Spatiotemporal Dynamics of Assyrtiko Grape Microbiota
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Next Generation Sequencing
2.2.1. DNA Extraction
2.2.2. Amplicon Amplification with PCR
2.2.3. Library Preparation and Sequencing
2.3. Data Analysis
3. Results
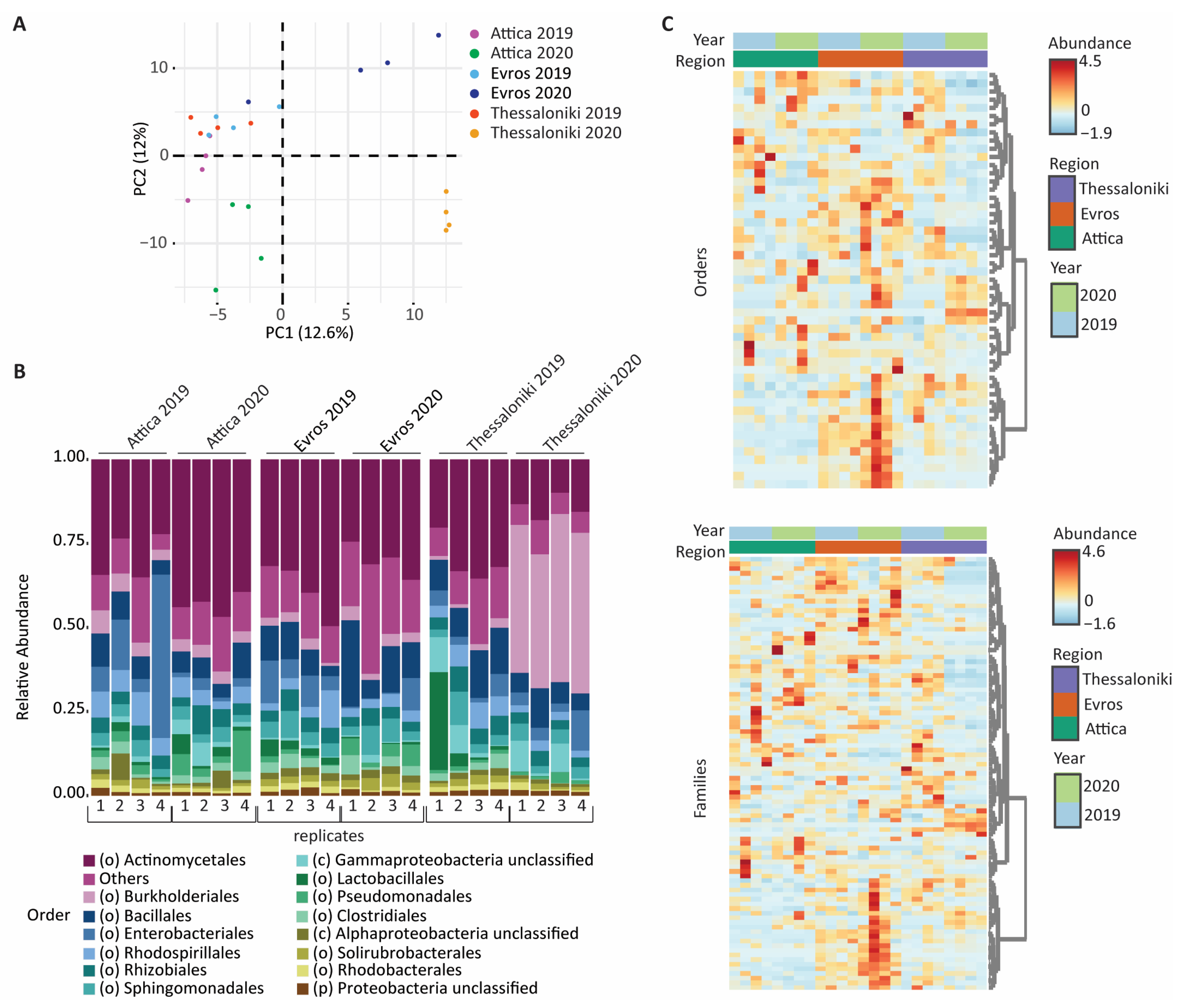
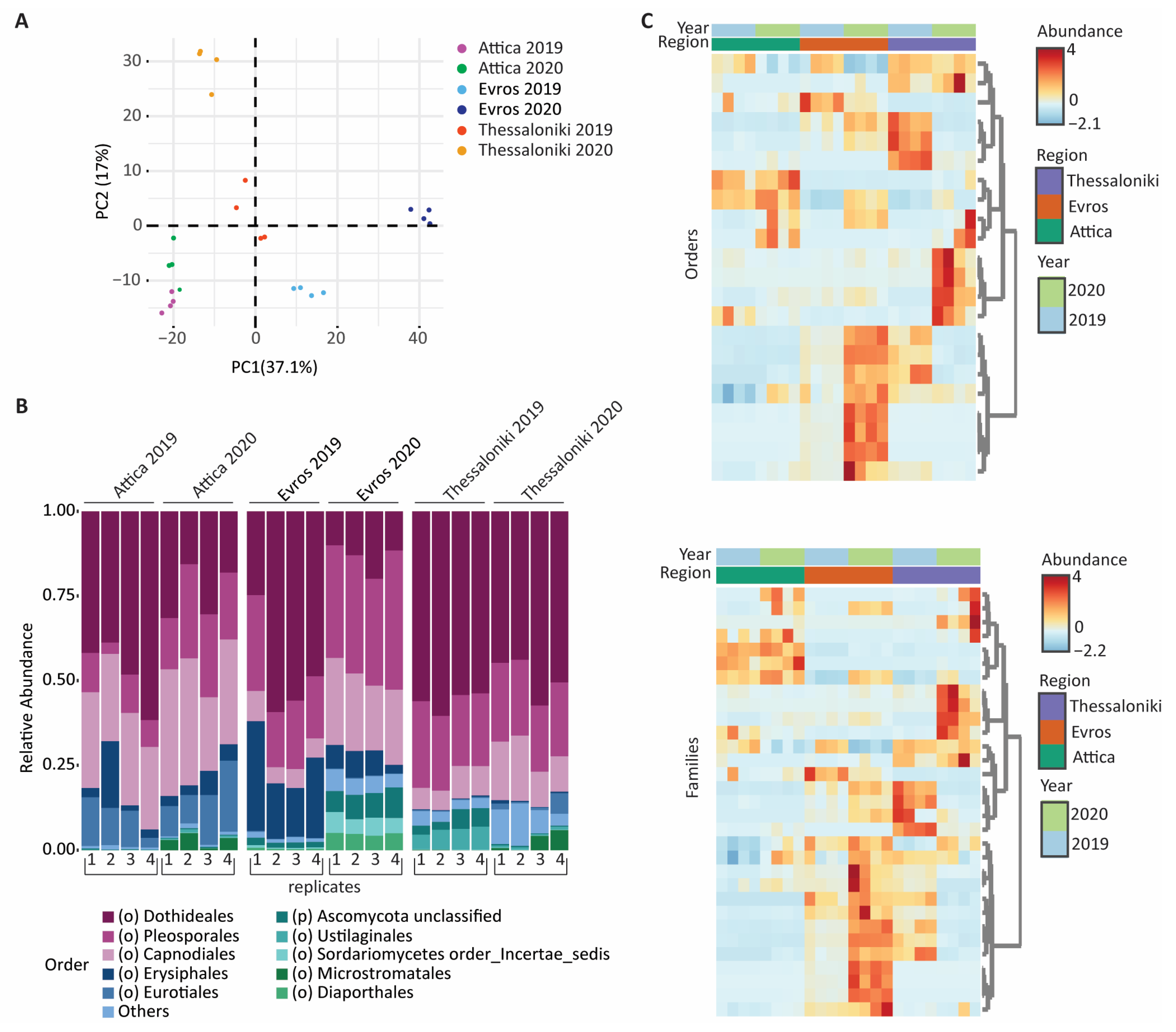
3.1. Geographic Distribution
3.2. Diversity Assessment
3.2.1. Bacterial Communities
3.2.2. Fungal Communities
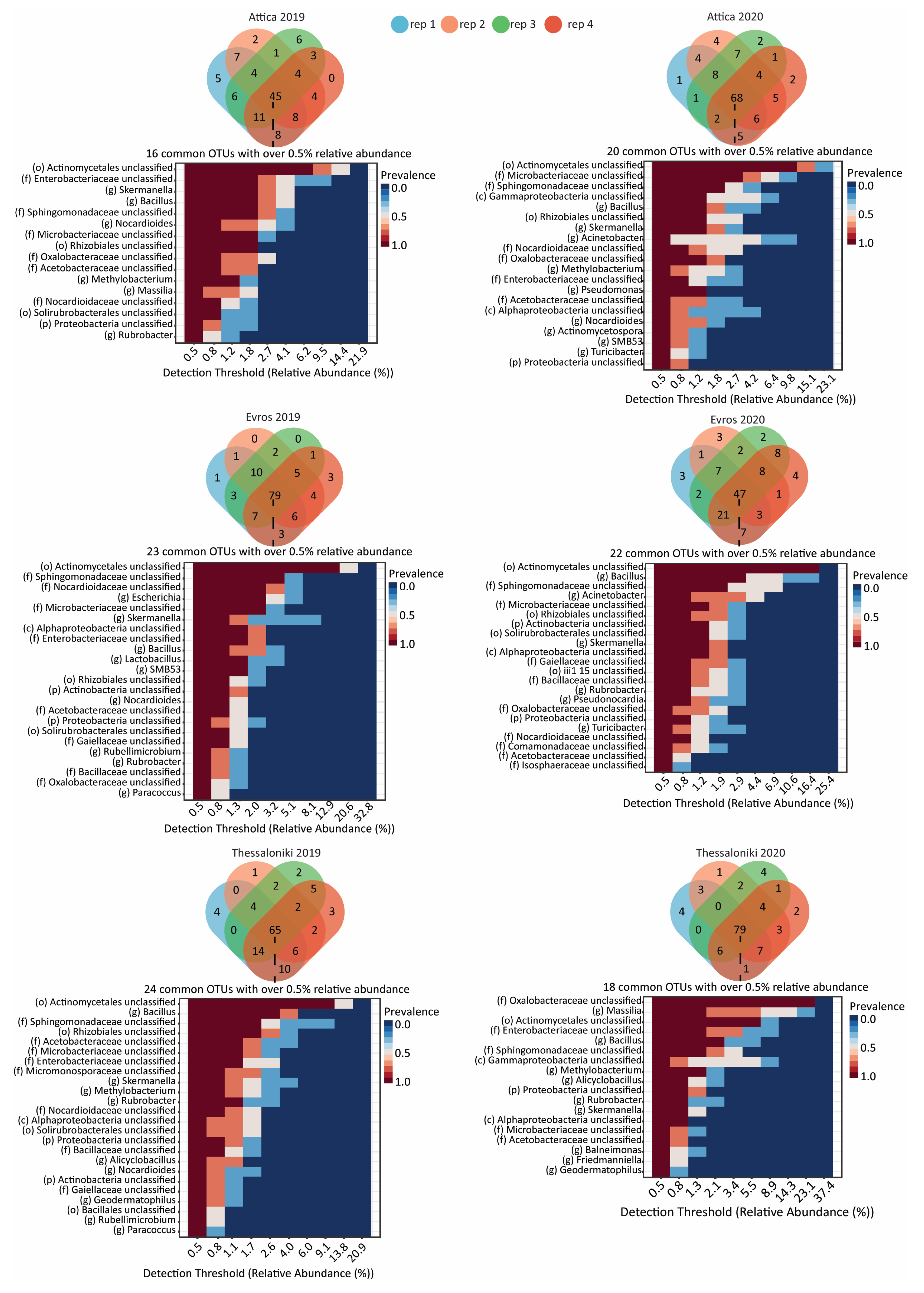
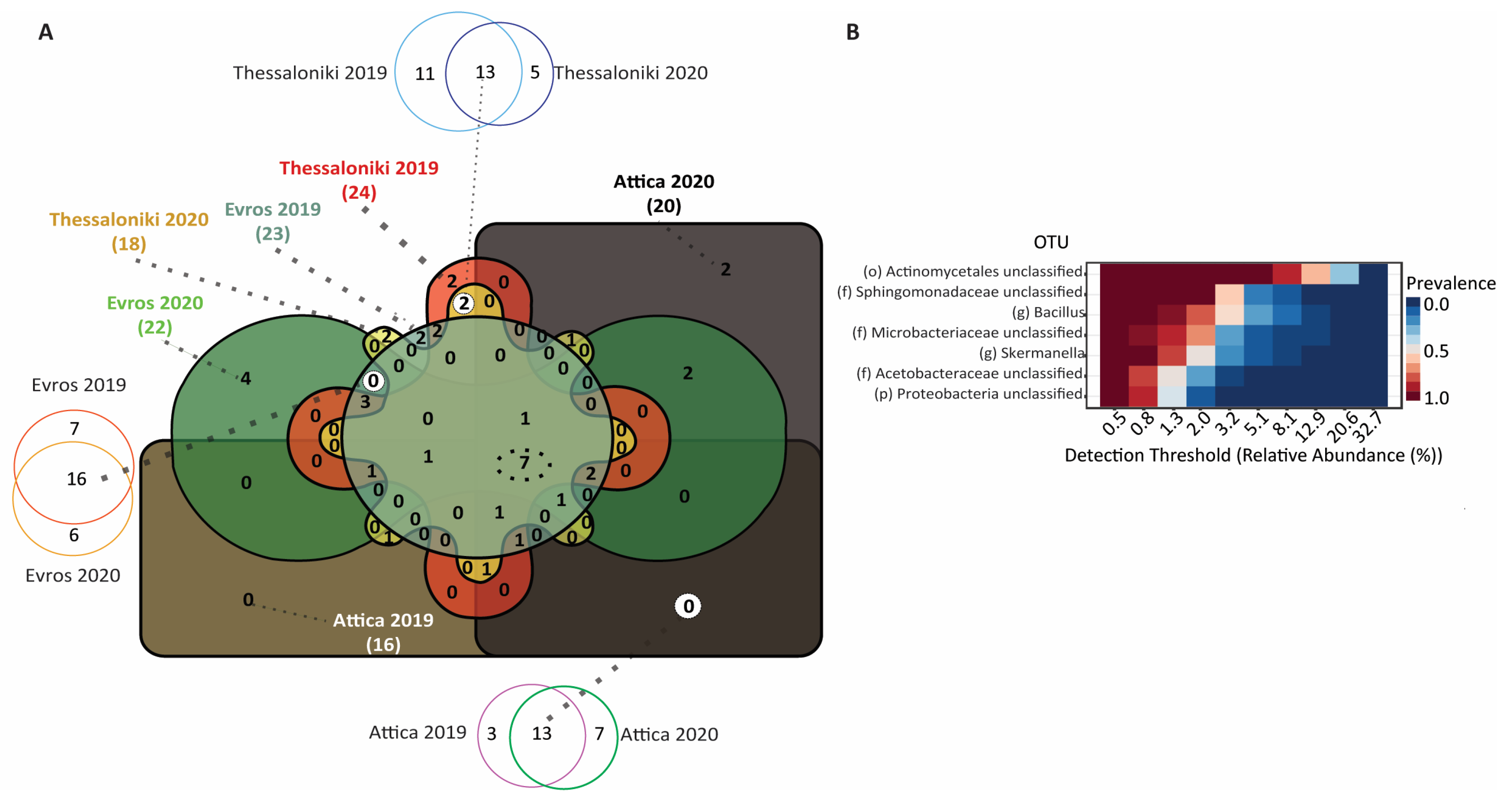
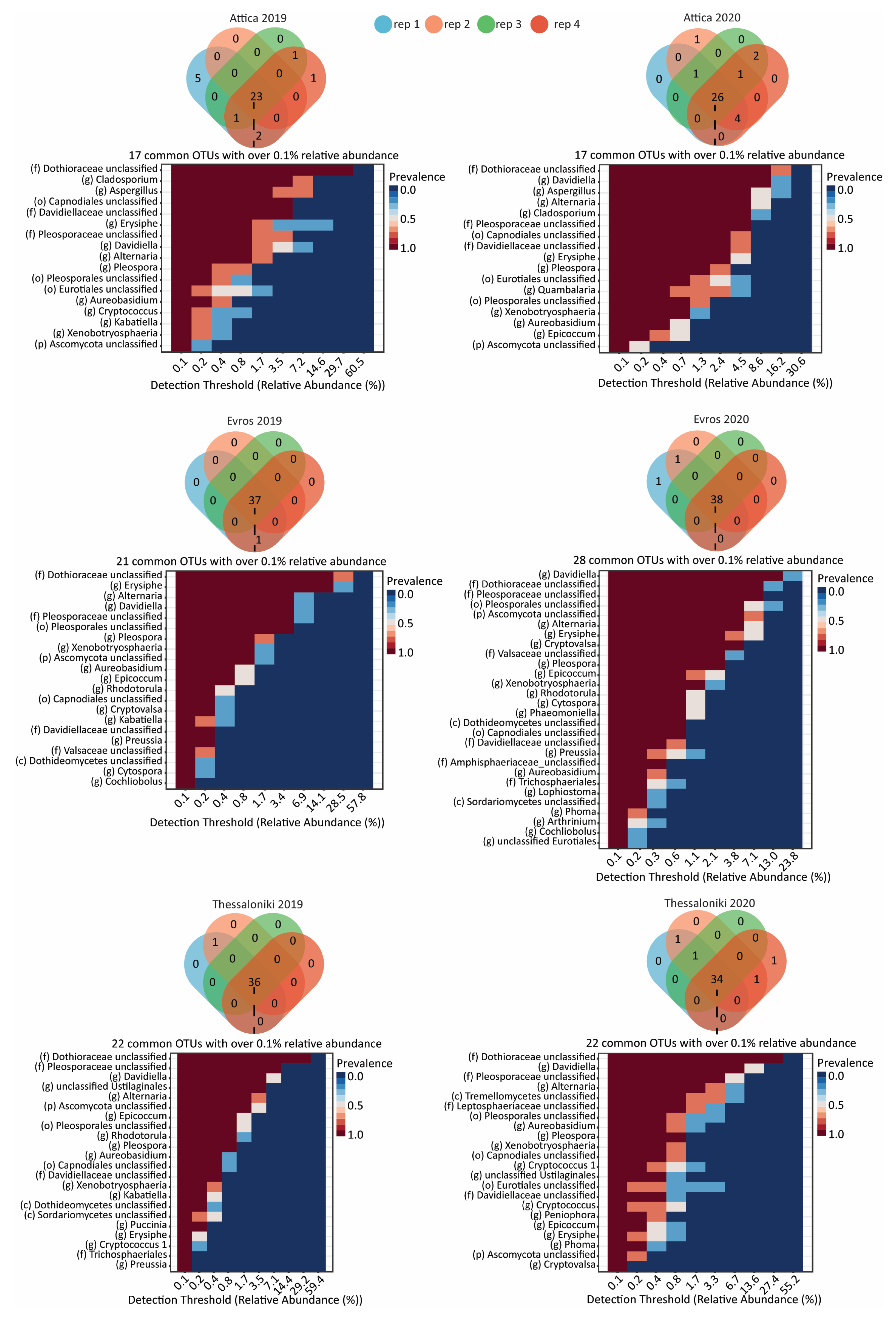
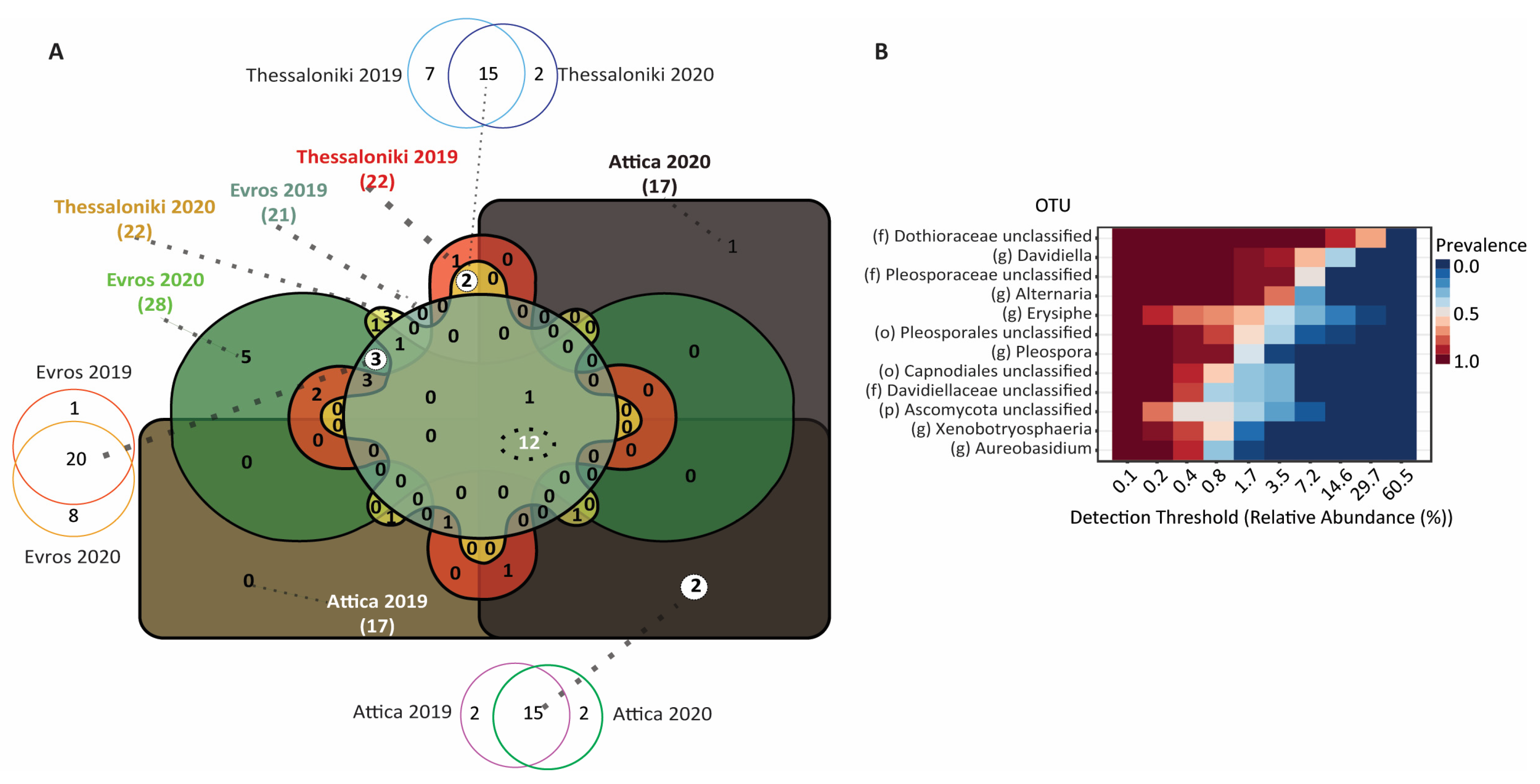
3.3. Taxonomic Distribution and Comparison
3.3.1. Bacterial Microbiome
3.3.2. Fungal Microbiome
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Savo, V.; Kumbaric, A.; Caneva, G. Grapevine (Vitis Vinifera L.) Symbolism in the Ancient Euro-Mediterranean Cultures. Econ. Bot. 2016, 70, 190–197. [Google Scholar] [CrossRef]
- Alston, J.M.; Sambucci, O. The Grape Genome; Springer: Berlin/Heidelberg, Germany, 2019; ISBN 978-3-030-18600-5. [Google Scholar]
- Bokulich, N.A.; Thorngate, J.H.; Richardson, P.M.; Mills, D.A. Microbial Biogeography of Wine Grapes Is Conditioned by Cultivar, Vintage, and Climate. Proc. Natl. Acad. Sci. USA 2014, 111, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Belda, I.; Ruiz, J.; Esteban-Fernández, A.; Navascués, E.; Marquina, D.; Santos, A.; Moreno-Arribas, M.V. Microbial Contribution to Wine Aroma and Its Intended Use for Wine Quality Improvement. Molecules 2017, 22, 189. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Miao, Y.; Wang, H.; Du, J.; Wang, C.; Shi, X.; Wang, B. Analysis of Microbial Community Diversity on the Epidermis of Wine Grapes in Manasi’s Vineyard, Xinjiang. Foods 2022, 11, 3174. