Investigation of Microbial Community of Korean Soy Sauce (Ganjang) Using Shotgun Metagenomic Sequencing and Its Relationship with Sensory Characteristics
Abstract
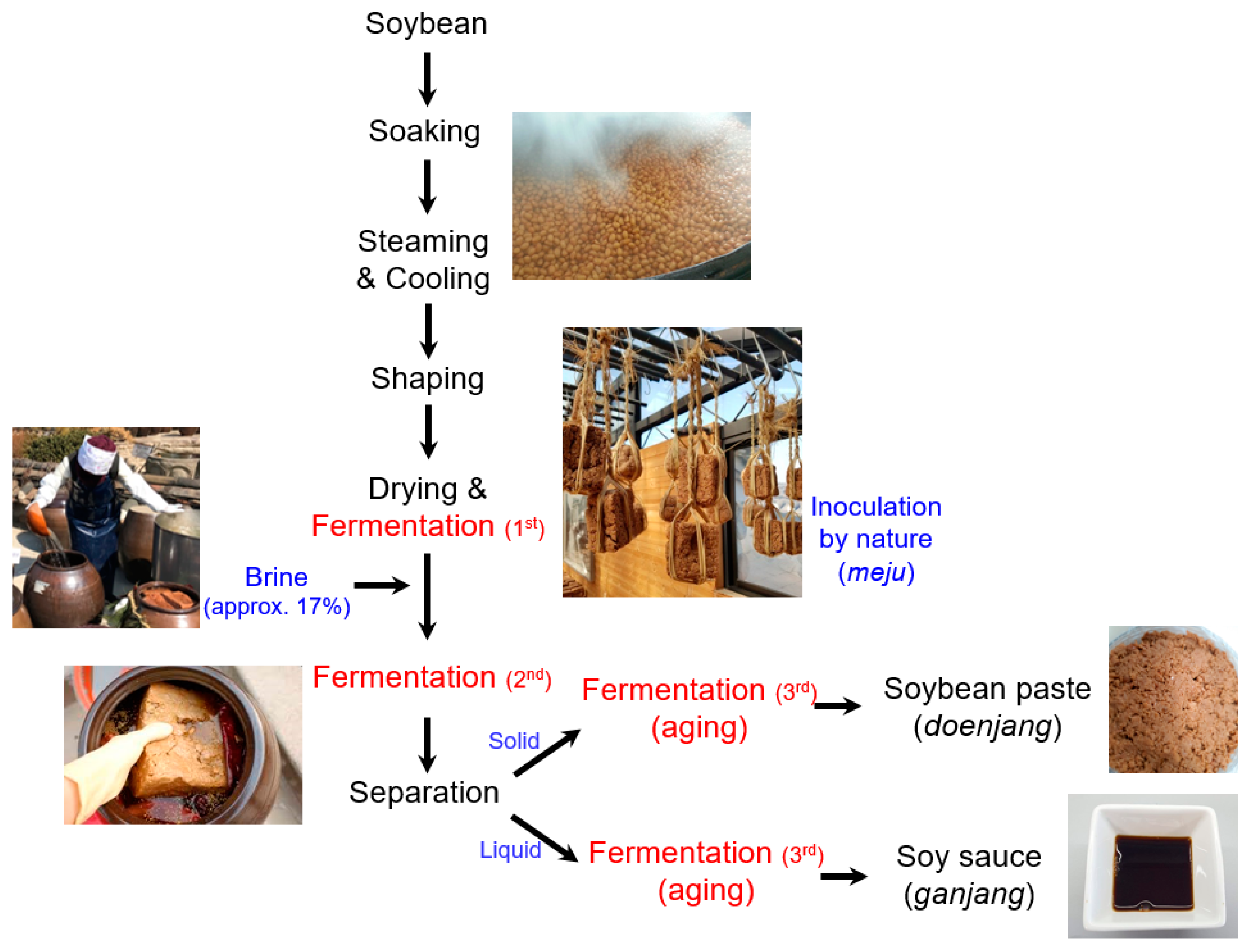
:1. Introduction
2. Materials and Methods
2.1. Soy Sauce Samples
2.2. Descriptive Sensory Analysis
2.3. Extraction of Metagenomic DNA
2.4. Metagenomic DNA Sequencing
2.5. De Novo Assembly of the Sequencing Results
2.6. Analysis of Microbial Communities and Annotation
2.7. Statistical Analysis
3. Results
3.1. Descriptive Sensory Analysis of Ganjang Samples
3.2. Microbial Communities of Ganjang Samples
3.2.1. Overview and Bacterial Community
3.2.2. Fungal Community
3.2.3. Viral Community
3.3. Correlation Between Microbial Communities and Sensory Characteristics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Lioe, H.N.; Selamat, J.; Yasuda, M. Soy sauce and its umami taste: A link from the past to current situation. J. Food Sci. 2010, 75, R71–R76. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-H.; Kim, S.-H.; Kwon, S.-W.; Lee, J.-K.; Hong, S.-B. Fungal diversity of rice straw for meju fermentation. J. Microbiol. Biotechnol. 2013, 23, 1654–1663. [Google Scholar] [CrossRef] [PubMed]
- Park, K.-Y.; Hwang, K.-M.; Jung, K.-O.; Lee, K.-B. Studies on the standardization of doenjang (Korean soybean paste): 1. Standardization of manufacturing method of doenjang by literatures. J. Korean Soc. Food Sci. Nutr. 2002, 31, 343–350. [Google Scholar]
- Sulaiman, J.; Gan, H.M.; Yin, W.-F.; Chan, K.-G. Microbial succession and the functional potential during the fermentation of Chinese soy sauce brine. Front. Microbiol. 2014, 5, 556. [Google Scholar] [CrossRef]
- Dugat-Bony, E.; Garnier, L.; Denonfoux, J.; Ferreira, S.; Sarthou, A.-S.; Bonnarme, P.; Irlinger, F. Highlighting the microbial diversity of 12 French cheese varieties. Int. J. Food Microbiol. 2016, 238, 265–273. [Google Scholar] [CrossRef]
- Nalbantoglu, U.; Cakar, A.; Dogan, H.; Abaci, N.; Ustek, D.; Sayood, K.; Can, H. Metagenomic analysis of the microbial community in kefir grains. Food Microbiol. 2014, 41, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Zhang, T.; Zhang, Q.; Luo, J.; Cai, C.; Mao, J. Microbial diversity and chemical analysis of the starters used in traditional Chinese sweet rice wine. Food Microbiol. 2018, 73, 319–326. [Google Scholar] [CrossRef]
