acid Profiles between Wild and Cultured Rio Grande Silvery Minnows. J. Fish Wildl. Manag. 2017, 8, 487–496. [Google Scholar] [CrossRef]
- Parma, L.; Badiani, A.; Bonaldo, A.; Viroli, C.; Farabegoli, F.; Silvi, M.; Bonvini, E.; Pirini, M.; Gatta, P.P. Farmed and wild common sole (Solea solea L.): Comparative assessment of morphometric parameters, processing yields, selected nutritional traits and sensory profile. Aquaculture 2019, 502, 63–71. [Google Scholar] [CrossRef]
- Kim, P.S.; Shin, N.R.; Lee, J.B.; Kim, M.S.; Whon, T.W.; Hyun, D.W.; Yun, J.H.; Jung, M.J.; Kim, J.Y.; Bae, J.W. Host habitat is the major determinant of the gut microbiome of fish. Microbiome 2021, 9, 166. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.; Geng, S.; Zhang, Y.; Qiu, H.; Zhou, J.; Zeng, Q.; Zhao, Y.; Wu, D.; Yu, G.; Gong, H.; et al. The impact of culture systems on the gut microbiota and gut metabolome of bighead carp (Hypophthalmichthys nobilis). Anim. Microbiome 2023, 5, 20. [Google Scholar] [CrossRef]
- Finegold, S.M.; Vaisanen, M.L.; Molitoris, D.R.; Tomzynski, T.J.; Song, Y.; Liu, C.; Collins, M.D.; Lawson, P.A. Cetobacterium somerae sp. nov. from human feces and emended description of the genus Cetobacterium. Syst. Appl. Microbiol. 2003, 26, 177–181. [Google Scholar] [CrossRef] [PubMed]
- Sugita, H.; Miyajima, C.; Deguchi, Y. The vitamin B12-producing ability of the intestinal microflora of freshwater fish. Aquaculture 1991, 92, 267–276. [Google Scholar] [CrossRef]
- Yuan, J.; Wang, Z.; Wang, B.; Mei, H.; Zhai, X.; Zhuang, Z.; Chen, M.; Zhang, Y. Non-specific immunity associated gut microbiome in Aristichthys nobilis under different rearing strategies. Genes 2021, 12, 916. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhang, P.; Li, W.; Liu, J.; Shang, X.; Cheng, Y.; Li, Y. Comparison of gut microbial communities, free amino acids or fatty acids contents in the muscle of wild Aristichthys nobilis from Xinlicheng reservoir and Chagan lake. BMC Microbiol. 2022, 22, 32. [Google Scholar] [CrossRef]
- Dinh-Hung, N.; Dong, H.T.; Senapin, S.; Pimsannil, K.; Thompson, K.D.; Shinn, A.P.; Soontara, C.; Sirimanapong, W.; Chatchaiphan, S.; Rodkhum, C. Insight into characteristics and pathogenicity of five rapidly growing nontuberculous Mycobacterium species isolated from the Siamese fighting fish, Betta splendens. Aquaculture 2023, 575, 73982. [Google Scholar] [CrossRef]
- Dinh-Hung, N.; Dong, H.T.; Senapin, S.; Linh, N.V.; Shinn, A.P.; Pirarat, N.; Hirono, I.; Chatchaiphan, S.; Rodkhum, C. Infection and histopathological consequences in Siamese fighting fish (Betta splendens) due to exposure to a pathogenic Mycobacterium chelonae via different routes. Aquaculture 2024, 579, 740191. [Google Scholar] [CrossRef]
- Balcázar, J.L.; De Blas, I.; Ruiz-Zarzuela, I.; Cunningham, D.; Vendrell, D.; Múzquiz, J.L. The role of probiotics in aquaculture. Vet. Microbiol. 2006, 114, 173–186. [Google Scholar] [CrossRef]