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.; Ding, Y.; Chen, N.; Wang, L.; Gao, F.; Zhang, L.; Song, R.; Liu, Y.; Li, H.; Wang, H. Diversity and Dynamics of Microbial Communities during Spontaneous Fermentation of Cabernet Sauvignon (Vitis Vinifera L.) from Different Regions of China and Their Relationship with the Volatile Components in the Wine. Food Res. Int. 2022, 156, 111372. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.T.; Chen, N.; Ding, Y.T.; Wang, L.; Liu, Y.H.; Gao, F.F.; Zhang, L.; Li, H.; Wang, H. Correlations between Microbiota with Physicochemical Properties and Volatile Compounds during the Spontaneous Fermentation of Cabernet Sauvignon (Vitis Vinifera L.) Wine. LWT 2022, 163, 113529. [Google Scholar] [CrossRef]
- Comitini, F.; Capece, A.; Ciani, M.; Romano, P. New Insights on the Use of Wine Yeasts. Curr. Opin. Food Sci. 2017, 13, 44–49. [Google Scholar] [CrossRef]
- Wei, T.L.; Zheng, Y.P.; Wang, Z.H.; Shang, Y.X.; Pei, M.S.; Liu, H.N.; Yu, Y.H.; Shi, Q.F.; Jiang, D.M.; Guo, D.L. Comparative Microbiome Analysis Reveals the Variation in Microbial Communities between ‘Kyoho’ Grape and Its Bud Mutant Variety. PLoS ONE 2023, 18, e0290853. [Google Scholar] [CrossRef]
- Wei, R.T.; Chen, N.; Ding, Y.T.; Wang, L.; Gao, F.F.; Zhang, L.; Liu, Y.H.; Li, H.; Wang, H. Diversity and Dynamics of Epidermal Microbes During Grape Development of Cabernet Sauvignon (Vitis Vinifera L.) in the Ecological Viticulture Model in Wuhai, China. Front. Microbiol. 2022, 13, 935647. [Google Scholar] [CrossRef]
- Barata, A.; Malfeito-Ferreira, M.; Loureiro, V. The Microbial Ecology of Wine Grape Berries. Int. J. Food Microbiol. 2012, 153, 243–259. [Google Scholar] [CrossRef]
- Nisiotou, A.A.; Rantsiou, K.; Iliopoulos, V.; Cocolin, L.; Nychas, G.J.E. Bacterial Species Associated with Sound and Botrytis-Infected Grapes from a Greek Vineyard. Int. J. Food Microbiol. 2011, 145, 432–436. [Google Scholar] [CrossRef]