- Nam, Y.-D.; Lee, S.-Y.; Lim, S.-I. Microbial community analysis of Korean soybean pastes by next-generation sequencing. Int. J. Food Microbiol. 2012, 155, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.Y.; Lee, S.H.; Jeon, C.O. Microbial community dynamics during fermentation of doenjang-meju, traditional Korean fermented soybean. Int. J. Food Microbiol. 2014, 185, 112–120. [Google Scholar] [CrossRef]
- Kim, H.; Hong, J.; Song, C.; Shin, H.; Kim, K. Sensory characteristics and consumer acceptability of fermented soybean paste (Doenjang). J. Food Sci. 2010, 75, S375–S383. [Google Scholar] [CrossRef]
- Lee, S.-J.; Ahn, B. Comparison of volatile components in fermented soybean pastes using simultaneous distillation and extraction (SDE) with sensory characterisation. Food Chem. 2009, 114, 600–609. [Google Scholar] [CrossRef]
- Kang, H.J.; Yang, H.J.; Kim, M.J.; Han, E.-S.; Kim, H.-J.; Kwon, D.Y. Metabolomic analysis of meju during fermentation by ultra performance liquid chromatography-quadrupole-time of flight mass spectrometry (UPLC-Q-TOF MS). Food Chem. 2011, 127, 1056–1064. [Google Scholar] [CrossRef]
- Lee, J.-E.; Yun, J.-H.; Lee, E.; Hong, S.P. Untargeted Metabolomics reveals Doenjang metabolites affected by manufacturing process and microorganisms. Food Res. Int. 2022, 157, 111422. [Google Scholar] [CrossRef] [PubMed]
- Han, D.M.; Chun, B.H.; Kim, H.M.; Jeon, C.O. Characterization and correlation of microbial communities and metabolite and volatile compounds in doenjang fermentation. Food Res. Int. 2021, 148, 110645. [Google Scholar] [CrossRef]
- Kim, M.J.; Kwak, H.S.; Jung, H.Y.; Kim, S.S. Microbial communities related to sensory attributes in Korean fermented soy bean paste (doenjang). Food Res. Int. 2016, 89, 724–732. [Google Scholar] [CrossRef] [PubMed]
- Chun, B.H.; Han, D.M.; Kim, H.M.; Park, D.; Jeong, D.M.; Kang, H.A.; Jeon, C.O. Metabolic features of ganjang (a Korean traditional soy sauce) fermentation revealed by genome-centered metatranscriptomics. Msystems 2021, 6, e0044121. [Google Scholar] [CrossRef]
- Han, D.M.; Chun, B.H.; Feng, T.; Kim, H.M.; Jeon, C.O. Dynamics of microbial communities and metabolites in ganjang, a traditional Korean fermented soy sauce, during fermentation. Food Microbiol. 2020, 92, 103591. [Google Scholar] [CrossRef]
- Ha, G.; Jeong, H.J.; Noh, Y.; Kim, J.; Jeong, S.-J.; Jeong, D.-Y.; Yang, H.-J. Comparative micro-biome analysis of and microbial biomarker discovery in two different fermented soy products, doenjang and ganjang, using next-generartion sequencing. J. Life Sci. 2022, 32, 803–811. [Google Scholar]
- Janda, J.M.; Abbott, S.L. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: Pluses, perils, and pitfalls. J. Clin. Microbiol. 2007, 45, 2761–2764. [Google Scholar] [CrossRef]
