Mining Biosynthetic Gene Clusters of Pseudomonas vancouverensis Utilizing Whole Genome Sequencing
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Isolation and Identification
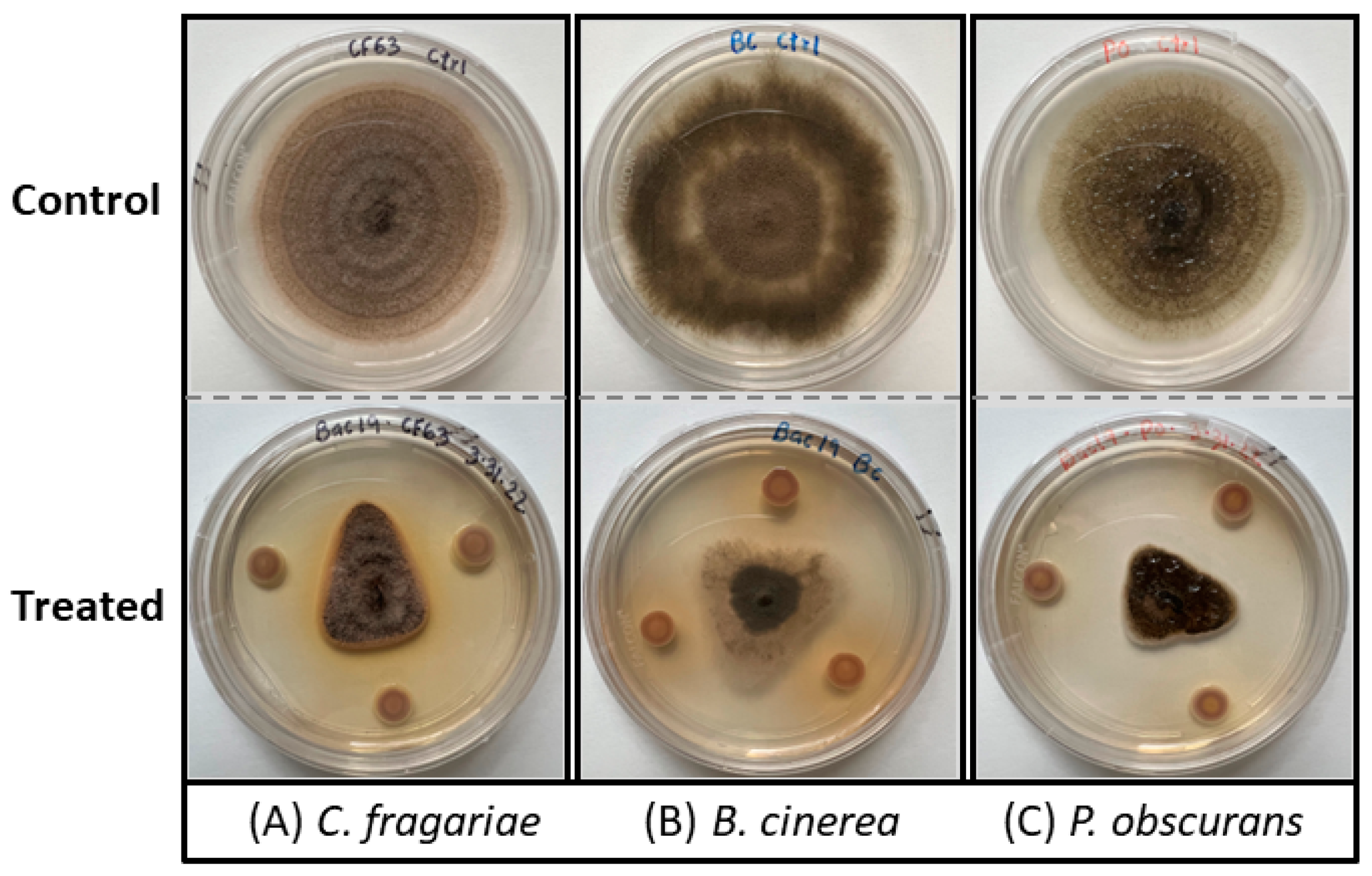
2.2. Antagonistic Activity Assay against Crop Fungal Pathogens
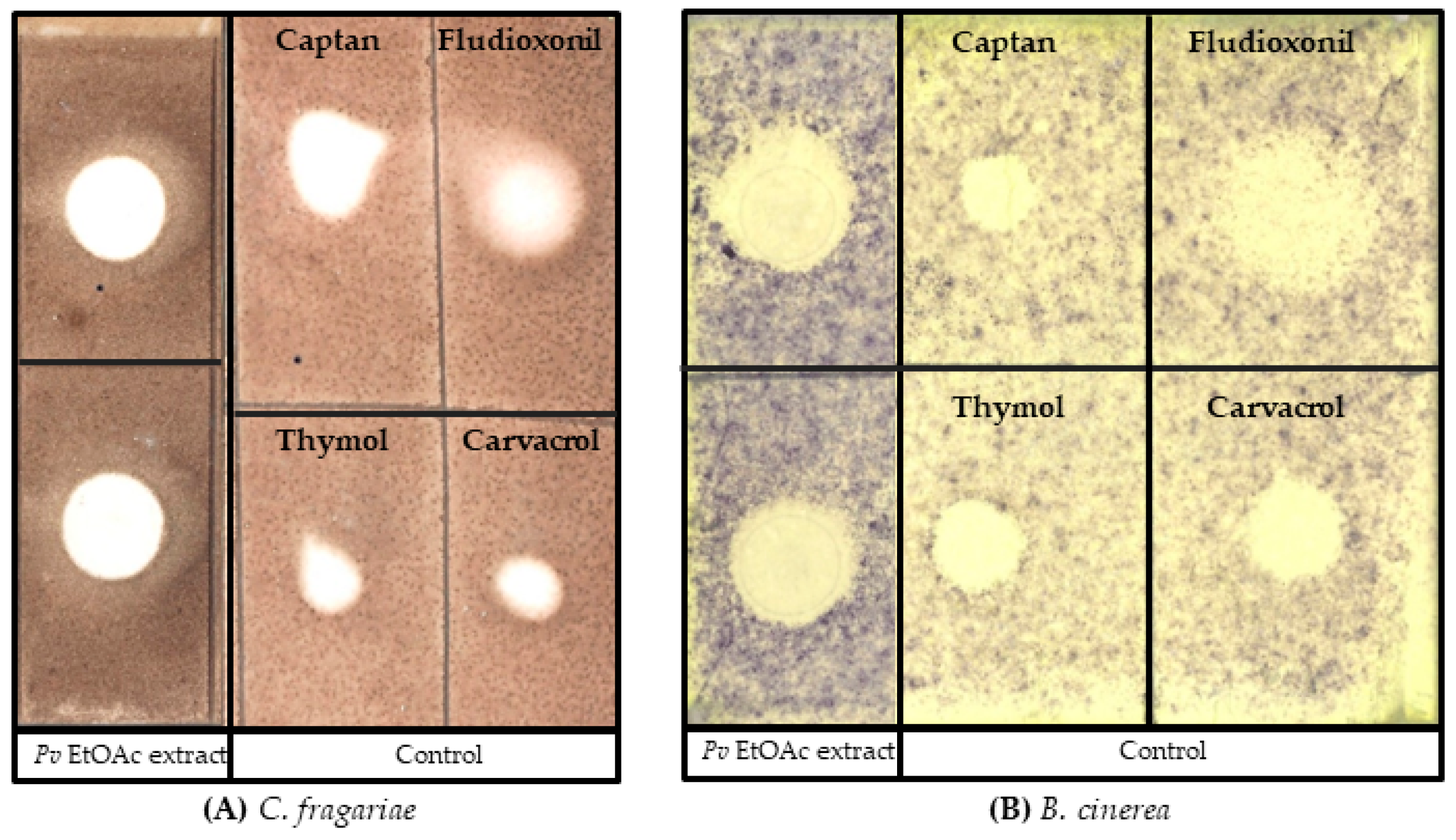
2.3. Bacterial Secondary Metabolite Crude Extraction
2.4. Antifungal Bioassay
2.5. Bacterial Culture, DNA Extraction, Library Construction, and Sequencing
2.6. Sequencing Read Quality Assessment and Curation
2.7. Genome Assembly, Annotation, and Phylogenetic Analysis
2.8. Phylogenetic Analysis
2.9. Secondary Metabolite Biosynthetic Gene Cluster (BGCs) Prediction
3. Results
3.1. Bacterial Identification and Antifungal Bioassay
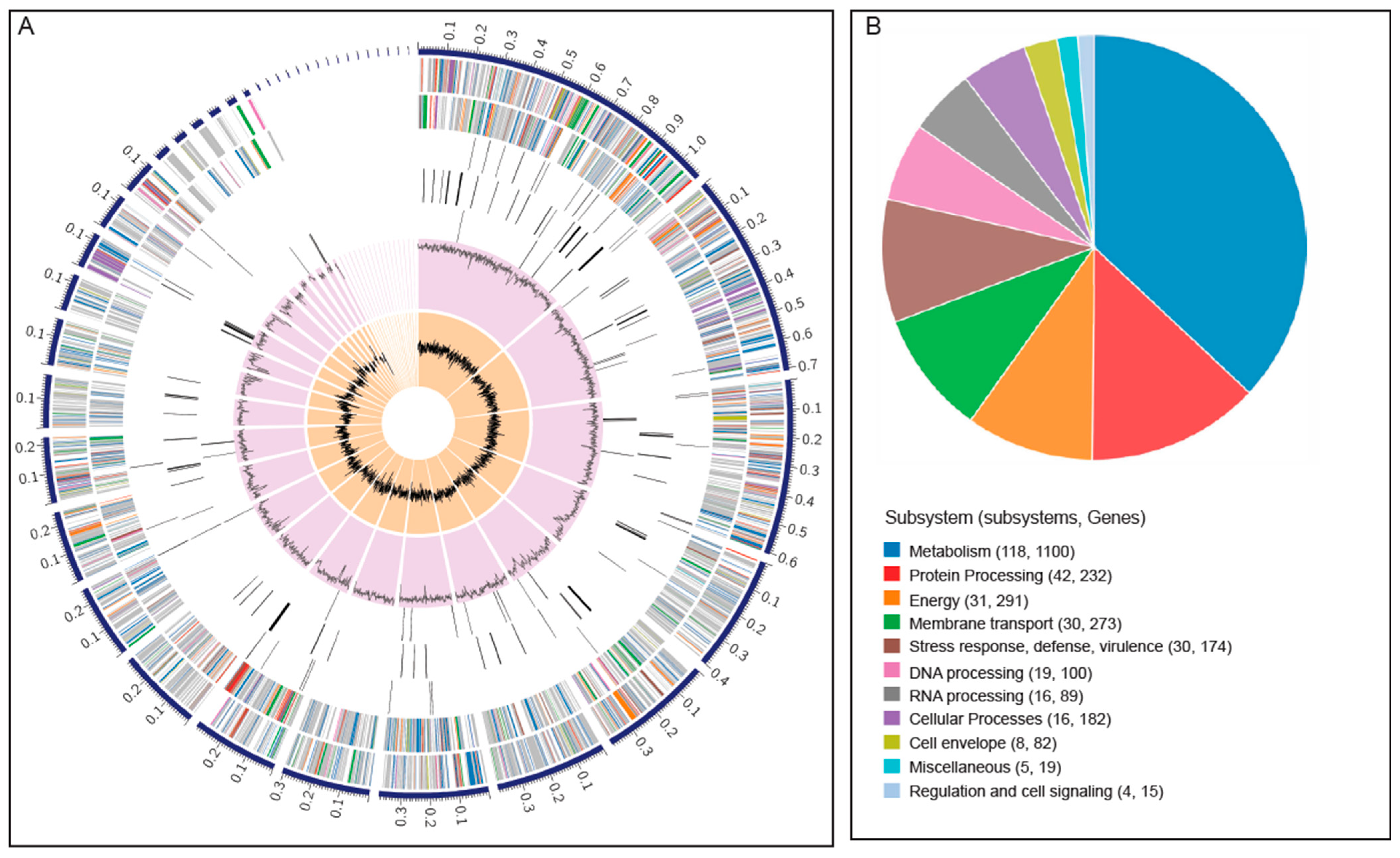
3.2. Genome Assembly, Sequencing Statistics, and Annotation
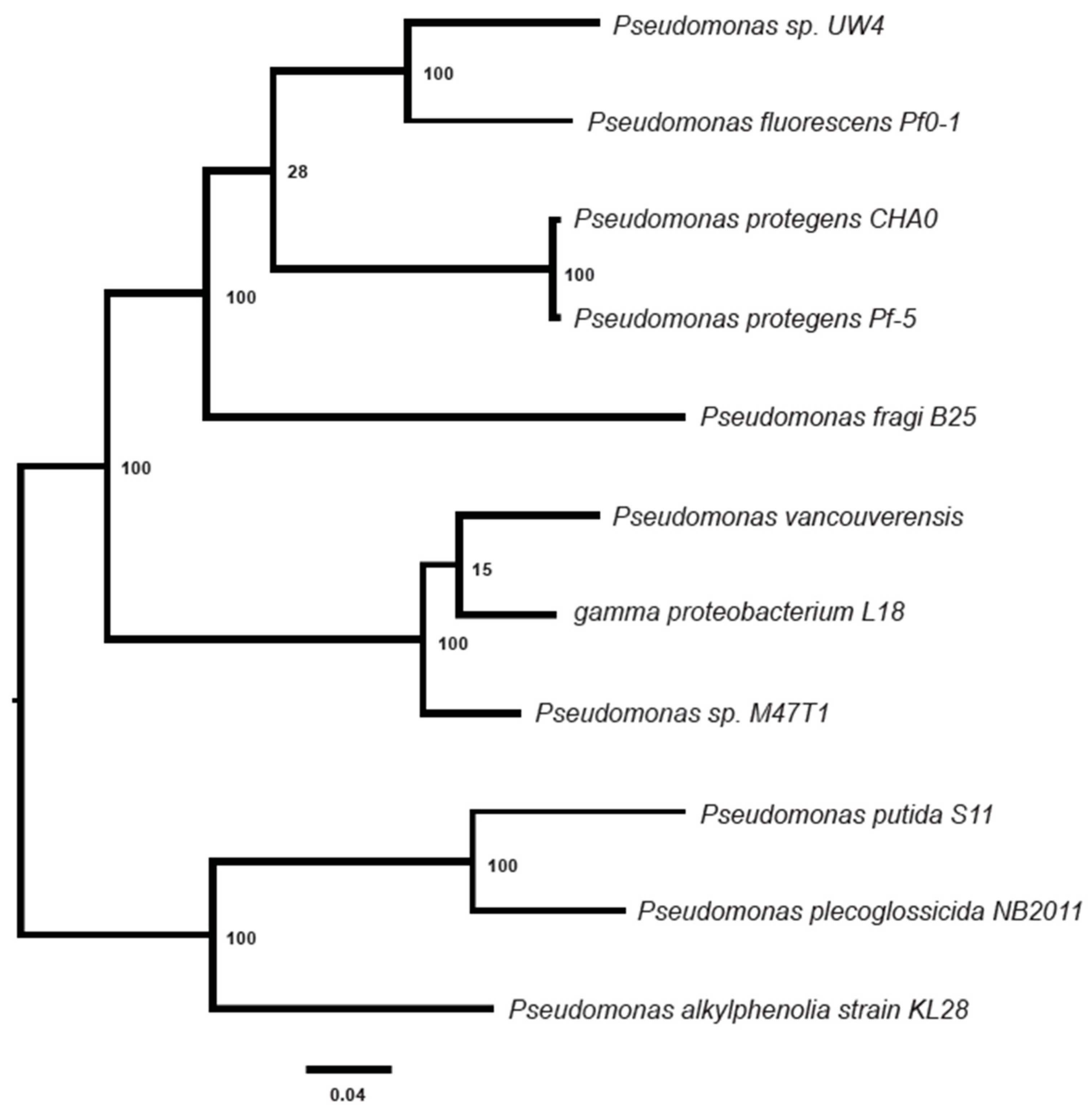
3.3. Phylogenetic Analysis
3.4. Prediction of BGCs in P. vancouverensis
3.4.1. Prediction of NP BGC with antiSMASH
3.4.2. Prediction of NP BGC with PRISM4
3.4.3. Prediction of NP BGC with BAGEL4
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Copping, L.G.; Duke, S.O. Natural products that have been used commercially as crop protection agents. Pest. Manag. Sci. 2007, 63, 524–554. [Google Scholar] [CrossRef]
- Dayan, F.E.; Cantrell, C.L.; Duke, S.O. Natural products in crop protection. Bioorganic Med. Chem. 2009, 17, 4022–4034. [Google Scholar] [CrossRef]
- Cantrell, C.L.; Dayan, F.E.; Duke, S.O. Natural products as sources for new pesticides. J. Nat. Prod. 2012, 75, 1231–1242. [Google Scholar] [CrossRef]
- Chuang, H.W.; Chang, P.Y.; Syu, Y.Y. Harpin Protein, an Elicitor of Disease Resistance, Acts as a Growth Promoter in Phalaenopsis Orchids. J. Plant Growth Regul. 2014, 33, 788–797. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, X.; Liu, W.; Liu, W.; Huang, J.; Liu, Q.; Sun, J.; Cai, X.; Miao, W. HpaXpm, a novel harpin of Xanthomonas phaseoli pv. manihotis, acts as an elicitor with high thermal stability, reduces disease, and promotes plant growth. BMC Microbiol. 2020, 20, 4. [Google Scholar] [CrossRef]
- Riseh, R.S.; Hassanisaadi, M.; Vatankhah, M.; Babaki, S.A.; Barka, E.A. Chitosan as a potential natural compound to manage plant diseases. Int. J. Biol. Macromol. 2022, 220, 998–1009. [Google Scholar] [CrossRef]