- Gao, F.; Chen, J.; Xiao, J.; Cheng, W.; Zheng, X.; Wang, B.; Shi, X. Microbial Community Composition on Grape Surface Controlled by Geographical Factors of Different Wine Regions in Xinjiang, China. Food Res. Int. 2019, 122, 348–360. [Google Scholar] [CrossRef]
- Kioroglou, D.; Kraeva-Deloire, E.; Schmidtke, L.M.; Mas, A.; Portillo, M.C. Geographical Origin Has a Greater Impact on Grape Berry Fungal Community than Grape Variety and Maturation State. Microorganisms 2019, 7, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Martins, G.; Vallance, J.; Mercier, A.; Albertin, W.; Stamatopoulos, P.; Rey, P.; Lonvaud, A.; Masneuf-Pomarède, I. Influence of the Farming System on the Epiphytic Yeasts and Yeast-like Fungi Colonizing Grape Berries during the Ripening Process. Int. J. Food Microbiol. 2014, 177, 21–28. [Google Scholar] [CrossRef]
- Perazzolli, M.; Antonielli, L.; Storari, M.; Puopolo, G.; Pancher, M.; Giovannini, O.; Pindo, M.; Pertot, I. Resilience of the Natural Phyllosphere Microbiota of the Grapevine to Chemical and Biological Pesticides. Appl. Environ. Microbiol. 2014, 80, 3585–3596. [Google Scholar] [CrossRef] [PubMed]
- Pinto, C.; Pinho, D.; Sousa, S.; Pinheiro, M.; Egas, C.; Gomes, A.C. Unravelling the Diversity of Grapevine Microbiome. PLoS ONE 2014, 9, e85622. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.J.; Wu, Y.; Yan, Y.Z.; Zou, W.; Xue, J.; Ma, W.R.; Wang, W.; Tian, G.; Wang, L.Y. High-Throughput Sequencing of Microbial Community Diversity in Soil, Grapes, Leaves, Grape Juice and Wine of Grapevine from China. PLoS ONE 2018, 13, e0193097. [Google Scholar] [CrossRef]
- Cocolin, L.; Heisey, A.; Mills, D.A. Direct Identification of the Indigenous Yeasts in Commercial Wine Fermentations. Am. J. Enol. Vitic. 2001, 52, 49–53. [Google Scholar] [CrossRef]