- Lee, J.-M.; Heo, S.; Kim, Y.-S.; Lee, J.-H.; Jeong, D.-W. Culture-dependent and-independent investigations of bacterial migration into doenjang from its components meju and solar salt. PLoS ONE 2020, 15, e0239971. [Google Scholar] [CrossRef]
- Liu, Y.-X.; Qin, Y.; Chen, T.; Lu, M.; Qian, X.; Guo, X.; Bai, Y. A practical guide to amplicon and metagenomic analysis of microbiome data. Protein Cell 2021, 12, 315–330. [Google Scholar] [CrossRef] [PubMed]
- Meilgaard, M.C.; Carr, B.T.; Civille, G.V. Sensory Evaluation Techniques; CRC Press: Boca Raton, FL, USA, 1999. [Google Scholar]
- Cox, M.P.; Peterson, D.A.; Biggs, P.J. SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinform. 2010, 11, 485. [Google Scholar] [CrossRef] [PubMed]
- Luo, R.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; He, G.; Chen, Y.; Pan, Q.; Liu, Y. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. Gigascience 2012, 1, 18. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Lee, K.-H.; Kim, N.-D.; Yoo, J.-Y. Survey on the manufacturing process of traditional meju for and of kanjang (Korean soy sauce). J. Korean Soc. Food Sci. Nutr. 1997, 26, 390–396. [Google Scholar]
- Sousa, M.; Ardö, Y.; McSweeney, P. Advances in the study of proteolysis during cheese ripening. Int. Dairy J. 2001, 11, 327–345. [Google Scholar] [CrossRef]
- Song, Y.R.; Jeong, D.Y.; Baik, S.H. Monitoring of yeast communities and volatile flavor changes during traditional Korean soy sauce fermentation. J. Food Sci. 2015, 80, M2005–M2014. [Google Scholar] [CrossRef]
- Liu, H.; Chen, Z.; Ma, M.; Zhang, Y.; Li, D.; Lu, J.; Chen, X. Metagenomic analysis of the relationship between microorganisms and flavor development during soy sauce fermentation. Food Biosci. 2023, 56, 103193. [Google Scholar] [CrossRef]
- Abdi, H.; Williams, L.J. Principal component analysis. Wiley Interdiscip. Rev. Comput. Stat. 2010, 2, 433–459. [Google Scholar] [CrossRef]
- Devanthi, P.V.P.; Linforth, R.; Onyeaka, H.; Gkatzionis, K. Effects of co-inoculation and sequential inoculation of Tetragenococcus halophilus and Zygosaccharomyces rouxii on soy sauce fermentation. Food Chem. 2018, 240, 1–8. [Google Scholar] [CrossRef]
