Impact of Swabbing Location, Self-Swabbing, and Food Intake on SARS-CoV-2 RNA Detection
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Study Population
2.3. Study Procedures and Sample Collection
2.4. Statistical Analysis
3. Results
3.1. Demographic and Clinical Characteristics
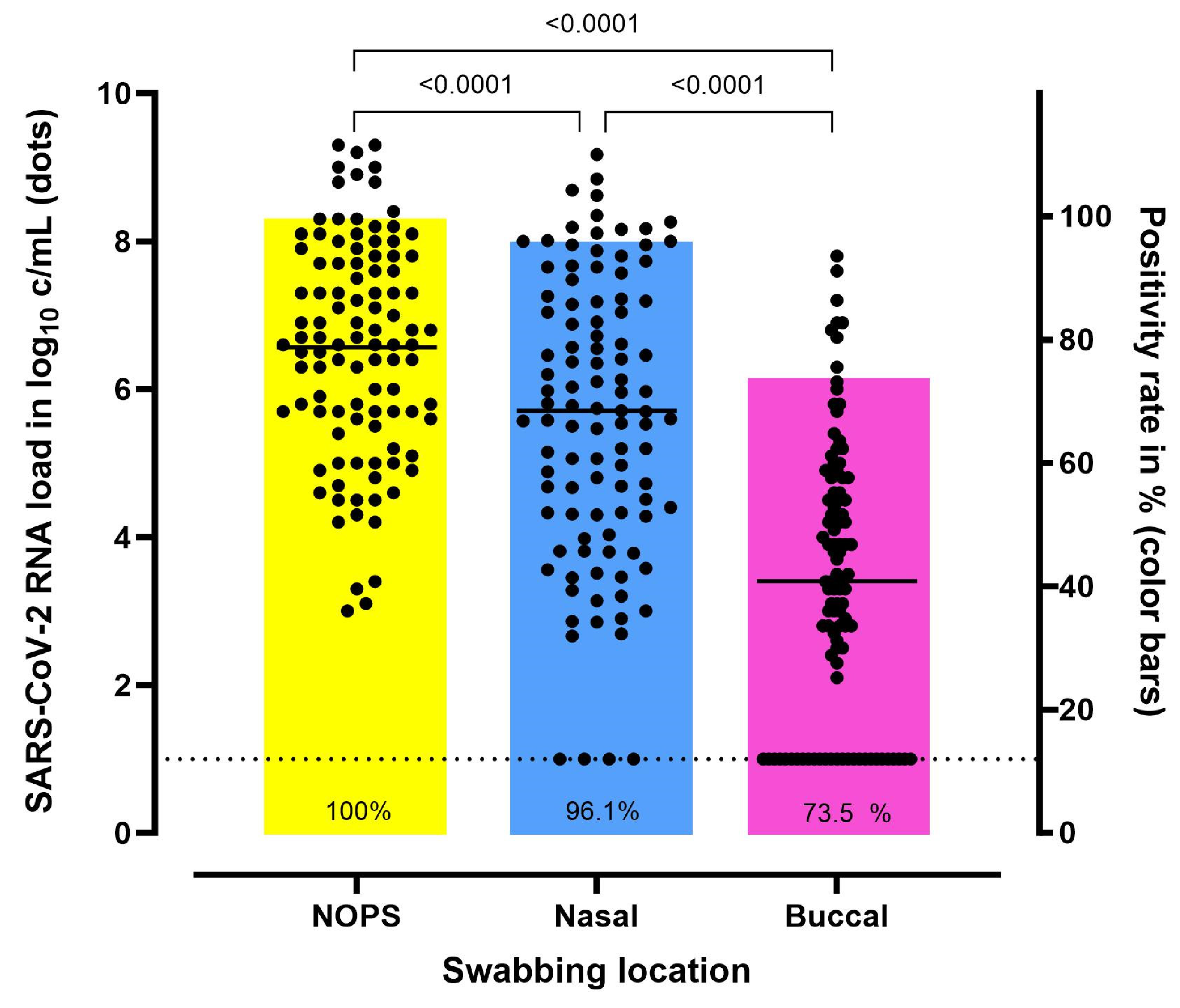
3.2. Comparison of SARS-CoV-2 RNA Load According to the Swabbing Location
3.3. Diagnostic Accuracy
3.4. The Impact of Food Intake on Buccal SARS-CoV-2 RNA Load
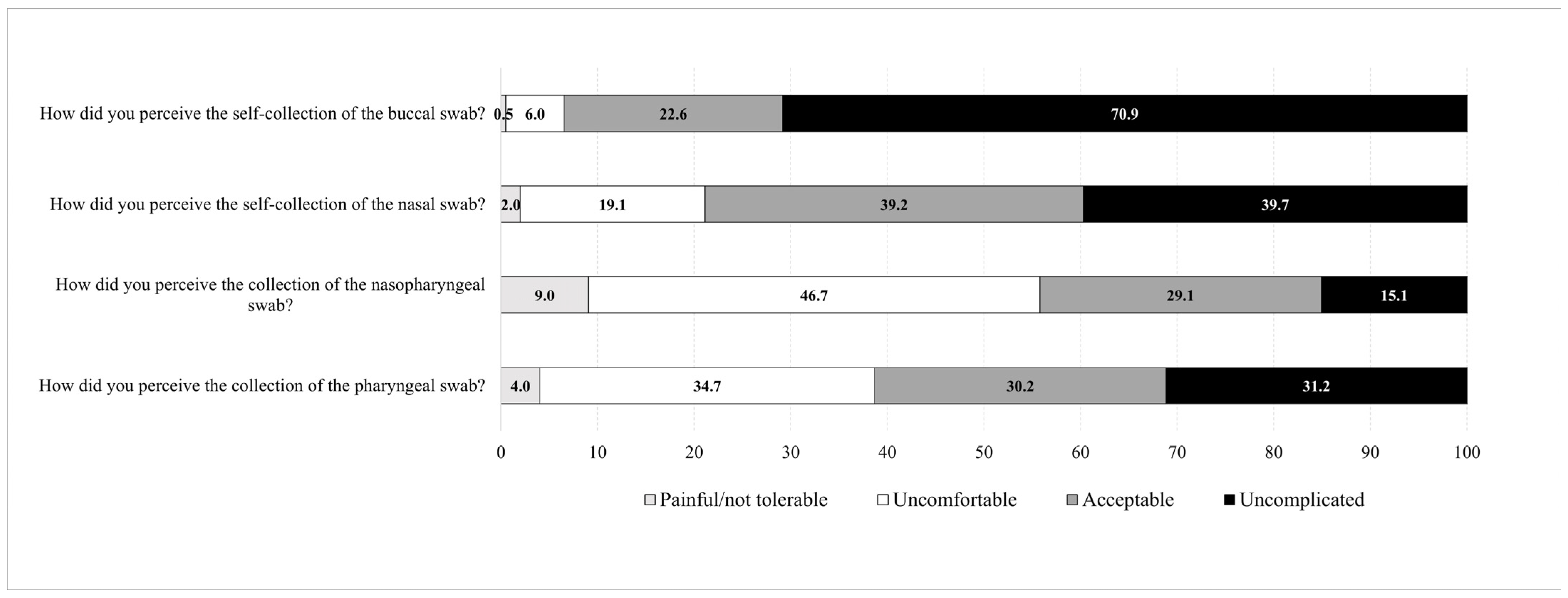
3.5. Patients’ Convenience Related to the Collection Method and Anatomical Site
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Atzrodt, C.L.; Maknojia, I.; McCarthy, R.D.P.; Oldfield, T.M.; Po, J.; Ta, K.T.L.; Stepp, H.E.; Clements, T.P. A Guide to COVID-19: A global pandemic caused by the novel coronavirus SARS-CoV-2. FEBS J. 2020, 287, 3633–3650. [Google Scholar] [CrossRef] [PubMed]