- Martins, G.; Miot-Sertier, C.; Lauga, B.; Claisse, O.; Lonvaud-Funel, A.; Soulas, G.; Masneuf-Pomarède, I. Grape Berry Bacterial Microbiota: Impact of the Ripening Process and the Farming System. Int. J. Food Microbiol. 2012, 158, 93–100. [Google Scholar] [CrossRef]
- Morgan, H.H.; du Toit, M.; Setati, M.E. The Grapevine and Wine Microbiome: Insights from High-Throughput Amplicon Sequencing. Front. Microbiol. 2017, 8, 249460. [Google Scholar] [CrossRef]
- Belda, I.; Zarraonaindia, I.; Perisin, M.; Palacios, A.; Acedo, A. From Vineyard Soil to Wine Fermentation: Microbiome Approximations to Explain the “Terroir” Concept. Front. Microbiol. 2017, 8, 247040. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Collins, T.; Masarweh, C.; Allen, G.; Heymann, H.; Ebeler, S.E.; Mills, D.A. Fermentation Behavior Suggest Microbial Contribution to Regional. mBio 2016, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Awad, M.; Giannopoulos, G.; Mylona, P.V.; Polidoros, A.N. Comparative Analysis of Grapevine Epiphytic Microbiomes among Different Varieties, Tissues, and Developmental Stages in the Same Terroir. Appl. Sci. 2023, 13, 102. [Google Scholar] [CrossRef]
- del Carmen Portillo, M.; Franquès, J.; Araque, I.; Reguant, C.; Bordons, A. Bacterial Diversity of Grenache and Carignan Grape Surface from Different Vineyards at Priorat Wine Region (Catalonia, Spain). Int. J. Food Microbiol. 2016, 219, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Vlachos, V.A. A Macroeconomic Estimation of Wine Production in Greece. Wine Econ. Policy 2017, 6, 3–13. [Google Scholar] [CrossRef]
- Nanou, E.; Mavridou, E.; Milienos, F.S.; Papadopoulos, G.; Tempère, S.; Kotseridis, Y. Odor Characterization of White Wines Produced from Indigenous Greek Grape Varieties Using the Trained Assessors. Foods 2020, 9, 1396. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing Mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhou, G.; Ewald, J.; Pang, Z.; Shiri, T.; Xia, J. MicrobiomeAnalyst 2.0: Comprehensive Statistical, Functional and Integrative Analysis of Microbiome Data. Nucleic Acids Res. 2023, 51, W310–W318. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Cordero-Bueso, G.; Arroyo, T.; Serrano, A.; Tello, J.; Aporta, I.; Vélez, M.D.; Valero, E. Influence of the Farming System and Vine Variety on Yeast Communities Associated with Grape Berries. Int. J. Food Microbiol. 2011, 145, 132–139. [Google Scholar] [CrossRef]
- Griggs, R.G.; Steenwerth, K.L.; Mills, D.A.; Cantu, D.; Bokulich, N.A. Sources and Assembly of Microbial Communities in Vineyards as a Functional Component of Winegrowing. Front. Microbiol. 2021, 12, 673810. [Google Scholar] [CrossRef]
- Canfora, L.; Vendramin, E.; Felici, B.; Tarricone, L.; Florio, A.; Benedetti, A. Vineyard Microbiome Variations during Different Fertilisation Practices Revealed by 16s RRNA Gene Sequencing. Appl. Soil Ecol. 2018, 125, 71–80. [Google Scholar] [CrossRef]
- Grangeteau, C.; Roullier-Gall, C.; Rousseaux, S.; Gougeon, R.D.; Schmitt-Kopplin, P.; Alexandre, H.; Guilloux-Benatier, M. Wine Microbiology Is Driven by Vineyard and Winery Anthropogenic Factors. Microb. Biotechnol. 2017, 10, 354–370. [Google Scholar] [CrossRef]
- Fernandes, P.; Afonso, I.M.; Pereira, J.; Rocha, R.; Rodrigues, A.S. Epiphitic Microbiome of Alvarinho Wine Grapes from Different Geographic Regions in Portugal. Biology 2023, 12, 146. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, E.T.; Singh, R.P.; Guo, C.; Shang, Y.; Chen, J.; Liu, C. Grape Berry Surface Bacterial Microbiome: Impact from the Varieties and Clones in the Same Vineyard from Central China. J. Appl. Microbiol. 2019, 126, 204–214. [Google Scholar] [CrossRef] [PubMed]
