Characterize the Growth and Metabolism of Acidithiobacillus ferrooxidans under Electroautotrophic and Chemoautotrophic Conditions
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strain and Growth Medium
2.2. Setup of Electrochemical Cultivation
2.3. Cell Growth Monitoring and Cell Concentration Measurements
2.4. Scanning Electron Microscopy and Raman Spectroscopy
2.5. RNA Extraction, Library Preparation, and Sequencing
2.6. Transcriptomic Analysis
2.7. Metabolite Extraction and UHPLC-MS/MS Analysis
2.8. Metabolomic Analysis
2.9. Joint Analysis of Transcriptome and Metabolome
3. Results and Discussions
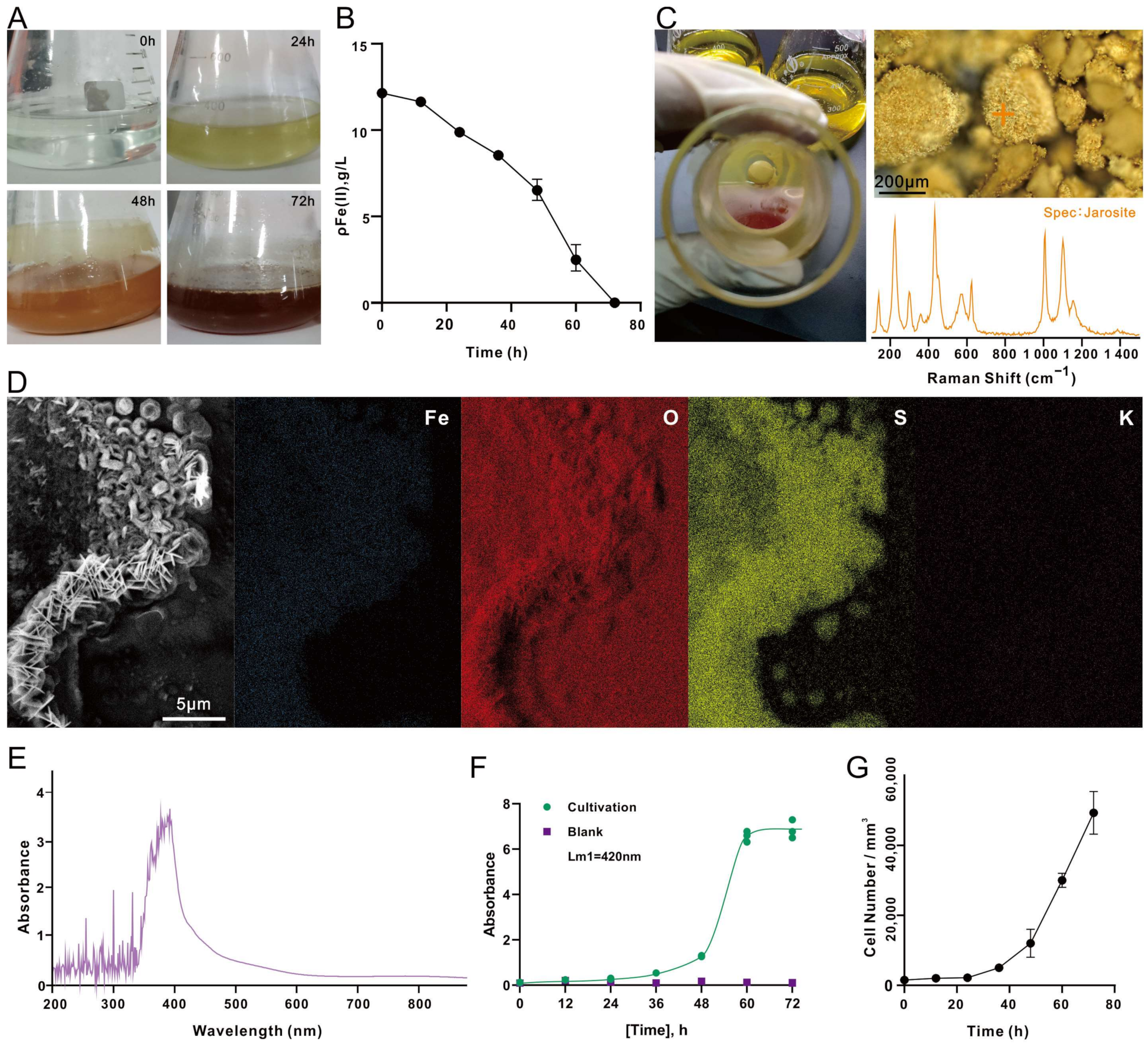
3.1. Avoiding Lausenite Formation in 9K Medium
3.2. Chemoautotrophic Growth of A. ferrooxidans
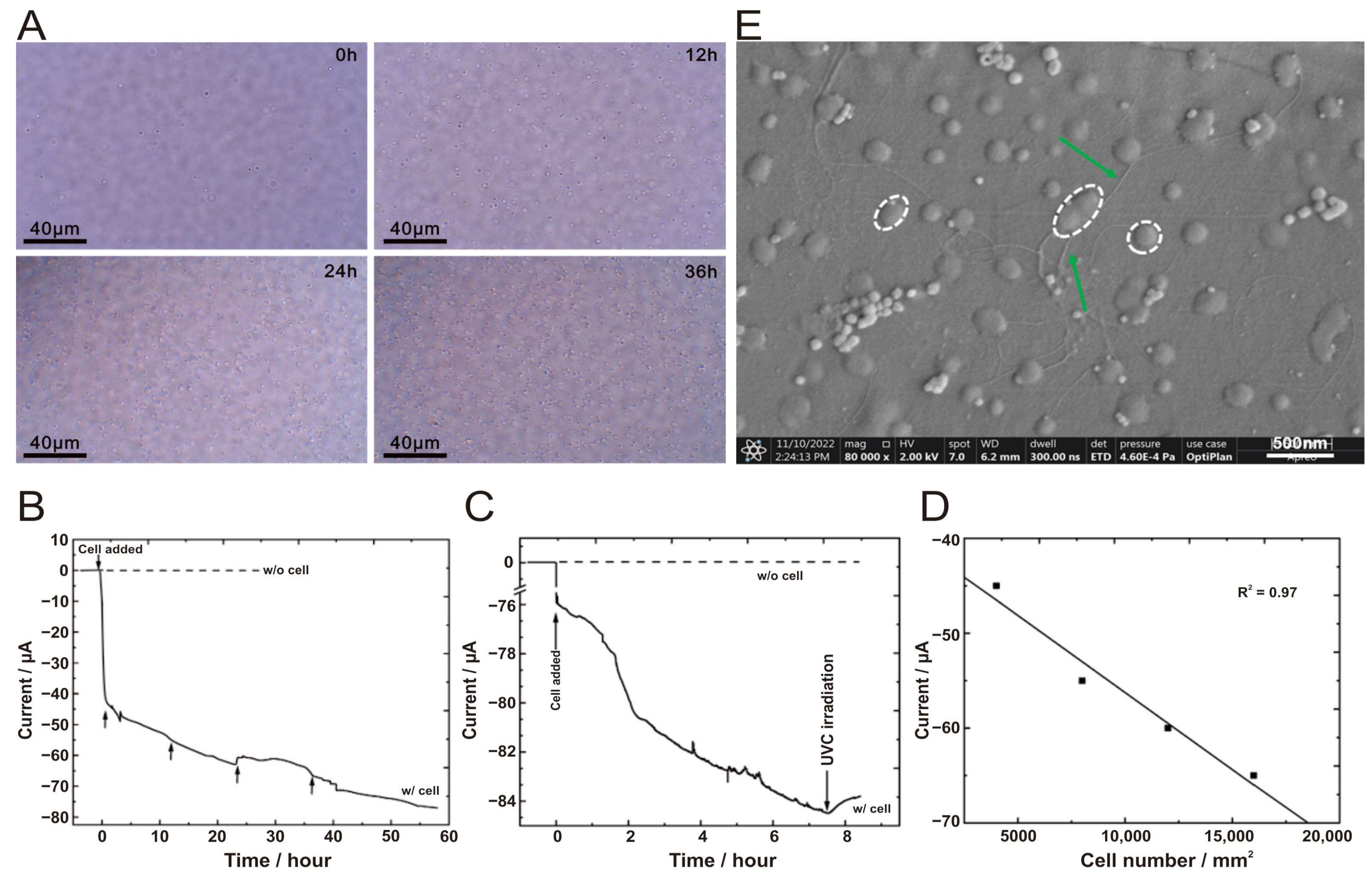
3.3. Electroautotrophic Growth of A. ferrooxidans
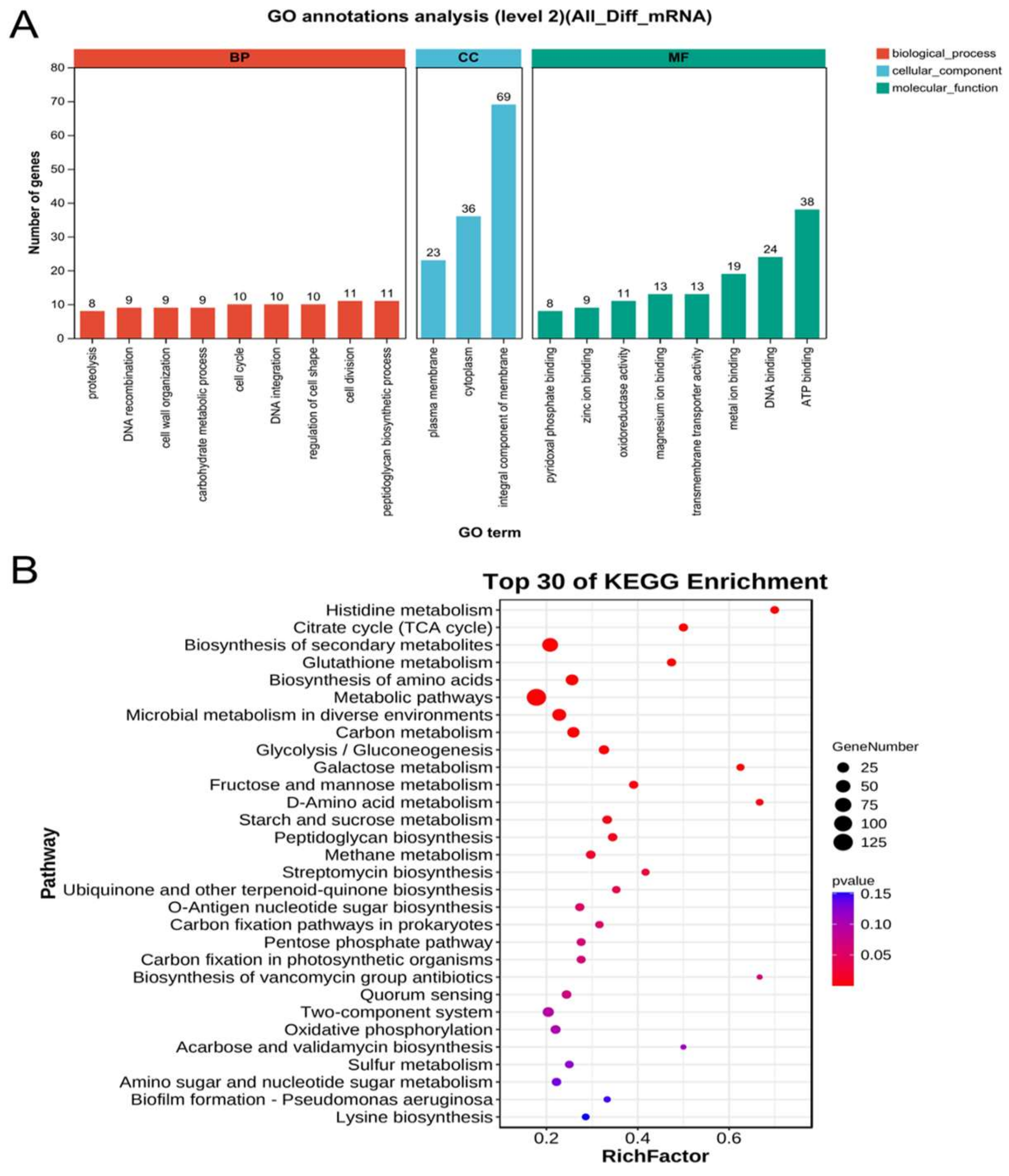
3.4. Transcriptomic Expression of A. ferrooxidans under Electroautotrophy vs. Chemoautotrophy Conditions
3.5. Joint Analysis of A. ferrooxidans Transcriptome and Metabolome under Mlectroautotrophy vs. Chemoautotrophy
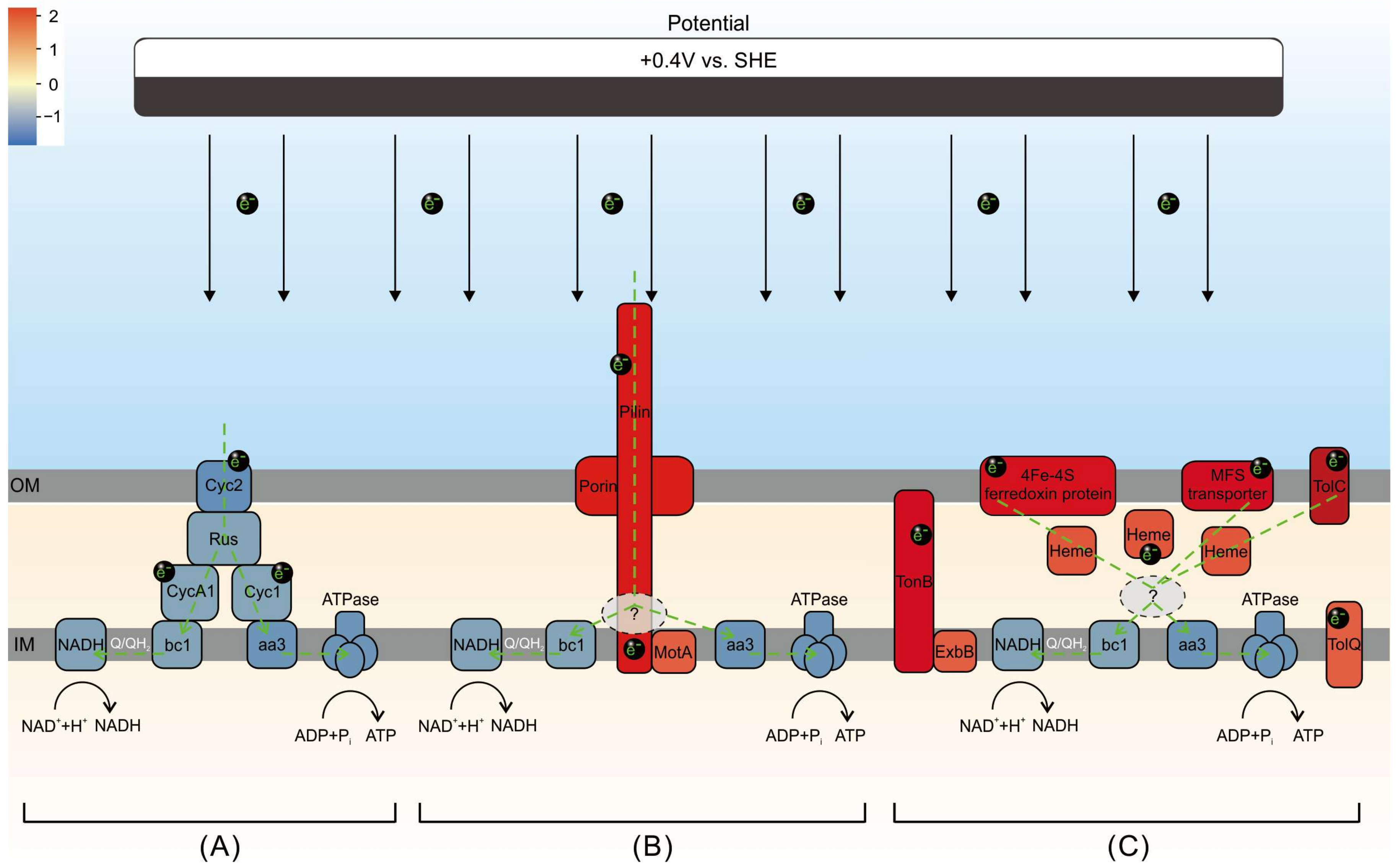
3.6. A Model of Extracellular Electron Uptake for A. ferrooxidans Electroautotrophy vs. Chemoautotrophy
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bobadilla Fazzini, R.A.; Levican, G.; Parada, P. Acidithiobacillus thiooxidans secretome containing a newly described lipoprotein Licanantase enhances chalcopyrite bioleaching rate. Appl. Microbiol. Biotechnol. 2010, 89, 771–780. [Google Scholar] [CrossRef] [PubMed]
- Sugio, T.; Ako, A.; Takeuchi, F. Sulfite oxidation catalyzed by aa3-Type cytochrome c oxidase in Acidithiobacillus ferrooxidans. Biosci. Biotechnol. Biochem. 2014, 74, 2242–2247. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Escuti, C.; Véliz, R.; Acosta, M.; Echeverría-Vega, A.; Araya, G.; Ayma, D.; Demergasso, C. The dynamics of two iron-oxidizing Acidithiobacillus strains in industrial copper sulfide heap-leaching. Res. Microbiol. 2023, 175, 104168. [Google Scholar] [CrossRef] [PubMed]
- Schippers, A.; Hedrich, S.; Vasters, J.; Drobe, M.; Sand, W.; Willscher, S. Biomining: Metal recovery from ores with microorganisms. In Geobiotechnology I: Metal-Related Issues; Schippers, A., Glombitza, F., Sand, W., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 1–47. [Google Scholar]
- Daoud, J.; Karamanev, D. Formation of jarosite during Fe2+ oxidation by Acidithiobacillus ferrooxidans. Miner. Eng. 2006, 19, 960–967. [Google Scholar] [CrossRef]
- Orellana, R.; Macaya, C.; Bravo, G.; Dorochesi, F.; Cumsille, A.; Valencia, R.; Rojas, C.; Seeger, M. Living at the frontiers of life: Extremophiles in Chile and their potential for bioremediation. Front. Microbiol. 2018, 9, 392041. [Google Scholar] [CrossRef] [PubMed]
- Micciche, A.C.; Barabote, R.D.; Dittoe, D.K.; Ricke, S.C. In silico genome analysis of an acid mine drainage species, Acidiphilium multivorum, for potential commercial acetic acid production and biomining. J. Environ. Sci. Health Part B 2020, 55, 447–454. [Google Scholar] [CrossRef] [PubMed]
- Temple, K.L.; Colmer, A.R. The autotrophic oxidation of iron by a new bacterium: Thiobacillus ferrooxidans. J. Bacteriol. 1951, 62, 605–611. [Google Scholar] [CrossRef]
- Kelly, D.P.; Wood, A.P. Reclassification of some species of Thiobacillus to the newly designated genera Acidithiobacillus gen. nov., Halothiobacillus gen. nov. and Thermithiobacillus gen. nov. Int. J. Syst. Evol. Microbiol. 2000, 50, 511–516. [Google Scholar] [CrossRef]