- Kritikos, A.; Caruana, G.; Brouillet, R.; Miroz, J.P.; Abed-Maillard, S.; Stieger, G.; Opota, O.; Croxatto, A.; Vollenweider, P.; Bart, P.A.; et al. Sensitivity of Rapid Antigen Testing and RT-PCR Performed on Nasopharyngeal Swabs versus Saliva Samples in COVID-19 Hospitalized Patients: Results of a Prospective Comparative Trial (RESTART). Microorganisms 2021, 9, 1910. [Google Scholar] [CrossRef] [PubMed]
- Rahbari, R.; Moradi, N.; Abdi, M. rRT-PCR for SARS-CoV-2: Analytical considerations. Clin. Chim. Acta 2021, 516, 1–7. [Google Scholar] [CrossRef]
- Tsang, N.N.Y.; So, H.C.; Ng, K.Y.; Cowling, B.J.; Leung, G.M.; Ip, D.K.M. Diagnostic performance of different sampling approaches for SARS-CoV-2 RT-PCR testing: A systematic review and meta-analysis. Lancet Infect. Dis. 2021, 21, 1233–1245. [Google Scholar] [CrossRef]
- WHO. Diagnostic Testing for SARS-CoV-2 Online: WHO. 2020. Available online: https://iris.who.int/bitstream/handle/10665/334254/WHO-2019-nCoV-laboratory-2020.6-eng.pdf?sequence=1 (accessed on 17 November 2023).