- Balba, H. Review of strobilurin fungicide chemicals. J. Environ. Sci. Health Part. B 2007, 42, 441–451. [Google Scholar] [CrossRef] [PubMed]
- Baltz, R.H. Genetic manipulation of secondary metabolite biosynthesis for improved production in Streptomyces and other actinomycetes. J. Ind. Microbiol. Biotechnol. 2016, 43, 343–370. [Google Scholar] [CrossRef] [PubMed]
- Katz, L.; Baltz, R.H. Natural product discovery: Past, present, and future. J. Ind. Microbiol. Biotechnol. 2016, 43, 155–176. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef] [PubMed]
- Demain, A.L. Importance of microbial natural products and the need to revitalize their discovery. Microbiol. Biotechnol. 2014, 41, 185–201. [Google Scholar] [CrossRef] [PubMed]
- Loiseleur, O. Natural Products in the Discovery of Agrochemicals. Chimia 2017, 1, 810–822. [Google Scholar] [CrossRef] [PubMed]
- Kardos, N.; Demain, A.L. Penicillin: The medicine with the greatest impact on therapeutic outcomes. Appl. Microbiol. Biotechnol. 2011, 92, 677–687. [Google Scholar] [CrossRef] [PubMed]
- Enderlin, G.; Morales, L.; Jacobs, R.F.; Cross, J.T. Streptomycin and Alternative Agents for the Treatment of Tularemia: Review of the Literature. Clin. Infect. Dis. 1994, 19, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, D.W.; Clough, J.M.; Godwin, J.R.; Hall, A.A.; Hamer, M.; Parr-Dobrzanski, B. The strobilurin fungicides. Pest. Manag. Sci. 2002, 58, 649–662. [Google Scholar] [CrossRef] [PubMed]
- Wohlleben, W.; Mast, Y.; Stegmann, E.; Ziemert, N. Antibiotic drug discovery. Microb. Biotechnol. 2016, 9, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, B.O.; Van Lanen, S.G.; Baltz, R.H. Microbial genome mining for accelerated natural products discovery: Is a renaissance in the making? J. Ind. Microbiol. Biotechnol. 2014, 41, 175–184. [Google Scholar] [CrossRef]
- Baltz, R.H. Natural product drug discovery in the genomic era: Realities, conjectures, misconceptions, and opportunities. J. Ind. Microbiol. Biotechnol. 2019, 46, 281–299. [Google Scholar] [CrossRef]
- Baltz, R.H. Genome mining for drug discovery: Cyclic lipopeptides related to daptomycin. J. Ind. Microbiol. Biotechnol. 2021, 48, kuab020. [Google Scholar] [CrossRef]
- Li, Z.; Zhu, D.; Shen, Y. Discovery of novel bioactive natural products driven by genome mining. Drug Discov. Ther. 2018, 12, 318–328. [Google Scholar] [CrossRef]