- Vargas-Straube, M.J.; Beard, S.; Norambuena, R.; Paradela, A.; Vera, M.; Jerez, C.A. High copper concentration reduces biofilm formation in Acidithiobacillus ferrooxidans by decreasing production of extracellular polymeric substances and its adherence to elemental sulfur. J. Proteom. 2020, 225, 103874. [Google Scholar] [CrossRef]
- Valdes, J.; Pedroso, I.; Quatrini, R.; Dodson, R.J.; Tettelin, H.; Blake, R., 2nd; Eisen, J.A.; Holmes, D.S. Acidithiobacillus ferrooxidans metabolism: From genome sequence to industrial applications. BMC Genom. 2008, 9, 597. [Google Scholar] [CrossRef]
- Osorio, H.c.; Mangold, S.; Denis, Y.; Nancucheo, I.; Esparza, M.; Johnson, D.B.; Bonnefoy, V.; Dopson, M.; Holmes, D.S. Anaerobic sulfur metabolism coupled to dissimilatory iron reduction in the extremophile Acidithiobacillus ferrooxidans. Appl. Environ. Microbiol. 2013, 79, 2172–2181. [Google Scholar] [CrossRef] [PubMed]
- Kucera, J.; Lochman, J.; Bouchal, P.; Pakostova, E.; Mikulasek, K.; Hedrich, S.; Janiczek, O.; Mandl, M.; Johnson, D.B. A model of aerobic and anaerobic metabolism of hydrogen in the extremophile Acidithiobacillus ferrooxidans. Front. Microbiol. 2020, 11, 610836. [Google Scholar] [CrossRef] [PubMed]
- Ishii, T.; Kawaichi, S.; Nakagawa, H.; Hashimoto, K.; Nakamura, R. From chemolithoautotrophs to electrolithoautotrophs: CO2 fixation by Fe (II)-oxidizing bacteria coupled with direct uptake of electrons from solid electron sources. Front. Microbiol. 2015, 6, 994. [Google Scholar] [CrossRef]
- Jung, H.; Inaba, Y.; Banta, S. Genetic engineering of the acidophilic chemolithoautotroph Acidithiobacillus ferrooxidans. Trends Biotechnol. 2022, 40, 677–692. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Y.; Yang, M.; Zhang, S.; Zhao, D.; Duan, J.; Wang, W.; Yan, L. Iron and sulfur oxidation pathways of Acidithiobacillus ferrooxidans. World J. Microbiol. Biotechnol. 2019, 35, 60. [Google Scholar] [CrossRef] [PubMed]
- Bauermeister, A.; Rettberg, P.; Flemming, H.-C. Growth of the acidophilic iron–sulfur bacterium Acidithiobacillus ferrooxidans under Mars-like geochemical conditions. Planet. Space Sci. 2014, 98, 205–215. [Google Scholar] [CrossRef]
- Limaye, S.S.; Mogul, R.; Smith, D.J.; Ansari, A.H.; Slowik, G.P.; Vaishampayan, P. Venus’ spectral signatures and the potential for life in the clouds. Astrobiology 2018, 18, 1181–1198. [Google Scholar] [CrossRef]
- Kaksonen, A.H.; Deng, X.; Morris, C.; Khaleque, H.N.; Zea, L.; Gumulya, Y. Potential of Acidithiobacillus ferrooxidans to grow on and bioleach metals from Mars and lunar regolith simulants under simulated microgravity conditions. Microorganisms 2021, 9, 2416. [Google Scholar] [CrossRef]
- Talla, E.; Hedrich, S.; Mangenot, S.; Ji, B.; Johnson, D.B.; Barbe, V.; Bonnefoy, V. Insights into the pathways of iron- and sulfur-oxidation, and biofilm formation from the chemolithotrophic acidophile Acidithiobacillus ferrivorans CF27. Res. Microbiol. 2014, 165, 753–760. [Google Scholar] [CrossRef]
- Brasseur, G.; Levicán, G.; Bonnefoy, V.; Holmes, D.; Jedlicki, E.; Lemesle-Meunier, D. Apparent redundancy of electron transfer pathways via bc1 complexes and terminal oxidases in the extremophilic chemolithoautotrophic Acidithiobacillus ferrooxidans. Biochim. Biophys. Acta BBA Bioenerg. 2004, 1656, 114–126. [Google Scholar] [CrossRef]