- Yan, Y.; Zhang, M.; Zhang, Y.; Zhang, X.; Zhang, X.; Zhao, X.; Xu, H.; Huang, Y. Correlation between bacterial diversity and flavor substances in Longgang soy sauce. Biosci. Biotechnol. Biochem. 2023, 87, 541–554. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Zhang, X.; Wei, L.; Wu, G.; Kumar, A.; Mao, T.; Liu, Z. A cold-adapted, solvent and salt tolerant esterase from marine bacterium Psychrobacter pacificensis. Int. J. Biol. Macromol. 2015, 81, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Du, F.; Zhang, X.; Gu, H.; Song, J.; Gao, X. Dynamic changes in the bacterial community during the fermentation of traditional Chinese fish sauce (TCFS) and their correlation with TCFS quality. Microorganisms 2019, 7, 371. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.Y.; Chun, B.H.; Jeon, C.O. Chromohalobacter is a causing agent for the production of organic acids and putrescine during fermentation of ganjang, a Korean traditional soy sauce. J. Food Sci. 2015, 80, M2853–M2859. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, L.; Xu, Y. Effects of Tetragenococcus halophilus and Candida versatilis on the production of aroma-active and umami-taste compounds during soy sauce fermentation. J. Sci. Food Agric. 2020, 100, 2782–2790. [Google Scholar] [CrossRef]
- Tan, G.; Wang, Y.; Hu, M.; Li, X.; Li, X.; Pan, Z.; Li, M.; Li, L.; Zheng, Z. Comparative evaluation of the microbial diversity and metabolite profiles of Japanese-style and Cantonese-style soy sauce fermentation. Front. Microbiol. 2022, 13, 976206. [Google Scholar] [CrossRef]
- Zhang, W.; Xiao, Z.; Gu, Z.; Deng, X.; Liu, J.; Luo, X.; Song, C.; Jiang, X. Fermentation-promoting effect of three salt-tolerant Staphylococcus and their co-fermentation flavor characteristics with Zygosaccharomyces rouxii in soy sauce brewing. Food Chem. 2024, 432, 137245. [Google Scholar] [CrossRef]
- Feng, Y.; Wu, W.; Chen, T.; Huang, M.; Zhao, M. Exploring the core functional microbiota related with flavor compounds in fermented soy sauce from different sources. Food Res. Int. 2023, 173, 113456. [Google Scholar] [CrossRef]
- Kubo, Y.; Rooney, A.P.; Tsukakoshi, Y.; Nakagawa, R.; Hasegawa, H.; Kimura, K. Phylogenetic analysis of Bacillus subtilis strains applicable to natto (fermented soybean) production. Appl. Environ. Microbiol. 2011, 77, 6463–6469. [Google Scholar] [CrossRef]
- An, F.; Wu, J.; Feng, Y.; Pan, G.; Ma, Y.; Jiang, J.; Yang, X.; Xue, R.; Wu, R.; Zhao, M. A systematic review on the flavor of soy-based fermented foods: Core fermentation microbiome, multisensory flavor substances, key enzymes, and metabolic pathways. Compr. Rev. Food Sci. Food Saf. 2023, 22, 2773–2801. [Google Scholar] [CrossRef]