- Bauman, K.D.; Butler, K.S.; Moore, B.S.; Chekan, J.R. Genome mining methods to discover bioactive natural products. Nat. Prod. Rep. 2021, 38, 2100–2129. [Google Scholar] [CrossRef] [PubMed]
- Esmaeel, Q.; Pupin, M.; Kieu, N.P.; Chataigné, G.; Béchet, M.; Deravel, J.; Krier, F.; Höfte, M.; Jacques, P.; Leclère, V. Burkholderia genome mining for nonribosomal peptide synthetases reveals a great potential for novel siderophores and lipopeptides synthesis. Microbiologyopen 2016, 5, 512–526. [Google Scholar] [CrossRef] [PubMed]
- Morishita, Y.; Aoki, Y.; Ito, M.; Hagiwara, D.; Torimaru, K.; Morita, D.; Kuroda, T.; Fukano, H.; Hoshino, Y.; Suzuki, M.; et al. Genome Mining-Based Discovery of Fungal Macrolides Modified by glycosylphosphatidylinositol (GPI)-Ethanolamine Phosphate Transferase Homologues. Org. Lett. 2020, 22, 5876–5879. [Google Scholar] [CrossRef] [PubMed]
- Singh, G.; Dal Grande, F.; Schmitt, I. Genome mining as a biotechnological tool for the discovery of novel biosynthetic genes in lichens. Front. Fungal Biol. 2022, 3, 993171. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ziemert, N.; Alanjary, M.; Weber, T. The evolution of genome mining in microbes—A review. Nat. Prod. Rep. 2016, 33, 988–1005. [Google Scholar] [CrossRef] [PubMed]
- Bentley, S.D.; Chater, K.F.; Cerdeño-Tárraga, A.M.; Challis, G.L.; Thomson, N.R.; James, K.D.; Harris, D.E.; Quail, M.A.; Kieser, H.; Harper, D.; et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 2002, 417, 141–147. [Google Scholar] [CrossRef]
- Challis, G. Exploitation of the Streptomyces coelicolor A3(2) genome sequence for discovery of new natural products and biosynthetic pathways. J. Ind. Microbiol. Biotechnol. 2014, 41, 219–232. [Google Scholar] [CrossRef]
- Van Der Voort, M.; Meijer, H.; Schmidt, Y.; Watrous, J.; Dekkers, E.; Mendes, R.; Dorrestein, P.C.; Gross, H.; Raaijmakers, J.M. Genome mining and metabolic profiling of the rhizosphere bacterium Pseudomonas sp. SH-C52 for antimicrobial compounds. Front. Microbiol. 2015, 6, 693. [Google Scholar] [CrossRef]
- Kirchner, N.; Cano-Prieto, C.; Schulz-Fincke, A.-C.; Gütschow, M.; Ortlieb, N.; Moschny, J.; Niedermeyer, T.H.J.; Horak, J.; Lämmerhofer, M.; Van Der Voort, M. Discovery of Thanafactin A, a Linear, Proline-Containing Octalipopeptide from Pseudomonas sp. SH-C52, Motivated by Genome Mining. J. Nat. Prod. 2020, 84, 101–109. [Google Scholar] [CrossRef]