- Yarzábal, A.S.; Brasseur, G.L.; Ratouchniak, J.; Lund, K.; Lemesle-Meunier, D.; DeMoss, J.A.; Bonnefoy, V. The high-molecular-weight cytochrome c Cyc2 of Acidithiobacillus ferrooxidans is an outer membrane protein. J. Bacteriol. 2002, 184, 313–317. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.; Geng, M.; Liu, Y.; Zhao, W.; Xia, L.; Liu, J.; Qiu, G. Expression, purification and molecular modelling of the Iro protein from Acidithiobacillus ferrooxidans Fe-1. Protein Expr. Purif. 2007, 52, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Lin, J.; Pang, X.; Cui, S.; Mi, S.; Lin, J. Overexpression of rusticyanin in Acidithiobacillus ferrooxidans ATCC19859 increased Fe (II) oxidation activity. Curr. Microbiol. 2011, 62, 320–324. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Yan, L.; Xing, W.; Chen, P.; Zhang, Y.; Wang, W. Acidithiobacillus ferrooxidans and its potential application. Extremophiles 2018, 22, 563–579. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Shen, L.; Yin, H.; Li, Q.; Chen, Q.; Luo, Y.; Liao, L.; Qiu, G.; Liu, X. Comparative genomic analysis of Acidithiobacillus ferrooxidans strains using the A. ferrooxidans ATCC 23270 whole-genome oligonucleotide microarray. Can. J. Microbiol. 2009, 55, 587–598. [Google Scholar] [CrossRef] [PubMed]
- Nakasono, S.; Matsumoto, N.; Saiki, H. Electrochemical cultivation of Thiobacillus ferrooxidans by potential control. Bioelectrochem. Bioenerg. 1997, 43, 61–66. [Google Scholar] [CrossRef]
- Thrash, J.C.; Coates, J.D. Direct and indirect electrical stimulation of microbial metabolism. Environ. Sci. Technol. 2008, 42, 3921–3931. [Google Scholar] [CrossRef] [PubMed]
- Carbajosa, S.; Malki, M.; Caillard, R.; Lopez, M.F.; Palomares, F.J.; Martin-Gago, J.A.; Rodriguez, N.; Amils, R.; Fernandez, V.M.; De Lacey, A.L. Electrochemical growth of Acidithiobacillus ferrooxidans on a graphite electrode for obtaining a biocathode for direct electrocatalytic reduction of oxygen. Biosens. Bioelectron. 2010, 26, 877–880. [Google Scholar] [CrossRef]
- Ai, C.; Liang, Y.; Miao, B.; Chen, M.; Zeng, W.; Qiu, G. Identification and analysis of a novel gene cluster involves in Fe2+ oxidation in Acidithiobacillus ferrooxidans ATCC 23270, a Typical Biomining Acidophile. Curr. Microbiol. 2018, 75, 818–826. [Google Scholar] [CrossRef]
- Kim, T.W.; Kim, C.J.; Chang, Y.K.; Ryu, H.W.; Cho, K.S. Development of an optimal medium for continuous ferrous iron oxidation by immobilized Acidothiobacillus ferrooxidans cells. Biotechnol. Prog. 2002, 18, 752–759. [Google Scholar] [CrossRef]
- Zhang, T.; Zhang, H. Electrochemical analysis for the rapid screening of copper-tolerant bacteria. Bioelectrochemistry 2022, 148, 108276. [Google Scholar] [CrossRef] [PubMed]
- Asakai, T.; Hioki, A. Reliability in standardization of sodium thiosulfate with potassium dichromate. Microchem. J. 2015, 123, 9–14. [Google Scholar] [CrossRef]
- Long, H.; Yang, H.; Qu, Y. Distinguishing geobiological signatures from organic matter in the Ediacaran chert nodules. Precambrian Res. 2023, 390, 107045. [Google Scholar] [CrossRef]
- Beyssac, O.; Goffé, B.; Petitet, J.-P.; Froigneux, E.; Moreau, M.; Rouzaud, J.-N. On the characterization of disordered and heterogeneous carbonaceous materials by Raman spectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2003, 59, 2267–2276. [Google Scholar] [CrossRef] [PubMed]
- Ammar, M.; Rouzaud, J.N. How to obtain a reliable structural characterization of polished graphitized carbons by Raman microspectroscopy. J. Raman Spectrosc. 2012, 43, 207–211. [Google Scholar] [CrossRef]
- Ribeiro, D.A.; Del Bem, L.E.; Vicentini, R.; Ferraz, L.F.; Murakami, M.T.; Ottoboni, L.M. The small heat shock proteins from Acidithiobacillus ferrooxidans: Gene expression, phylogenetic analysis, and structural modeling. BMC Microbiol. 2011, 11, 1471–2180. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Nan, X.; Zhao, Y.; Jiang, L.; Wang, H.; Zhang, F.; Hua, D.; Liu, J.; Yang, L.; Yao, J. Discrepancies among healthy, subclinical mastitic, and clinical mastitic cows in fecal microbiome and metabolome and serum metabolome. J. Dairy Sci. 2022, 105, 7668–7688. [Google Scholar] [CrossRef]