- Van der Moeren, N.; Zwart, V.F.; Louise van Leest, M.; Thijssen, M.; Groenewegen, R.; Heer, M.K.; Murk, J.L.; Tjhie, J.T.; Diederen, B.M.W.; Stohr, J. A SARS-CoV-2 and influenza rapid antigen test-based hospital isolation policy awaiting RT-PCR, a prospective observational study. Clin. Microbiol. Infect. 2023, 29, 1595–1599. [Google Scholar] [CrossRef]
- Wehrhahn, M.C.; Robson, J.; Brown, S.; Bursle, E.; Byrne, S.; New, D.; Chong, S.; Newcombe, J.P.; Siversten, T.; Hadlow, N. Self-collection: An appropriate alternative during the SARS-CoV-2 pandemic. J. Clin. Virol. 2020, 128, 104417. [Google Scholar] [CrossRef] [PubMed]
- Goyal, S.; Prasert, K.; Praphasiri, P.; Chittaganpitch, M.; Waicharoen, S.; Ditsungnoen, D.; Jaichuang, S.; Lindblade, K.A. The acceptability and validity of self-collected nasal swabs for detection of influenza virus infection among older adults in Thailand. Influenza Other Respir. Viruses 2017, 11, 412–417. [Google Scholar] [CrossRef]
- Laferl, H.; Seitz, T.; Baier-Grabner, S.; Kelani, H.; Scholz, E.; Heger, F.; Gotzinger, F.; Frischer, P.T.; Wenisch, C.; Allerberger, P.F. Evaluation of RT-qPCR of mouthwash and buccal swabs for detection of SARS-CoV-2 in children and adults. Am. J. Infect. Control 2022, 50, 176–181. [Google Scholar] [CrossRef]
- Baker, J.M.; Nakayama, J.Y.; O’Hegarty, M.; McGowan, A.; Teran, R.A.; Bart, S.M.; Mosack, K.; Roberts, N.; Campos, B.; Paegle, A.; et al. SARS-CoV-2 B.1.1.529 (Omicron) Variant Transmission Within Households—Four U.S. Jurisdictions, November 2021–February 2022. MMWR Morb. Mortal. Wkly. Rep. 2022, 71, 341–346. [Google Scholar] [CrossRef]
- Jamal, A.J.; Mozafarihashjin, M.; Coomes, E.; Anceva-Sami, S.; Barati, S.; Crowl, G.; Faheem, A.; Farooqi, L.; Kandel, C.E.; Khan, S.; et al. Sensitivity of midturbinate versus nasopharyngeal swabs for the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Infect. Control Hosp. Epidemiol. 2021, 42, 1001–1003. [Google Scholar] [CrossRef]
- Landry, M.L.; Criscuolo, J.; Peaper, D.R. Challenges in use of saliva for detection of SARS CoV-2 RNA in symptomatic outpatients. J. Clin. Virol. 2020, 130, 104567. [Google Scholar] [CrossRef] [PubMed]
- Wolfl-Duchek, M.; Bergmann, F.; Jorda, A.; Weber, M.; Muller, M.; Seitz, T.; Zoufaly, A.; Strassl, R.; Zeitlinger, M.; Herkner, H.; et al. Sensitivity and Specificity of SARS-CoV-2 Rapid Antigen Detection Tests Using Oral, Anterior Nasal, and Nasopharyngeal Swabs: A Diagnostic Accuracy Study. Microbiol. Spectr. 2022, 10, e0202921. [Google Scholar] [CrossRef] [PubMed]
- Kojima, N.; Turner, F.; Slepnev, V.; Bacelar, A.; Deming, L.; Kodeboyina, S.; Klausner, J.D. Self-Collected Oral Fluid and Nasal Swabs Demonstrate Comparable Sensitivity to Clinician Collected Nasopharyngeal Swabs for Coronavirus Disease 2019 Detection. Clin. Infect. Dis. 2021, 73, e3106–e3109. [Google Scholar] [CrossRef] [PubMed]