- Gu, Y.; Ma, Y.N.; Wang, J.; Xia, Z.; Wei, H.L. Genomic insights into a plant growth-promoting Pseudomonas koreensis strain with cyclic lipopeptide-mediated antifungal activity. MicrobiologyOpen 2020, 9, e1092. [Google Scholar] [CrossRef]
- Jahanshah, G.; Yan, Q.; Gerhardt, H.; Pataj, Z.; Lämmerhofer, M.; Pianet, I.; Josten, M.; Sahl, H.-G.; Silby, M.W.; Loper, J.E. Discovery of the cyclic lipopeptide gacamide A by genome mining and repair of the defective GacA regulator in Pseudomonas fluorescens Pf0-1. J. Nat. Prod. 2019, 82, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Chlebek, D.; Pinski, A.; Żur, J.; Michalska, J.; Hupert-Kocurek, K. Genome Mining and Evaluation of the Biocontrol Potential of Pseudomonas fluorescens BRZ63, a New Endophyte of Oilseed Rape (Brassica napus L.) against Fungal Pathogens. Int. J. Mol. Sci. 2020, 21, 8740. [Google Scholar] [CrossRef] [PubMed]
- Murata, K.; Suenaga, M.; Kai, K. Genome Mining Discovery of Protegenins A–D, Bacterial Polyynes Involved in the Antioomycete and Biocontrol Activities of Pseudomonas protegens. ACS Chem. Biol. 2022, 17, 3313–3320. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, K.; Someya, N.; Morohoshi, T. Genome-mining approaches for the evaluation of Pseudomonas protegens and related strains isolated from the rhizosphere in Japan. Physiol. Mol. Plant Pathol. 2023, 125, 101981. [Google Scholar] [CrossRef]
- Sun, C.; Yang, Z.; Zhang, C.; Liu, Z.; He, J.; Liu, Q.; Zhang, T.; Ju, J.; Ma, J. Genome Mining of Streptomyces atratus SCSIO ZH16: Discovery of Atratumycin and Identification of Its Biosynthetic Gene Cluster. Org. Lett. 2019, 21, 1453–1457. [Google Scholar] [CrossRef] [PubMed]
- Siupka, P.; Piński, A.; Babicka, D.; Piotrowska-Seget, Z. Genome Mining Revealed a High Biosynthetic Potential for Antifungal Streptomyces sp. S-2 Isolated from Black Soot. Int. J. Mol. Sci. 2020, 21, 2558. [Google Scholar] [CrossRef]
- Nguyen, C.T.; Dhakal, D.; Phan, V.T.T.; Nguyen, H.T.; Sohng, J.K. Recent advances in strategies for activation and discovery/characterization of cryptic biosynthetic gene clusters in Streptomyces. Microorganisms 2020, 8, 616. [Google Scholar] [CrossRef]