- Hafeez, M.; Ahmad, I.; Qureshi, S.; Kashoo, Z.; Farooq, S.; Asmi, O.; Shah, F.; Razak, N. Clostridium perfringens from fresh water fish of Kashmir Himalaya and their aquatic environment: Toxinotyping and phylogenetic analysis. Anaerobe 2022, 77, 102619. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Chen, H.; Zhao, L.; Liu, X.; Xiang, P.; Deng, L.; Song, Z. Host habitats influence the gut microbial assembly mechanisms of cold-water fish. Aquac. Rep. 2024, 36, 102094. [Google Scholar] [CrossRef]
- Ticinesi, A.; Lauretani, F.; Milani, C.; Nouvenne, A.; Tana, C.; Del Rio, D.; Maggio, M.; Ventura, M.; Meschi, T. Aging gut microbiota at the cross-road between nutrition, physical frailty, and sarcopenia: Is there a gut–muscle axis? Nutrients 2017, 9, 1303. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.; Dong, Y.; Hou, Q.; He, Y.; Lai, Y.; Liao, C.; Kawamura, Y.; Li, J.; Zhang, B. Intestinal microbiota regulate certain meat quality parameters in chicken. Front. Nutr. 2022, 9, 747705. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; Wang, C.; Huang, K.; Yang, X.; Luo, Y.; Huang, J.; Wang, D.; Li, J.; Jie, B.; Tang, Z.; et al. Exploring the muscle-hardening mechanisms via the muscle-gut axis in tilapia (Oreochromis niloticus) fed with faba bean (Vicia faba L.) supplementary diets. Aquac. Rep. 2024, 37, 102268. [Google Scholar] [CrossRef]
- Wade, A.M.; Tucker, H.N. Antioxidant characteristics of L-histidine. J. Nutr. Biochem. 1998, 9, 308–315. [Google Scholar] [CrossRef]
- Zhou, L.; Chen, C.; Xie, J.; Xu, C.; Zhao, Q.; Qin, J.G.; Chen, L.; Li, E. Intestinal bacterial signatures of the “cotton shrimp-like” disease explain the change of growth performance and immune responses in Pacific white shrimp (Litopenaeus vannamei). Fish Shellfish Immunol. 2019, 92, 629–636. [Google Scholar] [CrossRef]




| Culture Method | Significance | ||
|---|---|---|---|
| Index | Cultured | Wild | (p-Value) |
| Moisture (%) | 74.95 ± 0.71 | 75.91 ± 0.55 | ns |
| Crude protein (%) | 21.11 ± 0.41 | 20.45 ± 0.28 | ns |
| Lipid (%) | 2.12 ± 0.64 | 1.82 ± 0.06 | * |
| Ash (%) | 1.64 ± 0.07 | 1.58 ± 0.07 | ns |
| Hardness (g) | 1852.80 ± 211.49 | 1761.15 ± 128.88 | ns |
| Springiness | 0.385 ± 0.02 | 0.383 ±0.02 | ns |
| Cohesiveness | 0.402 ± 0.03 | 0.440 ± 0.034 | ns |
| Gumminess | 761.56 ± 112.27 | 780.54 ± 92.71 | ns |
| Chewiness | 304.87 ± 53.17 | 308.29 ± 49.66 | ns |
| Resilience | 0.16 ± 0.01 | 0.19 ± 0.01 | * |
| Shear force (N) | 1.26 ± 0.13 | 1.33 ± 0.22 | ns |
| Drip loss (%) | 15.54 ± 1.58 | 12.21 ± 0.91 | * |
| Cooking loss (%) | 13.84 ± 1.17 | 13.71 ± 2.23 | ns |
| pH | 5.71 ± 0.05 | 5.84 ± 0.07 | ns |
| Lactic acid (µmol/mgrot) | 49.53 ± 4.19 | 46.89 ± 3.88 | ns |
| Glucose (mmol/grot) | 4.06 ± 0.45 | 3.36 ± 0.46 | ns |
| LDH (U/gprot) | 7114.04 ± 86.12 | 6692.01 ± 179.41 | * |
| PK (U/gprot) | 169.94 ± 20.99 | 140.34 ± 11.56 | ns |
| PFK (U/mgrot) | 4.95 ± 0.49 | 4.06 ± 0.45 | ns |
| Amino Acid (g/100 g) | Cultured | Wild | Significance (p-Value) |
|---|---|---|---|
| Essential amino acids | |||
| Histidine | 0.56 ± 0.02 | 0.45 ± 0.01 | * |
| Threonine | 0.80 ± 0.02 | 0.79 ± 0.11 | ns |
| Arginine | 1.18 ± 0.02 | 1.18 ± 0.02 | ns |
| Valine | 0.88 ± 0.01 | 0.91 ± 0.02 | ns |
| Methionine | 0.52 ± 0.01 | 0.60 ± 0.01 | * |
| Phenylalanine | 0.83 ± 0.01 | 0.83 ± 0.01 | ns |