- Blanco, I.; Violan, C.; Suner, C.; Garcia-Prieto, J.; Argerich, M.J.; Rodriguez-Illana, M.; Moreno, N.; Cardona, P.J.; Blanco, A.; Toran-Monserrat, P.; et al. Comparison between mid-nasal swabs and buccal swabs for SARS-CoV-2 detection in mild COVID-19 patients. J. Infect. 2022, 84, e78–e79. [Google Scholar] [CrossRef] [PubMed]
- Hanson, K.E.; Barker, A.P.; Hillyard, D.R.; Gilmore, N.; Barrett, J.W.; Orlandi, R.R.; Shakir, S.M. Self-Collected Anterior Nasal and Saliva Specimens versus Health Care Worker-Collected Nasopharyngeal Swabs for the Molecular Detection of SARS-CoV-2. J. Clin. Microbiol. 2020, 58, e01824-20. [Google Scholar] [CrossRef] [PubMed]
- Leuzinger, K.; Gosert, R.; Sogaard, K.K.; Naegele, K.; Bielicki, J.; Roloff, T.; Bingisser, R.; Nickel, C.H.; Khanna, N.; Sutter, S.T.; et al. Epidemiology and precision of SARS-CoV-2 detection following lockdown and relaxation measures. J. Med. Virol. 2021, 93, 2374–2384. [Google Scholar] [CrossRef] [PubMed]
- Harris, P.A.; Taylor, R.; Thielke, R.; Payne, J.; Gonzalez, N.; Conde, J.G. Research electronic data capture (REDCap)—A metadata-driven methodology and workflow process for providing translational research informatics support. J. Biomed. Inform. 2009, 42, 377–381. [Google Scholar] [CrossRef]
- Harris, P.A.; Taylor, R.; Minor, B.L.; Elliott, V.; Fernandez, M.; O’Neal, L.; McLeod, L.; Delacqua, G.; Delacqua, F.; Kirby, J.; et al. The REDCap consortium: Building an international community of software platform partners. J. Biomed. Inform. 2019, 95, 103208. [Google Scholar] [CrossRef]
- Altamirano, J.; Govindarajan, P.; Blomkalns, A.L.; Kushner, L.E.; Stevens, B.A.; Pinsky, B.A.; Maldonado, Y. Assessment of Sensitivity and Specificity of Patient-Collected Lower Nasal Specimens for Severe Acute Respiratory Syndrome Coronavirus 2 Testing. JAMA Netw. Open 2020, 3, e2012005. [Google Scholar] [CrossRef]
- Kagan, R.M.; Rogers, A.A.; Borillo, G.A.; Clarke, N.J.; Marlowe, E.M. Performance of Unobserved Self-Collected Nasal Swabs for Detection of SARS-CoV-2 by RT-PCR Utilizing a Remote Specimen Collection Strategy. Open Forum. Infect. Dis. 2021, 8, ofab039. [Google Scholar] [CrossRef]
- Tu, Y.P.; Jennings, R.; Hart, B.; Cangelosi, G.A.; Wood, R.C.; Wehber, K.; Verma, P.; Vojta, D.; Berke, E.M. Swabs Collected by Patients or Health Care Workers for SARS-CoV-2 Testing. N. Engl. J. Med. 2020, 383, 494–496. [Google Scholar] [CrossRef] [PubMed]
- CDC. Interim Guidelines for Clinical Specimens for COVID-19 | CDC, (n.d.): CDC. 2021. Available online: https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specimens.html (accessed on 17 November 2023).