- Li, J.H.; Cho, W.; Hamchand, R.; Oh, J.; Crawford, J.M. A Conserved Nonribosomal Peptide Synthetase in Xenorhabdus bovienii Produces Citrulline-Functionalized Lipopeptides. J. Nat. Prod. 2021, 84, 2692–2699. [Google Scholar] [CrossRef]
- Alam, K.; Hao, J.; Zhong, L.; Fan, G.; Ouyang, Q.; Islam, M.M.; Islam, S.; Sun, H.; Zhang, Y.; Li, R.; et al. Complete genome sequencing and in silico genome mining reveal the promising metabolic potential in Streptomyces strain CS-7. Front. Microbiol. 2022, 13, 939919. [Google Scholar] [CrossRef]
- Alam, K.; Zhao, Y.; Lu, X.; Gong, K.; Zhong, L.; Hao, J.; Islam, M.M.; Islam, S.; Li, G.; Zhang, Y.; et al. Isolation, complete genome sequencing and in silico genome mining of Burkholderia for secondary metabolites. BMC Microbiol. 2022, 22, 323. [Google Scholar] [CrossRef] [PubMed]
- Aiman, S.; Shehroz, M.; Munir, M.; Gul, S.; Shah, M.; Khan, A. Species-wide genome mining of Pseudomonas putida for potential secondary metabolites and drug-like natural products characterization. J. Proteom. Bioinform. 2018, 11, 001–007. [Google Scholar] [CrossRef]
- Smith, B.J. Epidemiology and Pathology of Strawberry Anthracnose: A North American Perspective. HortScience Horts 2008, 43, 69–73. [Google Scholar] [CrossRef]
- Petrasch, S.; Knapp, S.J.; van Kan, J.A.L.; Blanco-Ulate, B. Grey mould of strawberry, a devastating disease caused by the ubiquitous necrotrophic fungal pathogen Botrytis cinerea. Mol. Plant Pathol. 2019, 20, 877–892. [Google Scholar] [CrossRef] [PubMed]
- Nita, M.; Ellis, M.A.; Madden, L.V. Effects of Temperature, Wetness Duration, and Leaflet Age on Infection of Strawberry Foliage by Phomopsis obscurans. Plant Dis. 2003, 87, 579–584. [Google Scholar] [CrossRef] [PubMed]
- Kandel, S.L.; Firrincieli, A.; Joubert, P.M.; Okubara, P.A.; Leston, N.D.; McGeorge, K.M.; Mugnozza, G.S.; Harfouche, A.; Kim, S.-H.; Doty, S.L. An In vitro Study of Bio-Control and Plant Growth Promotion Potential of Salicaceae Endophytes. Front. Microbiol. 2017, 8, 386. [Google Scholar] [CrossRef] [PubMed]
- Meepagala, K.M.; Sturtz, G.; Wedge, D.E. Antifungal constituents of the essential oil fraction of Artemisia dracunculus L. var. dracunculus. J. Agric. Food Chem. 2002, 50, 6989–6992. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 20 September 2023).