- Ren, Y.; Yu, G.; Shi, C.; Liu, L.; Guo, Q.; Han, C.; Zhang, D.; Zhang, L.; Liu, B.; Gao, H. Majorbio Cloud: A one-stop, comprehensive bioinformatic platform for multiomics analyses. iMeta 2022, 1, e12. [Google Scholar] [CrossRef]
- Sasaki, K.; Tanaike, O.; Konno, H. Distinction of jarosite-group compounds by Raman spectroscopy. Can. Mineral. 1998, 36, 1225–1235. [Google Scholar]
- Xie, H.; Liu, Z.; Zhou, E. Growth of Thiobacillus ferrooxidan in high concentration ferrous iron culture medium. Chin. J. Process Eng. 2004, 4, 43–46. (In Chinese) [Google Scholar]
- Yan, L.; Zhang, S.; Chen, P.; Wang, W.; Wang, Y.; Li, H. Magnetic properties of Acidithiobacillus ferrooxidans. Mater. Sci. Eng. C 2013, 33, 4026–4031. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.-D.; Sun, X.-M.; Guan, Y. Biogenic mineralization in the ferromanganese nodules and crusts from the South China Sea. J. Asian Earth Sci. 2019, 171, 46–59. [Google Scholar] [CrossRef]
- Schubert, B.A.; Lowenstein, T.K.; Timofeeff, M.N.; Parker, M.A. How do prokaryotes survive in fluid inclusions in halite for 30 ky? Geology 2009, 37, 1059–1062. [Google Scholar] [CrossRef]
- Novitsky, J.A.; Morita, R.Y. Morphological characterization of small cells resulting from nutrient starvation of a psychrophilic marine vibrio. Appl. Environ. Microbiol. 1976, 32, 617–622. [Google Scholar] [CrossRef] [PubMed]
- Winters, Y.D.; Lowenstein, T.K.; Timofeeff, M.N. Starvation-survival in Haloarchaea. Life 2015, 5, 1587–1609. [Google Scholar] [CrossRef] [PubMed]
- Lowenstein, T.K.; Schubert, B.A.; Timofeeff, M.N. Microbial communities in fluid inclusions and long-term survival in halite. GSA Today 2011, 21, 4–9. [Google Scholar] [CrossRef]
- Liu, H.; Wang, X.; Long, J.; Gao, Y.; Wan, Z.; Liu, C. Effect of Electric field on culture of Acidithiobacillus ferrooxidans. Nonferrous Met. Extr. Metall. 2022, 8, 76–81. (In Chinese) [Google Scholar] [CrossRef]
- Zheng, L.; Yu, H.; Fan, Y.; Shi, Z.-H.; Wu, P.; Wang, Z.-J.; Wang, H.-Y.; Ding, K.; Zhou, H. Enhancement in growth and iron oxidation of Acidithiobacillus ferrooxidans under extreme environment via applied electric potential. Environ. Eng. 2023, 11, 1–10. (In Chinese) [Google Scholar]
- Yang, Y.; Huang, Z.; Diao, M.; Qiu, G. Isolation and characterization of the petII promoter of Acidithiobacillus ferrooxidans. J. Food Agric. Environ. 2010, 8, 1383–1387. [Google Scholar]
- Bruscella, P.; Appia-Ayme, C.; Levicán, G.; Ratouchniak, J.; Jedlicki, E.; Holmes, D.S.; Bonnefoy, V. Differential expression of two bc1 complexes in the strict acidophilic chemolithoautotrophic bacterium Acidithiobacillus ferrooxidans suggests a model for their respective roles in iron or sulfur oxidation. Microbiology 2007, 153, 102–110. [Google Scholar] [CrossRef]
- Norris, P.R.; Laigle, L.; Slade, S. Cytochromes in anaerobic growth of Acidithiobacillus ferrooxidans. Microbiology 2018, 164, 383–394. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, P.; Guiliani, N.; Valenzuela, L.; Beard, S.; Jerez, C.A. Differential protein expression during growth of Acidithiobacillus ferrooxidans on ferrous iron, sulfur compounds, or metal sulfides. Appl. Environ. Microbiol. 2004, 70, 4491–4498. [Google Scholar] [CrossRef] [PubMed]
- Hedrich, S.; Schlömann, M.; Johnson, D.B. The iron-oxidizing proteobacteria. Microbiology 2011, 157, 1551–1564. [Google Scholar] [CrossRef] [PubMed]