- Dhiman, N.; Miller, R.M.; Finley, J.L.; Sztajnkrycer, M.D.; Nestler, D.M.; Boggust, A.J.; Jenkins, S.M.; Smith, T.F.; Wilson, J.W.; Cockerill, F.R., 3rd; et al. Effectiveness of patient-collected swabs for influenza testing. Mayo Clin. Proc. 2012, 87, 548–554. [Google Scholar] [CrossRef] [PubMed]
- Akmatov, M.K.; Gatzemeier, A.; Schughart, K.; Pessler, F. Equivalence of self- and staff-collected nasal swabs for the detection of viral respiratory pathogens. PLoS ONE 2012, 7, e48508. [Google Scholar] [CrossRef] [PubMed]
- Marx, G.E.; Biggerstaff, B.J.; Nawrocki, C.C.; Totten, S.E.; Travanty, E.A.; Burakoff, A.W.; Scott, T.; De Hey, J.C.; Carlson, J.J.; Wendel, K.A.; et al. Detection of Severe Acute Respiratory Syndrome Coronavirus 2 on Self-Collected Saliva or Anterior Nasal Specimens Compared With Healthcare Personnel-Collected Nasopharyngeal Specimens. Clin. Infect. Dis. 2021, 73, S65–S73. [Google Scholar] [CrossRef]
- Todsen, T.; Jakobsen, K.K.; Gronlund, M.P.; Callesen, R.E.; Folke, F.; Larsen, H.; Ersboll, A.K.; Benfield, T.; Gredal, T.; Klokker, M.; et al. COVID-19 Rapid Antigen Tests With Self-Collected vs. Health Care Worker-Collected Nasal and Throat Swab Specimens: A Randomized Clinical Trial. JAMA Netw. Open 2023, 6, e2344295. [Google Scholar] [CrossRef]
- Jung, E.J.; Lee, S.K.; Shin, S.H.; Kim, J.S.; Woo, H.; Cho, E.J.; Hyun, J.; Kim, J.S.; Kim, H.S. Comparison of Nasal Swabs, Nasopharyngeal Swabs, and Saliva Samples for the Detection of SARS-CoV-2 and other Respiratory Virus Infections. Ann. Lab. Med. 2023, 43, 434–442. [Google Scholar] [CrossRef]
- Viloria Winnett, A.; Akana, R.; Shelby, N.; Davich, H.; Caldera, S.; Yamada, T.; Reyna, J.R.B.; Romano, A.E.; Carter, A.M.; Kim, M.K.; et al. Daily SARS-CoV-2 Nasal Antigen Tests Miss Infected and Presumably Infectious People Due to Viral Load Differences among Specimen Types. Microbiol. Spectr. 2023, 11, e0129523. [Google Scholar] [CrossRef]
- Goodall, B.L.; LeBlanc, J.J.; Hatchette, T.F.; Barrett, L.; Patriquin, G. Investigating the Sensitivity of Nasal or Throat Swabs: Combination of Both Swabs Increases the Sensitivity of SARS-CoV-2 Rapid Antigen Tests. Microbiol. Spectr. 2022, 10, e0021722. [Google Scholar] [CrossRef]
- Teo, A.K.J.; Choudhury, Y.; Tan, I.B.; Cher, C.Y.; Chew, S.H.; Wan, Z.Y.; Cheng, L.T.E.; Oon, L.L.E.; Tan, M.H.; Chan, K.S.; et al. Saliva is more sensitive than nasopharyngeal or nasal swabs for diagnosis of asymptomatic and mild COVID-19 infection. Sci. Rep. 2021, 11, 3134. [Google Scholar] [CrossRef] [PubMed]
- Schrom, J.; Marquez, C.; Pilarowski, G.; Wang, C.Y.; Mitchell, A.; Puccinelli, R.; Black, D.; Rojas, S.; Ribeiro, S.; Tulier-Laiwa, V.; et al. Comparison of SARS-CoV-2 Reverse Transcriptase Polymerase Chain Reaction and BinaxNOW Rapid Antigen Tests at a Community Site During an Omicron Surge: A Cross-Sectional Study. Ann. Intern. Med. 2022, 175, 682–690. [Google Scholar] [CrossRef] [PubMed]