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Grigoriev, I.V.; Nordberg, H.; Shabalov, I.; Aerts, A.; Cantor, M.; Goodstein, D.; Kuo, A.; Minovitsky, S.; Nikitin, R.; Ohm, R.A.; et al. The genome portal of the department of energy joint genome institute. Nucleic Acids Res. 2012, 40, D26–D32. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Manni, M.; Berkeley, M.R.; Seppey, M.; Simão, F.A.; Zdobnov, E.M. BUSCO update: Novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol. Biol. Evol. 2021, 38, 4647–4654. [Google Scholar] [CrossRef]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef]
- Kent, W.J. BLAT—The BLAST-like alignment tool. Genome Res. 2002, 12, 656–664. [Google Scholar] [CrossRef]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef]
- Mao, C.; Abraham, D.; Wattam, A.R.; Wilson, M.J.; Shukla, M.; Yoo, H.S.; Sobral, B.W. Curation, integration and visualization of bacterial virulence factors in PATRIC. Bioinformatics 2015, 31, 252–258. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Chen, X.; Ji, Z.L.; Chen, Y.Z. Therapeutic Target Database. Nucleic Acids Res. 2002, 30, 412–415. [Google Scholar] [CrossRef]
- Saier Jr, M.H.; Reddy, V.S.; Moreno-Hagelsieb, G.; Hendargo, K.J.; Zhang, Y.; Iddamsetty, V. The transporter classification database (TCDB): 2021 update. Nucleic Acids Res. 2021, 49, D461–D467. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yang, J.; Yu, J.; Yao, Z.; Sun, L.; Shen, Y.; Jin, Q. VFDB: A reference database for bacterial virulence factors. Nucleic Acids Res. 2005, 33, D325–D328. [Google Scholar] [CrossRef] [PubMed]
- Sayers, S.; Li, L.; Ong, E.; Deng, S.; Fu, G.; Lin, Y.; Yang, B.; Zhang, S.; Fa, Z.; Zhao, B.; et al. Victors: A web-based knowledge base of virulence factors in human and animal pathogens. Nucleic Acids Res. 2019, 47, D693–D700. [Google Scholar] [CrossRef] [PubMed]
- Medema, M.H.; Blin, K.; Cimermancic, P.; de Jager, V.; Zakrzewski, P.; Fischbach, M.A.; Weber, T.; Takano, E.; Breitling, R. antiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011, 39, W339–W346. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.; et al. antiSMASH 7.0: New and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef]
- Skinnider, M.A.; Johnston, C.W.; Gunabalasingam, M.; Merwin, N.J.; Kieliszek, A.M.; MacLellan, R.J.; Li, H.; Ranieri, M.R.M.; Webster, A.L.H.; Cao, M.P.T.; et al. Comprehensive prediction of secondary metabolite structure and biological activity from microbial genome sequences. Nat. Commun. 2020, 11, 6058. [Google Scholar] [CrossRef]
- Van Heel, A.J.; De Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef]
- Yan, Y.; Liu, Q.; Jacobsen, S.E.; Tang, Y. The impact and prospect of natural product discovery in agriculture: New technologies to explore the diversity of secondary metabolites in plants and microorganisms for applications in agriculture. EMBO Rep. 2018, 19, e46824. [Google Scholar] [CrossRef]
- Elhamouly, N.A.; Hewedy, O.A.; Zaitoon, A.; Miraples, A.; Elshorbagy, O.T.; Hussien, S.; El-Tahan, A.; Peng, D. The hidden power of secondary metabolites in plant-fungi interactions and sustainable phytoremediation. Front. Plant Sci. 2022, 13, 1044896. [Google Scholar] [CrossRef] [PubMed]
- Vicente, M.F.; Basilio, A.; Cabello, A.; Peláez, F. Microbial natural products as a source of antifungals. Clin. Microbiol. Infect. 2003, 9, 15–32. [Google Scholar] [CrossRef] [PubMed]
- Vidaver, A.K. Uses of antimicrobials in plant agriculture. Clin. Infect. Dis. 2002, 34 (Suppl. 3), S107–S110. [Google Scholar] [CrossRef] [PubMed]
- Damalas, C.A.; Koutroubas, S.D. Current Status and Recent Developments in Biopesticide Use. Agriculture 2018, 8, 13. [Google Scholar] [CrossRef]
- Adnan, M.; Alshammari, E.; Patel, M.; Amir Ashraf, S.; Khan, S.; Hadi, S. Significance and potential of marine microbial natural bioactive compounds against biofilms/biofouling: Necessity for green chemistry. PeerJ 2018, 6, e5049. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.M.; Wong, F.T.; Wang, Y.; Luo, S.; Lim, Y.H.; Heng, E.; Yeo, W.L.; Cobb, R.E.; Enghiad, B.; Ang, E.L.; et al. CRISPR-Cas9 strategy for activation of silent Streptomyces biosynthetic gene clusters. Nat. Chem. Biol. 2017, 13, 607–609. [Google Scholar] [CrossRef] [PubMed]
- Mignard, S.; Flandrois, J.P. 16S rRNA sequencing in routine bacterial identification: A 30-month experiment. J. Microbiol. Methods 2006, 67, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Mohn, W.W.; Wilson, A.E.; Bicho, P.; Moore, E.R.B. Physiological and phylogenetic diversity of bacteria growing on resin acids. Syst. Appl. Microbiol. 1999, 22, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Inmaculada, S.; Rebecca, P.; Tino, K.; Jane, H. Pseudomonas chemotaxis. FEMS Microbiol. Rev. 2015, 39, 17–46. [Google Scholar] [CrossRef]