- Nasiri, S.S.; Sarabi, M.; Fatemi, F.; Dini, S. Investigating the rus and petI operon expression patterns in exposed Acidithiobacillus ferrooxidans sp. FJ2 to different doses of gamma irradiation. Appl. Radiat. Isot. 2021, 177, 109911. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Liu, Z.; Meng, D.; Liu, Y.; Liu, T.; Jiang, C.; Yin, H. Sequence similarity network and protein structure prediction offer insights into the evolution of microbial pathways for ferrous iron oxidation. mSystems 2023, 8, e00720–e00723. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-Q.; Wan, D.-S.; Huang, S.-S.; Leng, F.-F.; Yan, L.; Ni, Y.-Q.; Li, H.-Y. Type IV pili of Acidithiobacillus ferrooxidans are necessary for sliding, twitching motility, and adherence. Curr. Microbiol. 2010, 60, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Huang, S.; Zhang, X.; Huang, T.; Li, H. Cloning, expression, and functional analysis of molecular motor pilT and pilU genes of type IV pili in Acidithiobacillus ferrooxidans. Appl. Microbiol. Biotechnol. 2013, 97, 1251–1257. [Google Scholar] [CrossRef]
- Li, Y.; Li, H. Type IV pili of Acidithiobacillus ferrooxidans can transfer electrons from extracellular electron donors. J. Basic Microbiol. 2014, 54, 226–231. [Google Scholar] [CrossRef]
- Wall, D.; Kaiser, D. Type IV pili and cell motility. Mol. Microbiol. 1999, 32, 1–10. [Google Scholar] [CrossRef]
- Schirmer, T. General and specific porins from bacterial outer membranes. J. Struct. Biol. 1998, 121, 101–109. [Google Scholar] [CrossRef]
- Ligthart, K.; Belzer, C.; De Vos, W.M.; Tytgat, H.L. Bridging bacteria and the gut: Functional aspects of type IV pili. Trends Microbiol. 2020, 28, 340–348. [Google Scholar] [CrossRef] [PubMed]
- Andrews, J. Deletion and Characterization of a Putative Major Type IV Pilin Protein In Vibrio Parahaemolyticus. Bachelor’s Thesis, University of Delaware, Newark, DE, USA, 2022. [Google Scholar]
- Sticht, H.; Rösch, P. The structure of iron–sulfur proteins. Prog. Biophys. Mol. Biol. 1998, 70, 95–136. [Google Scholar] [CrossRef]
- Otaka, E.; Ooi, T. Examination of protein sequence homologies: IV. Twenty-seven bacterial ferredoxins. J. Mol. Evol. 1987, 26, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Santos, M.R.; Cosme, A.M.; Becker, J.D.; Medeiros, J.M.C.; Mata, M.F.; Moreira, L.M. Absence of functional TolC protein causes increased stress response gene expression in Sinorhizobium meliloti. BMC Microbiol. 2010, 10, 180. [Google Scholar] [CrossRef] [PubMed]
- Moussatova, A.; Kandt, C.; O’Mara, M.L.; Tieleman, D.P. ATP-binding cassette transporters in Escherichia coli. Biochim. Biophys. Acta BBA Biomembr. 2008, 1778, 1757–1771. [Google Scholar] [CrossRef] [PubMed]
- Pao, S.S.; Paulsen, I.T.; Saier, M.H., Jr. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 1998, 62, 1–34. [Google Scholar] [CrossRef] [PubMed]
- Quistgaard, E.M.; Löw, C.; Guettou, F.; Nordlund, P. Understanding transport by the major facilitator superfamily (MFS): Structures pave the way. Nat. Rev. Mol. Cell Biol. 2016, 17, 123–132. [Google Scholar] [CrossRef]
- Fujita, M.; Mori, K.; Hara, H.; Hishiyama, S.; Kamimura, N.; Masai, E. A TonB-dependent receptor constitutes the outer membrane transport system for a lignin-derived aromatic compound. Commun. Biol. 2019, 2, 432. [Google Scholar] [CrossRef]
- Ferguson, A.D.; Deisenhofer, J. TonB-dependent receptors-structural perspectives. Biochim. Biophys. Acta 2002, 1565, 318–332. [Google Scholar] [CrossRef]
- Braun, T.F.; Poulson, S.; Gully, J.B.; Empey, J.C.; Van Way, S.; Putnam, A.; Blair, D.F. Function of proline residues of MotA in torque generation by the flagellar motor of Escherichia coli. J. Bacteriol. 1999, 181, 3542–3551. [Google Scholar] [CrossRef]
- Wolfger, H.; Mamnun, Y.M.; Kuchler, K. Fungal ABC proteins: Pleiotropic drug resistance, stress response and cellular detoxification. Res. Microbiol. 2001, 152, 375–389. [Google Scholar] [CrossRef] [PubMed]