- Connor, M.C.; Copeland, M.; Curran, T. Investigation of saliva, tongue swabs and buccal swabs as alternative specimen types to nasopharyngeal swabs for SARS-CoV-2 testing. J. Clin. Virol. 2022, 146, 105053. [Google Scholar] [CrossRef]
- Ku, C.W.; Shivani, D.; Kwan, J.Q.T.; Loy, S.L.; Erwin, C.; Ko, K.K.K.; Ng, X.W.; Oon, L.; Thoon, K.C.; Kalimuddin, S.; et al. Validation of self-collected buccal swab and saliva as a diagnostic tool for COVID-19. Int. J. Infect. Dis. 2021, 104, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Hung, D.L.; Li, X.; Chiu, K.H.; Yip, C.C.; To, K.K.; Chan, J.F.; Sridhar, S.; Chung, T.W.; Lung, K.C.; Liu, R.W.; et al. Early-Morning vs. Spot Posterior Oropharyngeal Saliva for Diagnosis of SARS-CoV-2 Infection: Implication of Timing of Specimen Collection for Community-Wide Screening. Open Forum. Infect. Dis. 2020, 7, ofaa210. [Google Scholar] [CrossRef] [PubMed]
- Tng, D.J.H.; Yin, B.C.Y.; Cao, J.; Ko, K.K.K.; Goh, K.C.M.; Chua, D.X.W.; Zhang, Y.; Chua, M.L.K.; Low, J.G.H.; Ooi, E.E.; et al. Amplified parallel antigen rapid test for point-of-care salivary detection of SARS-CoV-2 with improved sensitivity. Mikrochim. Acta 2021, 189, 14. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.M.; Benoit, N.; Tissot-Dupont, H.; Million, M.; Pradines, B.; Granjeaud, S.; Almeras, L. Mouth Washing Impaired SARS-CoV-2 Detection in Saliva. Diagnostics 2021, 11, 1509. [Google Scholar] [CrossRef]
- Puyskens, A.; Michel, J.; Stoliaroff-Pepin, A.; Bayram, F.; Sesver, A.; Wichmann, O.; Harder, T.; Schaade, L.; Nitsche, A.; Peine, C. Direct comparison of clinical diagnostic sensitivity of saliva from buccal swabs versus combined oro-/nasopharyngeal swabs in the detection of SARS-CoV-2 B.1.1.529 Omicron. J. Clin. Virol. 2023, 165, 105496. [Google Scholar] [CrossRef]
- Wurstle, S.; Spinner, C.D.; Voit, F.; Hoffmann, D.; Hering, S.; Weidlich, S.; Schneider, J.; Zink, A.; Treiber, M.; Iakoubov, R.; et al. Self-sampling versus health care professional-guided swab collection for SARS-CoV-2 testing. Infection 2021, 49, 927–934. [Google Scholar] [CrossRef]
- Valentine-Graves, M.; Hall, E.; Guest, J.L.; Adam, E.; Valencia, R.; Shinn, K.; Hardee, I.; Sanchez, T.; Siegler, A.J.; Sullivan, P.S. At-home self-collection of saliva, oropharyngeal swabs and dried blood spots for SARS-CoV-2 diagnosis and serology: Post-collection acceptability of specimen collection process and patient confidence in specimens. PLoS ONE 2020, 15, e0236775. [Google Scholar] [CrossRef]


| Variable | COVID-19 Patients n = 103 | Non-COVID-19 Patients n = 96 | All Patients n = 199 |
|---|---|---|---|