- Contesini, F.J.; Melo, R.R.d.; Sato, H.H. An overview of Bacillus proteases: From production to application. Crit. Rev. Biotechnol. 2018, 38, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Kuda, T.; Izawa, Y.; Yoshida, S.; Koyanagi, T.; Takahashi, H.; Kimura, B. Rapid identification of Tetragenococcus halophilus and Tetragenococcus muriaticus, important species in the production of salted and fermented foods, by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS). Food Control 2014, 35, 419–425. [Google Scholar] [CrossRef]
- Li, C.; Li, W.; Li, L.; Chen, S.; Wu, Y.; Qi, B. Microbial community changes induced by a newly isolated salt-tolerant Tetragenococcus muriaticus improve the volatile flavor formation in low-salt fish sauce. Food Res. Int. 2022, 156, 111153. [Google Scholar] [CrossRef] [PubMed]
- Silvetti, T.; Capra, E.; Morandi, S.; Cremonesi, P.; Decimo, M.; Gavazzi, F.; Giannico, R.; De Noni, I.; Brasca, M. Microbial population profile during ripening of Protected Designation of Origin (PDO) Silter cheese, produced with and without autochthonous starter culture. LWT-Food Sci. Technol. 2017, 84, 821–831. [Google Scholar] [CrossRef]
- Oh, Y.; Kong, H.; Jeong, D.-W.; Lee, J.-H. Technological characteristics and safety of Enterococcus faecium isolates from Meju, a traditional Korean fermented soybean food. Microbiol. Biotechnol. Lett. 2021, 49, 255–263. [Google Scholar] [CrossRef]
- Yoon, M.Y.; Kim, Y.J.; Hwang, H.-J. Properties and safety aspects of Enterococcus faecium strains isolated from Chungkukjang, a fermented soy product. LWT-Food Sci. Technol. 2008, 41, 925–933. [Google Scholar] [CrossRef]
- Feng, Y.; Wu, W.; Huang, M.; Su, G.; Zhao, M.; Feng, Y. Mechanistic insights into soy sauce flavor enhancement via Co-culture of Limosilactobacillus fermentum and Zygosaccharomyces rouxii. Food Biosci. 2024, 61, 104979. [Google Scholar] [CrossRef]
- Fu, B.; Wang, J.; Xiao, T.; Liu, Z.; Fu, C.; Zhou, M.; Liu, Z.; Li, W.; Hu, Y.; Wang, C. Co-culture relationship between Zygosaccharomyces rouxii and Candida versatilis and its effect on the flavour of soy sauce. Int. J. Food Sci. 2024, 59, 228–240. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, Z.; Huang, J.; Zhou, R.; Wu, C. Co-culture of Tetragenococcus halophilus and Zygosaccharomyces rouxii to improve microbiota and metabolites in secondary fortified fermented soy sauce. Food Biosci. 2024, 61, 104850. [Google Scholar] [CrossRef]
- Hauck, T.; Brühlmann, F.; Schwab, W. Formation of 4-hydroxy-2,5-dimethyl-3[2H]-furanone by Zygosaccharomyces rouxii: Identification of an intermediate. Appl. Environ. Microbiol. 2003, 69, 3911–3918. [Google Scholar] [CrossRef]