- Verginer, M.; Leitner, E.; Berg, G. Production of Volatile Metabolites by Grape-Associated Microorganisms. J. Agric. Food Chem. 2010, 58, 8344–8350. [Google Scholar] [CrossRef]
- Gilbert, J.A.; Van Der Lelie, D.; Zarraonaindia, I. Microbial Terroir for Wine Grapes. Proc. Natl. Acad. Sci. USA 2014, 111, 5–6. [Google Scholar] [CrossRef] [PubMed]
- Mezzasalma, V.; Sandionigi, A.; Bruni, I.; Bruno, A.; Lovicu, G.; Casiraghi, M.; Labra, M. Grape Microbiome as a Reliable and Persistent Signature of Field Origin and Environmental Conditions in Cannonau Wine Production. PLoS ONE 2017, 12, e0184615. [Google Scholar] [CrossRef]
- De Filippis, F.; La Storia, A.; Blaiotta, G. Monitoring the Mycobiota during Greco Di Tufo and Aglianico Wine Fermentation by 18S RRNA Gene Sequencing. Food Microbiol. 2017, 63, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulou, E.; Bekris, F.; Vasileiadis, S.; Krokida, A.; Rouvali, T.; Veskoukis, A.S.; Liadaki, K.; Kouretas, D.; Karpouzas, D.G. Vineyard-Mediated Factors Are Still Operative in Spontaneous and Commercial Fermentations Shaping the Vinification Microbial Community and Affecting the Antioxidant and Anticancer Properties of Wines. Food Res. Int. 2023, 173, 113359. [Google Scholar] [CrossRef]
- Taylor, M.W.; Tsai, P.; Anfang, N.; Ross, H.A.; Goddard, M.R. Pyrosequencing Reveals Regional Differences in Fruit-Associated Fungal Communities. Environ. Microbiol. 2014, 16, 2848–2858. [Google Scholar] [CrossRef]
- Kamilari, E.; Mina, M.; Karallis, C.; Tsaltas, D. Metataxonomic Analysis of Grape Microbiota during Wine Fermentation Reveals the Distinction of Cyprus Regional Terroirs. Front. Microbiol. 2021, 12, 726483. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Yang, S.; Lin, M.; Guo, S.; Han, X.; Ren, M.; Du, L.; Song, Y.; You, Y.; Zhan, J.; et al. The Biogeography of Fungal Communities across Different Chinese Wine-Producing Regions Associated with Environmental Factors and Spontaneous Fermentation Performance. Front. Microbiol. 2022, 12, 636639. [Google Scholar] [CrossRef] [PubMed]