- Liang, F.; Wu, R.; Cao, C.; Zheng, Y.; Yang, Z.; Zhao, F. Research on extracellular electron transfer of Acidithiobacillus ferrooxidans. Chem. J. Chin. Univ. 2014, 35, 372–376. (In Chinese) [Google Scholar] [CrossRef]
- Wang, J.; Tao, L.; Zhao, H.; Hu, M.; Zheng, X.; Peng, H.; Gan, X.; Xiao, W.; Cao, P.; Qin, W. Cooperative effect of chalcopyrite and bornite interactions during bioleaching by mixed moderately thermophilic culture. Miner. Eng. 2016, 95, 116–123. [Google Scholar] [CrossRef]
- Li, H.; Qiu, G.; Hu, Y.; Cang, D.; Wang, D. Electrochemical behavior of chalcopyrite in presence of Thiobacillus ferrooxidans. Trans. Nonferrous Met. Soc. China 2006, 16, 1240–1245. (In Chinese) [Google Scholar] [CrossRef]
- Quatrini, R.; Appia-Ayme, C.; Denis, Y.; Jedlicki, E.; Holmes, D.S.; Bonnefoy, V. Extending the models for iron and sulfur oxidation in the extreme acidophile Acidithiobacillus ferrooxidans. BMC Genom. 2009, 10, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Su, R.; Cao, K.; Guan, J. Low current density on Thiobacillus ferrooxidans under technology of microbial leaching metals from waste printed circuit boards. J. Shanghai Second Polytech. Univ. 2015, 32, 7–11. (In Chinese) [Google Scholar] [CrossRef]
- Vardanyan, N.; Badalyan, H.; Markosyan, L.; Vardanyan, A.; Zhang, R.; Sand, W. Newly isolated Acidithiobacillus sp. ksh from kashen copper ore: Peculiarities of EPS and colloidal exopolysaccharide. Front. Microbiol. 2020, 11, 550223. [Google Scholar] [CrossRef] [PubMed]
- Saavedra, A.; Aguirre, P.; Gentina, J.C. Biooxidation of iron by Acidithiobacillus ferrooxidans in the presence of D-galactose: Understanding its influence on the production of EPS and cell tolerance to high concentrations of iron. Front. Microbiol. 2020, 11, 517326. [Google Scholar] [CrossRef]
- Aguirre, P.; Guerrero, K.; Sanchez-Rodriguez, A.; Gentina, J.C.; Schippers, A. Making sticky cells: Effect of galactose and ferrous iron on the attachment of Leptospirillum ferrooxidans to mineral surfaces. Res. Microbiol. 2018, 169, 569–575. [Google Scholar] [CrossRef]
- Gehrke, T.; Telegdi, J.; Thierry, D.; Sand, W. Importance of extracellular polymeric substances from Thiobacillus ferrooxidans for bioleaching. Appl. Environ. Microbiol. 1998, 64, 2743–2747. [Google Scholar] [CrossRef]
- Yang, Y.; Diao, M.; Liu, K.; Qian, L.; Nguyen, A.V.; Qiu, G. Column bioleaching of low-grade copper ore by Acidithiobacillus ferrooxidans in pure and mixed cultures with a heterotrophic acidophile Acidiphilium sp. Hydrometallurgy 2013, 131, 93–98. [Google Scholar] [CrossRef]
- Bellenberg, S.; Huynh, D.; Poetsch, A.; Sand, W.; Vera, M. Proteomics reveal enhanced oxidative stress responses and metabolic adaptation in Acidithiobacillus ferrooxidans biofilm cells on pyrite. Front. Microbiol. 2019, 10, 434788. [Google Scholar] [CrossRef] [PubMed]
- Vardanyan, A.; Vardanyan, N.; Khachatryan, A.; Zhang, R.; Sand, W. Adhesion to mineral surfaces by cells of Leptospirillum, Acidithiobacillus and Sulfobacillus from Armenian sulfide ores. Minerals 2019, 9, 69. [Google Scholar] [CrossRef]
- Das, S.; Diels, L.; Pant, D.; Patil, S.A.; Ghangrekar, M. Microbial electrosynthesis: A way towards the production of electro-commodities through carbon sequestration with microbes as biocatalysts. J. Electrochem. Soc. 2020, 167, 155510. [Google Scholar] [CrossRef]
- Karbelkar, A.A.; Rowe, A.R.; El-Naggar, M.Y. An electrochemical investigation of interfacial electron uptake by the sulfur oxidizing bacterium Thioclava electrotropha ElOx9. Electrochim. Acta 2019, 324, 134838. [Google Scholar] [CrossRef]