| Sex: female | 40 (38.8) | 43 (44.8) | 83 (42) |
| Age in years, median | 55 (43–71) | 51 (34–65) | 54 (38–68) |
| Length of hospital stay in days | 6 (3–8) | 8 (5–13) | 6 (4–9) |
| Inpatients | 101 (98.1) | 37 (38.5) | 138 (69) |
| Main diagnosis for admission: | |||
| 92 (89) | - | 92 (46) |
| 1 (1) | 1 (1) | 2 (1) |
| - | 8 (8) | 8 (4) |
| - | 8 (8) | 8 (4) |
| 8 (8) | 21 (22) | 29 (15) |
| 2 (2) | 58 (60) | 60 (30) |
| Comorbidities: | |||
| 24 (23.3) | 21 (21.9) | 45 (23) |
| 24 (23.3) | 15 (15.6) | 39 (20) |
| 20 (19.4) | 14 (14.6) | 34 (17) |
| 12 (11.7) | 10 (10.4) | 22 (11) |
| 11 (10.7) | 11 (11.5) | 22 (11) |
| 10 (7.5) | 20 (20.8) | 30 (15) |
| 9 (9.7) | 18 (18.8) | 27 (14) |
| Pathogen identified (other than SARS-CoV-2): | |||
| - | 1 (1) | 1 (1) |
| - | 4 (4) | 4 (2) |
| - | 1 (1) | 1 (1) |
| - | 7 (7) | 7 (4) |
| - | 2 (2) | 2 (1) |
| - | 1 (1) | 1 (1) |
| Vaccinated against SARS-CoV-2: | |||
| 38 (36.9) | 19 (19.8) | 57 (29) |
| 32 (31.1) | 16 (16.7) | 48 (24) |
| 33 (32.0) | 61(63.5) | 94 (47) |
| Swabbing Location | n | Sensitivity (95% CI) | Specificity (95% CI) | PLR (95% CI) | NLR (95% CI) | PPV (95% CI) | NPV (95% CI) |
|---|---|---|---|---|---|---|---|
| Nasal | 103 | 96.1% (90.4–98.9) | 100% (96.2–100) | - | 0.04 (0–0.10) | 100% (96.3–100) | 96.0% (90.2–98.4) |
| Buccal | 102 * | 75.5% (63.9–81.8) | 99.0% (94.3–100.0) | 70.6 (10.0–497.8) | 0.27 (0.2–0.4) | 98.7% (91.4–99.8) | 77.9% (71.8–83.0) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dräger, S.; Bruni, F.; Bernasconi, M.; Hammann-Hänni, A.; Jirasko, V.; Tanno, A.; Blickenstorfer, Y.; Leuzinger, K.; Hirsch, H.H.; Osthoff, M. Impact of Swabbing Location, Self-Swabbing, and Food Intake on SARS-CoV-2 RNA Detection. Microorganisms 2024, 12, 591. https://doi.org/10.3390/microorganisms12030591
Dräger S, Bruni F, Bernasconi M, Hammann-Hänni A, Jirasko V, Tanno A, Blickenstorfer Y, Leuzinger K, Hirsch HH, Osthoff M. Impact of Swabbing Location, Self-Swabbing, and Food Intake on SARS-CoV-2 RNA Detection. Microorganisms. 2024; 12(3):591. https://doi.org/10.3390/microorganisms12030591
Chicago/Turabian StyleDräger, Sarah, Flavio Bruni, Melina Bernasconi, Anya Hammann-Hänni, Vlastimil Jirasko, Alexander Tanno, Yves Blickenstorfer, Karoline Leuzinger, Hans H. Hirsch, and Michael Osthoff. 2024. "Impact of Swabbing Location, Self-Swabbing, and Food Intake on SARS-CoV-2 RNA Detection" Microorganisms 12, no. 3: 591. https://doi.org/10.3390/microorganisms12030591
APA StyleDräger, S., Bruni, F., Bernasconi, M., Hammann-Hänni, A., Jirasko, V., Tanno, A., Blickenstorfer, Y., Leuzinger, K., Hirsch, H. H., & Osthoff, M. (2024). Impact of Swabbing Location, Self-Swabbing, and Food Intake on SARS-CoV-2 RNA Detection. Microorganisms, 12(3), 591. https://doi.org/10.3390/microorganisms12030591