- Steenwerth, K.L.; Morelan, I.; Stahel, R.; Figueroa-Balderas, R.; Cantu, D.; Lee, J.; Runnebaum, R.C.; Poret-Peterson, A.T. Fungal and Bacterial Communities of “Pinot Noir” Must: Effects of Vintage, Growing Region, Climate, and Basic Must Chemistry. PeerJ 2021, 9, e10836. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Howell, K. Community Succession of the Grapevine Fungal Microbiome in the Annual Growth Cycle. Environ. Microbiol. 2020, 23, 1842–1857. [Google Scholar] [CrossRef]
- Setati, M.E.; Jacobson, D.; Bauer, F.F. Sequence-Based Analysis of the Vitis Vinifera L. Cv Cabernet Sauvignon Grape Must Mycobiome in Three South African Vineyards Employing Distinct Agronomic Systems. Front. Microbiol. 2015, 6, 159533. [Google Scholar] [CrossRef]
- Martins, G.; Soulas, L.; Soulas, G.; Masneuf-pomare, I. Characterization of Epiphytic Bacterial Communities from Grapes, Leaves, Bark and Soil of Grapevine Plants Grown, and Their Relations. PLoS ONE 2013, 8, e73013. [Google Scholar] [CrossRef] [PubMed]
- Leveau, J.H.J.; Tech, J.J. Grapevine Microbiomics: Bacterial Diversity on Grape Leaves and Berries Revealed by High-Throughput Sequence Analysis of 16S RRNA Amplicons. Acta Hortic. 2011, 905, 31–42. [Google Scholar] [CrossRef]
- Kačániová, M.; Kunová, S.; Felsöciová, S.; Ivanišová, E.; Kántor, A.; Puchalski, C.; Terentjeva, M. Microbiota of Different Wine Grape Berries. J. Food Sci. 2019, 13, 174–181. [Google Scholar] [CrossRef]
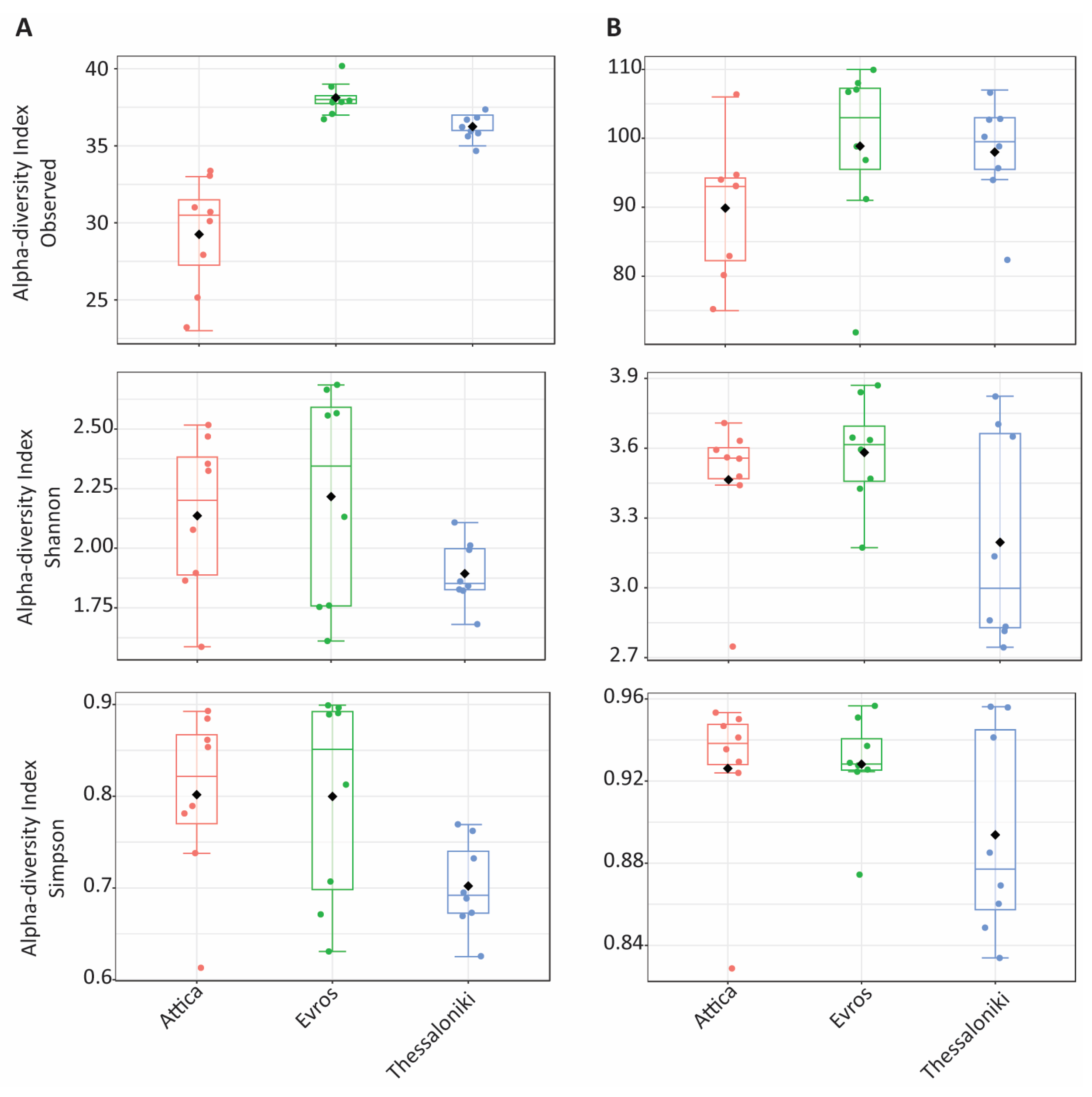
| Region | Year | OTUs | Observed OTUs | Shannon | Simpson |
|---|---|---|---|---|---|
| Attica | 2019 | 114 | 83.00 ± 8.04 | 3.40 ± 0.44 | 0.92 ± 0.06 |
| Attica | 2020 | 119 | 96.75 ± 6.24 | 3.53 ± 0.08 | 0.94 ± 0.01 |