- Hecquet, L.; Sancelme, M.; Bolte, J.; Demuynck, C. Biosynthesis of 4-hydroxy-2,5-dimethyl-3(2H)-furanone by Zygosaccharomyces rouxii. J. Agric. Food Chem. 1996, 44, 1357–1360. [Google Scholar] [CrossRef]
- Qi, W.; Zhang, W.-T.; Lu, F.-P. Effect of temperature, NaCl and ferulic acid concentration on bioconversion of ferulic acid to 4-vinylguaiacol and 4-ethylguaiacol by halotolerant yeasts Candida versatilis. In Proceedings of the International Conference on Applied Biotechnology, Tianjin, China, 25–27 November 2016; pp. 289–297. [Google Scholar]
- Jeong, D.M.; Yoo, S.J.; Jeon, M.-S.; Chun, B.H.; Han, D.M.; Jeon, C.O.; Eyun, S.-i.; Seo, Y.-J.; Kang, H.A. Genomic features, aroma profiles, and probiotic potential of the Debaryomyces hansenii species complex strains isolated from Korean soybean fermented food. Food Microbiol. 2022, 105, 104011. [Google Scholar] [CrossRef] [PubMed]
- Maske, B.L.; de Melo Pereira, G.V.; da Silva Vale, A.; Souza, D.S.M.; Lindner, J.D.D.; Soccol, C.R. Viruses in fermented foods: Are they good or bad? Two sides of the same coin. Food Microbiol. 2021, 98, 103794. [Google Scholar] [CrossRef]
- Jung, M.-J.; Kim, J.; Lee, S.H.; Whon, T.W.; Sung, H.; Bae, J.-W.; Choi, Y.-E.; Roh, S.W. Role of combinated lactic acid bacteria in bacterial, viral, and metabolite dynamics during fermentation of vegetable food, kimchi. Food Res. Int. 2022, 157, 111261. [Google Scholar] [CrossRef] [PubMed]
- Kumar, J.; Sharma, N.; Kaushal, G.; Samurailatpam, S.; Sahoo, D.; Rai, A.K.; Singh, S.P. Metagenomic insights into the taxonomic and functional features of kinema, a traditional fermented soybean product of Sikkim Himalaya. Front. Microbiol. 2019, 10, 1744. [Google Scholar] [CrossRef]
- Bandara, N.; Jo, J.; Ryu, S.; Kim, K.-P. Bacteriophages BCP1-1 and BCP8-2 require divalent cations for efficient control of Bacillus cereus in fermented foods. Food Microbiol. 2012, 31, 9–16. [Google Scholar] [CrossRef]
- Park, E.-J.; Kim, K.-H.; Abell, G.C.; Kim, M.-S.; Roh, S.W.; Bae, J.-W. Metagenomic analysis of the viral communities in fermented foods. Appl. Environ. Microbiol. 2011, 77, 1284–1291. [Google Scholar] [CrossRef]





| G1 | G2 | G3 | G4 | G5 | G6 | G7 | G8 | G9 | G10 | G11 | G12 | G13 | G14 | G15 | G16 | G17 | G18 | G19 | G20 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (P) Appearance | 5.7 defg | 4.6 h | 6.3 abc | 4.1 i | 5.5 efg | 6.4 ab | 4.9 g | 6.2 abcd | 4.8 g | 6.0 bcde | 5.9 cdef | 6.6 a | 5.8 defg | 5.4 g | 5.6 efg | 4.7 h | 5.7 efg | 4.5 hi | 5.5 fg | 6.7 a |