- Kernan, T.; Majumdar, S.; Li, X.; Guan, J.; West, A.C.; Banta, S. Engineering the iron-oxidizing chemolithoautotroph Acidithiobacillus ferrooxidans for biochemical production. Biotechnol. Bioeng. 2016, 113, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; West, A.C.; Banta, S. Enhancing isobutyric acid production from engineered Acidithiobacillus ferrooxidans cells via media optimization. Biotechnol. Bioeng. 2015, 113, 790–796. [Google Scholar] [CrossRef]
- Inaba, Y.; Kernan, T.; Bantaa, S. Transposase-mediated chromosomal integration of exogenous genes in Acidithiobacillus ferrooxidans. Appl. Environ. Microbiol. 2018, 84, e01381-18. [Google Scholar] [CrossRef]
- Kai, M.; Yano, T.; Fukumori, Y.; Yamanaka, T. Cytochrome oxidase of an acidophilic iron-oxidizing bacterium, Thiobacillus ferrooxidans, functions at pH 3.5. Biochem. Biophys. Res. Commun. 1989, 160, 839–843. [Google Scholar] [CrossRef]
- Malarte, G.; Leroy, G.; Lojou, E.; Abergel, C.; Bruschi, M.; Giudici-Orticoni, M.T. Insight into molecular stability and physiological properties of the diheme cytochrome CYC41 from the acidophilic bacterium Acidithiobacillus ferrooxidans. Biochemistry 2005, 44, 6471–6481. [Google Scholar] [CrossRef]
- Shi, L.; Dong, H.; Reguera, G.; Beyenal, H.; Lu, A.; Liu, J.; Yu, H.-Q.; Fredrickson, J.K. Extracellular electron transfer mechanisms between microorganisms and minerals. Nat. Rev. Microbiol. 2016, 14, 651–662. [Google Scholar] [CrossRef] [PubMed]
- White, G.F.; Edwards, M.J.; Gomez-Perez, L.; Richardson, D.J.; Butt, J.N.; Clarke, T.A. Mechanisms of bacterial extracellular electron exchange. Adv. Microb. Physiol. 2016, 68, 87–138. [Google Scholar]
- Reguera, G.; McCarthy, K.D.; Mehta, T.; Nicoll, J.S.; Tuominen, M.T.; Lovley, D.R. Extracellular electron transfer via microbial nanowires. Nature 2005, 435, 1098–1101. [Google Scholar] [CrossRef] [PubMed]
- Gorby, Y.A.; Yanina, S.; McLean, J.S.; Rosso, K.M.; Moyles, D.; Dohnalkova, A.; Beveridge, T.J.; Chang, I.S.; Kim, B.H.; Kim, K.S. Electrically conductive bacterial nanowires produced by Shewanella oneidensis strain MR-1 and other microorganisms. Proc. Natl. Acad. Sci. USA 2006, 103, 11358–11363. [Google Scholar] [CrossRef]
- Pirbadian, S.; Barchinger, S.E.; Leung, K.M.; Byun, H.S.; Jangir, Y.; Bouhenni, R.A.; Reed, S.B.; Romine, M.F.; Saffarini, D.A.; Shi, L. Shewanella oneidensis MR-1 nanowires are outer membrane and periplasmic extensions of the extracellular electron transport components. Proc. Natl. Acad. Sci. USA 2014, 111, 12883–12888. [Google Scholar] [CrossRef]
- He, S.; Barco, R.A.; Emerson, D.; Roden, E.E. Comparative genomic analysis of neutrophilic Iron(II) Oxidizer genomes for candidate genes in extracellular electron transfer. Front. Microbiol. 2017, 8, 285775. [Google Scholar] [CrossRef] [PubMed]




| Gene_id | Gene Name | Gene Description | Log2FC (ea/ca) | Pathways |
|---|---|---|---|---|
| AFE_RS16265 | AFE_RS16265 | NAD(P)-dependent oxidoreductase | −2.25 | uphill |
| AFE_RS03815 | AFE_RS03815 | FAD binding domain-containing protein | −2.05 | uphill |
| AFE_RS14440 | cyc2 | c-type cytochrome | −1.57 | downhill/uphill |
| AFE_RS04465 | cydB | cytochrome d ubiquinol oxidase subunit II | −1.54 | uphill |
| AFE_RS14290 | AFE_RS14290 | FAD/NAD(P)-binding oxidoreductase | −1.54 | uphill |
| AFE_RS00620 | AFE_RS00620 | cytochrome c oxidase assembly protein | −1.52 | downhill |
| AFE_RS12010 | nuoL | NADH-quinone oxidoreductase subunit L | −1.44 | uphill |
| AFE_RS14425 | AFE_RS14425 | cbb3-type cytochrome c oxidase subunit I | −1.43 | downhill |
| AFE_RS12615 | hslU | ATP-dependent protease ATPase subunit HslU | −1.38 | downhill |