- Mishra, P.K.; Mishra, S.; Selvakumar, G.; Bisht, S.C.; Bisht, J.K.; Kundu, S.; Gupta, H.S. Characterisation of a psychrotolerant plant growth promoting Pseudomonas sp. strain PGERs17 (MTCC 9000) isolated from Northwestern Indian Himalayas. Ann. Microbiol. 2008, 58, 561–568. [Google Scholar] [CrossRef]
- Mikiciński, A.; Puławska, J.; Molzhigitova, A.; Sobiczewski, P. Bacterial species recognized for the first time for its biocontrol activity against fire blight (Erwinia amylovora). Eur. J. Plant Pathol. 2020, 156, 257–272. [Google Scholar] [CrossRef]
- Castaldi, S.; Masi, M.; Sautua, F.; Cimmino, A.; Isticato, R.; Carmona, M.; Tuzi, A.; Evidente, A. Pseudomonas fluorescens Showing Antifungal Activity against Macrophomina phaseolina, a Severe Pathogenic Fungus of Soybean, Produces Phenazine as the Main Active Metabolite. Biomolecules 2021, 11, 1728. [Google Scholar] [CrossRef]
- Saati-Santamaría, Z.; Selem-Mojica, N.; Peral-Aranega, E.; Rivas, R.; García-Fraile, P. Unveiling the genomic potential of Pseudomonas type strains for discovering new natural products. Microb. Genom. 2022, 8, 000758. [Google Scholar] [CrossRef]
- Kirienko, N.V.; Kirienko, D.R.; Larkins-Ford, J.; Wählby, C.; Ruvkun, G.; Ausubel, F.M. Pseudomonas aeruginosa disrupts Caenorhabditis elegans iron homeostasis, causing a hypoxic response and death. Cell Host Microbe 2013, 13, 406–416. [Google Scholar] [CrossRef]
- Kirienko, N.V.; Ausubel, F.M.; Ruvkun, G. Mitophagy confers resistance to siderophore-mediated killing by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2015, 112, 1821–1826. [Google Scholar] [CrossRef]
- Batista, B.B.; Santos, R.E.R.D.S.; Ricci-Azevedo, R.; da Silva Neto, J.F. Production and uptake of distinct endogenous catecholate-type siderophores are required for iron acquisition and virulence in Chromobacterium violaceum. Infect. Immun. 2019, 87, e00577-19. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, K.; Yamazaki, T.; Takeuchi, Y.; Oishi, T. Studies on T-2636 antibiotics. IV. In vitro and in vivo antibacterial activity of T-2636 antibiotics. J. Antibiot. 1971, 24, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Ootsu, K.; Matsumoto, T. Effects of Lankacidin group (T2636) antibiotics on the tumor growth and immune response against sheep erythrocytes in mice. GANN Jpn. J. Cancer Res. 1973, 64, 481–492. [Google Scholar]
- Michelsen, C.F.; Stougaard, P. Hydrogen cyanide synthesis and antifungal activity of the biocontrol strain Pseudomonas fluorescens In5 from Greenland is highly dependent on growth medium. Can. J. Microbiol. 2012, 58, 381–390. [Google Scholar] [CrossRef]
- Anand, A.; Chinchilla, D.; Tan, C.; Mène-Saffrané, L.; L′Haridon, F.; Weisskopf, L. Contribution of hydrogen cyanide to the antagonistic activity of Pseudomonas strains against Phytophthora infestans. Microorganisms 2020, 8, 1144. [Google Scholar] [CrossRef]
- Fidor, A.; Konkel, R.; Mazur-Marzec, H. Bioactive Peptides Produced by Cyanobacteria of the Genus Nostoc: A Review. Mar. Drugs 2019, 17, 561. [Google Scholar] [CrossRef]
- Bhat, J.M.; Narayanan, M.S. Antifungal activity (invitro) of certain polyene macrolide antibiotics against various plant pathogens. Hindustan Antibiot. Bull. 1996, 38, 32–36. [Google Scholar]
- Dalhoff, A. Does the use of antifungal agents in agriculture and food foster polyene resistance development? A reason for concern. J. Global Antimicrob. Resist. 2018, 13, 40–48. [Google Scholar] [CrossRef]
- Park, H.S.; Nah, H.J.; Kang, S.H.; Choi, S.S.; Kim, E.S. Screening and Isolation of a Novel Polyene-Producing Streptomyces Strain Inhibiting Phytopathogenic Fungi in the Soil Environment. Front. Bioeng. Biotechnol. 2021, 9, 692340. [Google Scholar] [CrossRef]
- Subramanian, S.; Smith, D.L. Bacteriocins from the rhizosphere microbiome—From an agriculture perspective. Front. Plant Sci. 2015, 6, 909. [Google Scholar] [CrossRef]
- Hudson, G.A.; Mitchell, D.A. RiPP antibiotics: Biosynthesis and engineering potential. Curr. Opin. Microbiol. 2018, 45, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Cesa-Luna, C.; Alatorre-Cruz, J.M.; Carreño-López, R.; Quintero-Hernández, V.; Baez, A. Emerging Applications of Bacteriocins as Antimicrobials, Anticancer Drugs, and Modulators of The Gastrointestinal Microbiota. Pol. J. Microbiol. 2021, 70, 143–159. [Google Scholar] [CrossRef] [PubMed]