| (P) Smell | 5.3 bcd | 3.9 gh | 6.0 a | 3.5 h | 4.8 def | 6.1 a | 4.3 fg | 5.1 cde | 4.4 fg | 5.4 bc | 4.5 f | 5.7 ab | 3.0 i | 3.9 gh | 4.7 ef | 3.4 hi | 4.8 def | 3.9 gh | 5.4 bc | 5.7 ab |
| (P) Taste | 4.7 fg | 4.3 g | 6.1 bc | 3.7 h | 5.3 de | 6.3 ab | 4.7 fg | 5.1 ef | 5.4 de | 5.7 cd | 4.7 fg | 6.1 bc | 3.3 h | 4.2 g | 4.7 fg | 4.3 g | 5.0 ef | 4.2 g | 6.0 bc | 6.7 a |
| (P) Aftertaste | 4.5 ghi | 4.2 hij | 5.8 abc | 3.7 j | 5.0 efg | 6.1 a | 4.4 ghi | 4.8 fgh | 5.1 def | 5.4 cde | 4.4 ghi | 5.5 bcd | 3.1 k | 4.0 ij | 4.6 ghi | 4.1 ij | 4.7 fgh | 4.1 ij | 5.9 ab | 6.1 a |
| (P) Whole preference | 4.7 ghi | 4.2 jk | 6.0 bc | 3.6 l | 5.4 def | 6.2 ab | 4.6 ghij | 5.3 ef | 5.1 fg | 5.6 cde | 4.8 gh | 6.1 abc | 3.3 l | 4.4 hijk | 4.7 ghi | 4.1 k | 5.0 fg | 4.2 ijk | 5.8 bcd | 6.6 a |
| (A) Brown color | 4.7 f | 3.6 h | 6.3 a | 2.5 i | 4.3 g | 5.9 b | 3.8 h | 5.0 de | 2.0 j | 5.9 b | 5.2 cd | 5.2 cd | 4.9 ef | 3.5 h | 4.6 fg | 2.1 j | 5.4 c | 2.7 i | 2.5 i | 5.5 c |
| (A) Turbidity | 4.5 de | 3.6 fg | 5.8 a | 2.7 h | 4.1 e | 5.1 b | 3.7 f | 4.4 de | 2.0 i | 5.2 b | 4.6 cd | 4.5 de | 4.3 de | 3.3 g | 4.3 de | 2.1 i | 4.9 bc | 2.8 h | 2.2 i | 4.5 de |
| (S) Roasted soybean | 3.7 defg | 3.1 ij | 4.3 ab | 3.0 j | 4.0 bcde | 4.4 a | 3.6 efg | 4.3 ab | 4.1 abc | 4.0 abcd | 3.4 ghi | 4.3 ab | 3.1 hij | 3.5 fgh | 3.6 defg | 3.0 j | 3.7 cdefg | 3.4 ghij | 3.9 bcdef | 4.4 a |
| (S) Sweet | 3.6 ef | 3.2 ghi | 4.4 ab | 3.2 ghi | 3.8 de | 4.5 a | 3.6 ef | 4.1 bcd | 3.9 cde | 4.0 cde | 3.7 def | 4.2 abc | 3.0 hi | 3.5 efg | 3.8 cde | 2.9 i | 3.8 def | 3.4 fgh | 4.0 cde | 4.6 a |
| (S) Sour | 3.8 abc | 3.7 abcd | 3.5 bcde | 3.9 ab | 3.8 abc | 3.3 cdef | 3.8 abc | 3.1 ef | 3.0 f | 3.5 bcde | 3.8 abc | 3.5 bcde | 3.5 bcde | 4.0 a | 3.7 abcd | 3.7 abcd | 3.6 abcd | 3.7 abc | 3.2 def | 3.5 bcde |
| (S) Meju | 4.7 cde | 4.8 bcde | 4.0 gh | 4.4 defg | 5.2 ab | 4.4 efg | 4.8 abcd | 4.8 bcde | 5.2 ab | 4.4 defg | 4.5 def | 4.8 bcde | 4.8 bcde | 5.0 abc | 4.2 fgh | 4.2 fgh | 5.0 abc | 5.3 a | 3.9 h | 3.9 h |
| (S) Alcohol | 3.4 bcd | 3.3 cde | 3.2 cde | 3.5 bcd | 3.5 bc | 3.2 cde | 3.5 bc | 3.0 de | 2.9 e | 3.5 bc | 3.3 cde | 3.2 cde | 3.6 bc | 3.9 ab | 3.4 bcd | 4.0 a | 3.2 cde | 3.3 cde | 3.3 cde | 3.8 ab |
| (S) Musty | 3.4 ghij | 4.3 bc | 3.0 j | 4.2 cd | 3.8 defg | 3.0 j | 3.8 defg | 3.4 ghij | 3.6 fghi | 3.4 ghij | 4.0 cdef | 3.1 ij | 5.2 a | 4.2 cd | 3.6 fghi | 4.6 b | 3.7 efgh | 4.1 cde | 3.1 ij | 3.2 hij |
| (T) Sweet | 3.2 hi | 3.3 hi | 4.1 cd | 3.2 i | 3.9 cde | 4.2 bc | 3.5 fghi | 3.7 defg | 3.9 cdef | 4.1 cd | 3.4 ghi | 4.2 bc | 3.4 ghi | 3.4 ghi | 3.4 ghi | 3.4 ghi | 3.6 efgh | 3.4 ghi | 4.5 b | 5.0 a |