| AFE_RS04470 | AFE_RS04470 | cytochrome ubiquinol oxidase subunit I | −1.36 | uphill |
| AFE_RS00650 | AFE_RS00650 | FAD-binding protein | −1.34 | uphill |
| AFE_RS01260 | AFE_RS01260 | FAD-dependent oxidoreductase | −1.26 | uphill |
| AFE_RS12210 | apbC | NADH:ubiquinone oxidoreductase (complex I) | −1.25 | uphill |
| AFE_RS04275 | clpX | ATP-dependent Clp protease ATP-binding subunit ClpX | −1.24 | downhill |
| AFE_RS14500 | AFE_RS14500 | FMN-binding negative transcriptional regulator | −1.22 | uphill |
| AFE_RS02000 | icd | NADP-dependent isocitrate dehydrogenase | −1.22 | uphill |
| AFE_RS14415 | rus | Rusticyanin | −1.21 | downhill/uphill |
| AFE_RS14430 | AFE_RS14430 | cytochrome c oxidase subunit II | −1.17 | downhill |
| AFE_RS12530 | AFE_RS12530 | cytochrome bc complex cytochrome b subunit | −1.16 | uphill |
| AFE_RS13940 | hisG | ATP phosphoribosyltransferase | −1.12 | downhill |
| AFE_RS10975 | AFE_RS10975 | FAD-dependent monooxygenase | −1.07 | uphill |
| AFE_RS03470 | thyX | FAD-dependent thymidylate synthase | −1.05 | uphill |
| AFE_RS14650 | AFE_RS14650 | ATP-binding protein | −1.01 | downhill |
| Gene_id | Gene Name | Gene Description | Log2FC (ea/ca) |
|---|---|---|---|
| novel0441 | - | TolC family protein, partial | 3.32 |
| novel0021 | - | membrane protein, putative | 3.06 |
| novel0531 | - | putative ABC transporter ATP-binding protein | 2.59 |
| novel0233 | - | type IV pilin biogenesis protein, putative, partial | 2.50 |
| AFE_RS00795 | AFE_RS00795 | MFS transporter | 2.38 |
| novel0534 | - | TonB-dependent receptor | 2.37 |
| AFE_RS10580 | AFE_RS10580 | biopolymer transporter ExbD | 2.36 |
| novel0363 | - | MULTISPECIES: 4Fe-4S dicluster domain-containing protein | 2.27 |
| AFE_RS10585 | AFE_RS10585 | energy transducer TonB | 2.20 |
| AFE_RS13360 | AFE_RS13360 | porin | 2.19 |
| novel0120 | - | MULTISPECIES: MFS transporter | 2.09 |
| novel0582 | - | MULTISPECIES: TonB-dependent receptor | 2.01 |
| novel0397 | - | TonB family protein | 1.91 |
| novel0535 | - | MULTISPECIES: energy transducer TonB | 1.89 |
| novel0185 | - | heme-utilization protein HutZ, putative | 1.75 |
| AFE_RS10575 | AFE_RS10575 | MotA/TolQ/ExbB proton channel family protein | 1.66 |
| novel0005 | - | MULTISPECIES: ferredoxin family protein | 1.57 |
| AFE_RS09710 | AFE_RS09710 | ABC-2 transporter permease | 1.49 |
| AFE_RS10630 | AFE_RS10630 | sugar porter family MFS transporter | 1.28 |
| AFE_RS13730 | AFE_RS13730 | peptide ABC transporter substrate-binding protein | 1.19 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Q.; Long, H.; Wang, H.; Lau Vetter, M.C.Y. Characterize the Growth and Metabolism of Acidithiobacillus ferrooxidans under Electroautotrophic and Chemoautotrophic Conditions. Microorganisms 2024, 12, 590. https://doi.org/10.3390/microorganisms12030590
Wang Q, Long H, Wang H, Lau Vetter MCY. Characterize the Growth and Metabolism of Acidithiobacillus ferrooxidans under Electroautotrophic and Chemoautotrophic Conditions. Microorganisms. 2024; 12(3):590. https://doi.org/10.3390/microorganisms12030590
Chicago/Turabian StyleWang, Quansheng, Haijun Long, Huiqi Wang, and Maggie C. Y. Lau Vetter. 2024. "Characterize the Growth and Metabolism of Acidithiobacillus ferrooxidans under Electroautotrophic and Chemoautotrophic Conditions" Microorganisms 12, no. 3: 590. https://doi.org/10.3390/microorganisms12030590
APA StyleWang, Q., Long, H., Wang, H., & Lau Vetter, M. C. Y. (2024). Characterize the Growth and Metabolism of Acidithiobacillus ferrooxidans under Electroautotrophic and Chemoautotrophic Conditions. Microorganisms, 12(3), 590. https://doi.org/10.3390/microorganisms12030590