| Evros | 2019 | 125 | 108.00 ± 1.41 | 3.63 ± 0.32 | 0.93 ± 0.04 |
| Evros | 2020 | 119 | 89.75 ± 12.31 | 3.53 ± 0.10 | 0.93 ± 0.01 |
| Thessaloniki | 2019 | 120 | 96.50 ± 11.09 | 3.58 ± 0.30 | 0.93 ± 0.03 |
| Thessaloniki | 2020 | 117 | 99.50 ± 2.89 | 2.81 ± 0.05 | 0.85 ± 0.02 |
| Region | Year | OTUs | Observed OTUs | Shannon | Simpson |
|---|---|---|---|---|---|
| Attica | 2019 | 33 | 26.75 ± 3.50 | 1.86 ± 0.20 | 0.73 ± 0.08 |
| Attica | 2020 | 35 | 31.75 ± 1.50 | 2.42 ± 0.09 | 0.87 ± 0.02 |
| Evros | 2019 | 38 | 37.50 ± 0.58 | 1.81 ± 0.22 | 0.71 ± 0.08 |
| Evros | 2020 | 40 | 38.75 ± 0.96 | 2.62 ± 0.07 | 0.89 ± 0.01 |
| Thessaloniki | 2019 | 37 | 36.50 ± 0.58 | 1.80 ± 0.08 | 0.67 ± 0.03 |
| Thessaloniki | 2020 | 38 | 36.00 ± 0.82 | 1.99 ± 0.11 | 0.73 ± 0.04 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tegopoulos, K.; Tsirka, T.; Stekas, C.; Gerasimidi, E.; Skavdis, G.; Kolovos, P.; Grigoriou, M.E. Spatiotemporal Dynamics of Assyrtiko Grape Microbiota. Microorganisms 2024, 12, 577. https://doi.org/10.3390/microorganisms12030577
Tegopoulos K, Tsirka T, Stekas C, Gerasimidi E, Skavdis G, Kolovos P, Grigoriou ME. Spatiotemporal Dynamics of Assyrtiko Grape Microbiota. Microorganisms. 2024; 12(3):577. https://doi.org/10.3390/microorganisms12030577
Chicago/Turabian StyleTegopoulos, Konstantinos, Theodora Tsirka, Christos Stekas, Eleni Gerasimidi, George Skavdis, Petros Kolovos, and Maria E. Grigoriou. 2024. "Spatiotemporal Dynamics of Assyrtiko Grape Microbiota" Microorganisms 12, no. 3: 577. https://doi.org/10.3390/microorganisms12030577
APA StyleTegopoulos, K., Tsirka, T., Stekas, C., Gerasimidi, E., Skavdis, G., Kolovos, P., & Grigoriou, M. E. (2024). Spatiotemporal Dynamics of Assyrtiko Grape Microbiota. Microorganisms, 12(3), 577. https://doi.org/10.3390/microorganisms12030577