| Isoleucine | 0.81 ± 0.02 | 0.86 ± 0.01 | ns |
| Leucine | 1.59 ± 0.02 | 1.59 ± 0.03 | ns |
| Lysine | 1.87 ± 0.03 | 1.85 ± 0.04 | ns |
| Non-essential amino acids | |||
| Aspartic acid | 2.35 ± 0.04 | 2.28 ± 0.05 | ns |
| Glutamic acid | 3.41 ± 0.06 | 3.35 ± 0.06 | ns |
| Serine | 0.82 ± 0.01 | 0.79 ± 0.01 | ns |
| Glycine | 0.89 ± 0.01 | 0.88 ± 0.03 | ns |
| Alanine | 1.11 ± 0.01 | 1.11 ± 0.02 | ns |
| Tyrosine | 0.59 ± 0.01 | 0.58 ± 0.01 | ns |
| Cystine-s | 0.09 ± 0.00 | 0.07 ± 0.00 | ns |
| Proline | 0.72 ± 0.01 | 0.72 ± 0.04 | ns |
| Taste AA | 10.79 ± 1.15 | 10.60 ± 0.75 | ns |
| ∑EAA | 9.04 ± 0.19 | 9.06 ± 0.19 | ns |
| ∑NEAA | 9.98 ± 0.15 | 9.78 ± 0.21 | ns |
| ∑TAA | 19.02 ± 0.34 | 18.84 ± 0.41 | ns |
| Fatty Acid (%) | Cultured | Wild | Significance (p-Value) |
|---|---|---|---|
| C14:0 | 2.09 ± 0.42 | 2.59 ± 0.33 | ns |
| C16:0 | 18.54 ± 0.39 | 21.05 ± 0.44 | * |
| C16:1 | 2.88 ± 0.35 | 4.92 ± 0.45 | * |
| C18:0 | 7.01 ± 0.32 | 8.75 ± 0.34 | * |
| C18:1n9c | 16.92 ± 1.14 | 12.31 ± 1.11 | * |
| C18:2n6c | 15.47 ± 1.34 | 3.01 ± 0.27 | * |
| C20:1 | 3.85 ± 0.60 | 4.23 ± 0.65 | * |
| C20:2 | 1.23 ± 0.10 | - | ns |
| C20:3n6 | 1.82 ± 0.18 | - | ns |
| C20:4n6 (ARA) | 9.22 ± 0.49 | 9.44 ± 0.54 | * |
| C20:5n3 (EPA) | 7.57 ± 1.00 | 14.52 ± 1.19 | * |
| C22:1n9 | 0.62 ± 0.09 | 0.50 ± 0.05 | * |
| C22:6n3 (DHA) | 12.77 ± 0.83 | 18.02 ± 1.21 | * |
| ∑SFA | 27.64 ± 0.60 | 32.39 ± 0.91 | * |
| ∑MUFA | 24.27 ± 2.00 | 21.96 ± 2.92 | ns |
| ∑PUFA | 48.08 ± 1.78 | 45.00 ± 3.41 | ns |
| ∑n − 3 PUFA | 20.34 ± 2.55 | 32.54 ± 3.67 | * |
| ∑n − 6 PUFA | 26.51 ± 1.85 | 12.45 ± 0.53 | * |
| ∑n − 3 PUFA/n − 6 PUFA | 0.76 ± 0.13 | 2.61 ± 0.35 | * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muhammad, A.M.; Yang, C.; Liu, B.; Sun, C.; Miao, L.; Zheng, X.; Pan, L.; Xia, D.; Zhou, Q.-L. Comparative Analysis of Meat Quality and Hindgut Microbiota of Cultured and Wild Bighead Carp (Hypophthalmichthys nobilis, Richardson 1845) from the Yangtze River Area. Microorganisms 2025, 13, 20. https://doi.org/10.3390/microorganisms13010020
Muhammad AM, Yang C, Liu B, Sun C, Miao L, Zheng X, Pan L, Xia D, Zhou Q-L. Comparative Analysis of Meat Quality and Hindgut Microbiota of Cultured and Wild Bighead Carp (Hypophthalmichthys nobilis, Richardson 1845) from the Yangtze River Area. Microorganisms. 2025; 13(1):20. https://doi.org/10.3390/microorganisms13010020
Chicago/Turabian StyleMuhammad, Abdullateef Mukhtar, Chang Yang, Bo Liu, Cunxin Sun, Linghong Miao, Xiaochuan Zheng, Liangkun Pan, Dong Xia, and Qun-Lan Zhou. 2025. "Comparative Analysis of Meat Quality and Hindgut Microbiota of Cultured and Wild Bighead Carp (Hypophthalmichthys nobilis, Richardson 1845) from the Yangtze River Area" Microorganisms 13, no. 1: 20. https://doi.org/10.3390/microorganisms13010020
APA StyleMuhammad, A. M., Yang, C., Liu, B., Sun, C., Miao, L., Zheng, X., Pan, L., Xia, D., & Zhou, Q.-L. (2025). Comparative Analysis of Meat Quality and Hindgut Microbiota of Cultured and Wild Bighead Carp (Hypophthalmichthys nobilis, Richardson 1845) from the Yangtze River Area. Microorganisms, 13(1), 20. https://doi.org/10.3390/microorganisms13010020