| (T) Umami | 3.9 fg | 3.7 gh | 4.5 bcd | 3.4 h | 4.5 bcd | 4.6 bc | 4.0 efg | 4.2 def | 4.3 cde | 4.5 bcd | 3.9 efg | 4.9 ab | 3.7 gh | 3.7 gh | 4.0 efg | 3.8 fg | 4.0 efg | 3.7 gh | 4.7 bc | 5.1 a |
| (T) Roasted soybean | 3.3 fg | 3.1 fg | 3.9 bc | 3.0 g | 3.7 cde | 4.1 bc | 3.4 def | 3.8 bcd | 3.8 bcd | 3.8 bcd | 3.4 efg | 4.1 b | 3.3 efg | 3.3 fg | 3.4 def | 3.3 fg | 3.4 def | 3.1 fg | 4.1 bc | 4.5 a |
| (T) Salty | 5.6 a | 5.3 ab | 5.2 abc | 5.4 ab | 5.4 ab | 5.3 abc | 5.5 a | 5.4 ab | 5.0 bcd | 5.3 ab | 5.5 a | 5.3 ab | 4.9 cd | 5.1 bcd | 5.3 ab | 5.2 abc | 5.4 ab | 5.4 ab | 4.8 d | 4.3 e |
| (T) Astringent | 3.8 ab | 3.5 bc | 3.2 cd | 3.8 ab | 3.8 ab | 3.0 de | 3.4 bc | 3.8 ab | 3.4 bcd | 3.2 cd | 4.0 a | 3.2 cd | 3.8 ab | 3.9 ab | 3.6 abc | 3.5 abc | 3.8 ab | 3.9 ab | 2.7 e | 2.7 e |
| (T) Bitter | 3.6 abc | 3.4 abcd | 3.0 def | 3.6 ab | 3.3 bcd | 2.8 efg | 3.2 bcde | 3.3 abcd | 2.8 efg | 3.2 bcde | 3.7 a | 2.8 egf | 3.8 a | 3.8 a | 3.4 abcd | 3.1 cde | 3.4 abcd | 3.4 abcd | 2.6 fg | 2.5 g |
| (T) Musty | 3.4 defg | 3.8 bc | 2.7 ijk | 4.1 b | 3.2 fghi | 2.4 k | 3.3 fgh | 3.3 efg | 3.0 ghij | 2.8 hijk | 3.4 defg | 2.6 jk | 4.9 a | 3.8 bcde | 3.4 dfg | 3.8 bcd | 3.3 fg | 3.5 cdef | 2.4 k | 2.4 k |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chin, Y.-W.; Hong, S.-P.; Lim, S.-D.; Yi, S.-H. Investigation of Microbial Community of Korean Soy Sauce (Ganjang) Using Shotgun Metagenomic Sequencing and Its Relationship with Sensory Characteristics. Microorganisms 2024, 12, 2559. https://doi.org/10.3390/microorganisms12122559
Chin Y-W, Hong S-P, Lim S-D, Yi S-H. Investigation of Microbial Community of Korean Soy Sauce (Ganjang) Using Shotgun Metagenomic Sequencing and Its Relationship with Sensory Characteristics. Microorganisms. 2024; 12(12):2559. https://doi.org/10.3390/microorganisms12122559
Chicago/Turabian StyleChin, Young-Wook, Sang-Pil Hong, Sang-Dong Lim, and Sung-Hun Yi. 2024. "Investigation of Microbial Community of Korean Soy Sauce (Ganjang) Using Shotgun Metagenomic Sequencing and Its Relationship with Sensory Characteristics" Microorganisms 12, no. 12: 2559. https://doi.org/10.3390/microorganisms12122559
APA StyleChin, Y.-W., Hong, S.-P., Lim, S.-D., & Yi, S.-H. (2024). Investigation of Microbial Community of Korean Soy Sauce (Ganjang) Using Shotgun Metagenomic Sequencing and Its Relationship with Sensory Characteristics. Microorganisms, 12(12), 2559. https://doi.org/10.3390/microorganisms12122559