- Rooney, W.M.; Grinter, R.W.; Correia, A.; Parkhill, J.; Walker, D.C.; Milner, J.J. Engineering bacteriocin-mediated resistance against the plant pathogen Pseudomonas syringae. Plant Biotechnol. J. 2020, 18, 1296–1306. [Google Scholar] [CrossRef] [PubMed]

| Features a | P. vancouverensis |
|---|---|
| Sequencing statistics | |
| Number of raw sequencing reads | 17,268,958 |
| Average length of raw reads (bp) | 150 |
| Average coverage with raw reads (x) | 390 |
| Quality sequencing reads | 14,402,604 |
| Reads retained after normalization | 5,989,222 |
| Assembly statistics | |
| Number of contigs | 41 |
| Total length (bp) | 6,646,214 |
| Largest contig (bp) | 1,079,378 |
| N50 (bp) | 384,424 |
| N90 (bp) | 127,873 |
| L50 | 6 |
| L90 | 16 |
| GC (%) | 63.27 |
| Busco completeness (%) | 99.3 |
| Genome features | |
| CDS | 6052 |
| tRNA | 67 |
| rRNA | 2 |
| Partial CDS | 0 |
| Misc. RNA | 0 |
| Repeat Regions | 0 |
| Protein features | |
| Protein-encoding genes (PEGs) | 6052 |
| PEGs with functional assignment | 4678 |
| Hypothetical proteins | 1374 |
| Proteins with GO assignments | 1148 |
| Proteins with pathway assignments | 1018 |
| Specialty Genes | Database | Number of Genes |
|---|---|---|
| Antibiotic Resistance | CARD | 3 |
| PATRIC | 85 | |
| Drug Target | DrugBank | 24 |
| TTD | 6 | |
| Transporter | TCDB | 59 |
| Virulence Factor | VFDB | 27 |
| Victors | 18 |
| Region | Gene Type a | Span (nt) | Most Similar BGCs | Type | Similarity b | |
|---|---|---|---|---|---|---|
| From | To | |||||
| 1.1 | NAGGN | 665,212 | 679,985 | - | - | - |
| 2.1 | NRPS-like | 242,243 | 285,626 | Pyoverdine | NRP | 7% |
| 2.2 | redox-cofactor | 612,182 | 634,341 | Lankacidin C | NRP + Polyketide | 13% |
| 4.1 | RiPP-like | 63,468 | 75672 | - | ||
| 5.1 | NRPS | 179,999 | 224,264 | Viobactin | NRP | 15% |
| 6.1 | NRPS | 190,093 | 257,271 | MA026 | NRP | 10% |
| 7.1 | Aryl polyene | 36,509 | 80,104 | APE Vf | Other | 40% |
| 8.1 | Hydrogen-cyanide | 225,474 | 238,306 | - | - | - |
| 10.1 | NRPS, hserlactone | 119,551 | 178,652 | Frederiksenibactin | NRP | 15% |
| 11.1 | hserlactone | 51,665 | 72,261 | - | - | - |
| 11.2 | CDPS | 77,532 | 98,269 | - | - | - |
| 12.1 | NRPS | 185,164 | 229,126 | - | - | - |
| 14.1 | NRP-metallophore, NRPS | 143,663 | 182,106 | Pyoverdine SMX-1 | NRP | 35% |
| 22.1 | NRP-metallophore, NRPS | 1 | 38,911 | Pf-5 pyoverdine | NRP | 9% |
| 26.1 | NRPS | 1 | 4959 | Nostopeptolide A2 | Polyketide + NRP: Cyclic depsipeptide | 37% |
| 31.1 | NRPS | 1 | 2237 | - | - | - |
| 34.1 | NRPS-like | 1 | 1903 | Bovienimide A | NRP:Lipopeptide | 100% |
| Gene Cluster Type | Contigs | Clusters | Predicted Structure |
|---|---|---|---|
| PK | 1 | 1 | No |
| NRP | 2 | 2 | Yes |
| NRP | 3 | 3 | Yes |
| NRP | 4 | 4 | Yes |
| Acyl Homoserine Lactone | 5 | No | |
| Acyl Homoserine Lactone | 5 | 6 | No |
| Cyclopeptide (XYP family) | 7 | Yes | |
| NRP | 6 | 8 | Yes |
| NRP | 7 | 9 | Yes |
| NRP | 8 | 10 | Yes |
| NRP | 9 | 11 | Yes |
| NRP | 10 | 12 | Yes |
| Areas of Interest | Annotation |
|---|---|
| Orf00003 | Large-conductance mechanosensitive channel OS = Pelobacter propionicus |
| Orf00011 | HTH-type transcriptional regulator for conjugative element SXT OS = Vibrio cholerae |
| Orf00019 | Uncharacterized protein HI_1412 OS = Haemophilus influenzae |
| Orf00026 | P21 prophage-derived protein NinB OS = Escherichia coli O6:H1 |
| Orf00036 | Putidacin_L1 family lectin-like bacteriocin * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tamang, P.; Upadhaya, A.; Paudel, P.; Meepagala, K.; Cantrell, C.L. Mining Biosynthetic Gene Clusters of Pseudomonas vancouverensis Utilizing Whole Genome Sequencing. Microorganisms 2024, 12, 548. https://doi.org/10.3390/microorganisms12030548
Tamang P, Upadhaya A, Paudel P, Meepagala K, Cantrell CL. Mining Biosynthetic Gene Clusters of Pseudomonas vancouverensis Utilizing Whole Genome Sequencing. Microorganisms. 2024; 12(3):548. https://doi.org/10.3390/microorganisms12030548
Chicago/Turabian StyleTamang, Prabin, Arjun Upadhaya, Pradeep Paudel, Kumudini Meepagala, and Charles L. Cantrell. 2024. "Mining Biosynthetic Gene Clusters of Pseudomonas vancouverensis Utilizing Whole Genome Sequencing" Microorganisms 12, no. 3: 548. https://doi.org/10.3390/microorganisms12030548
APA StyleTamang, P., Upadhaya, A., Paudel, P., Meepagala, K., & Cantrell, C. L. (2024). Mining Biosynthetic Gene Clusters of Pseudomonas vancouverensis Utilizing Whole Genome Sequencing. Microorganisms, 12(3), 548. https://doi.org/10.3390/microorganisms12030548







