Microbiological Evaluation of Local and Imported Raw Beef Meat at Retail Sites in Oman with Emphasis on Spoilage and Pathogenic Psychrotrophic Bacteria
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Sample Preparation and Microbial Counts
2.3. Identification of Bacteria and Biochemical Analysis
2.4. Data Analysis
3. Results
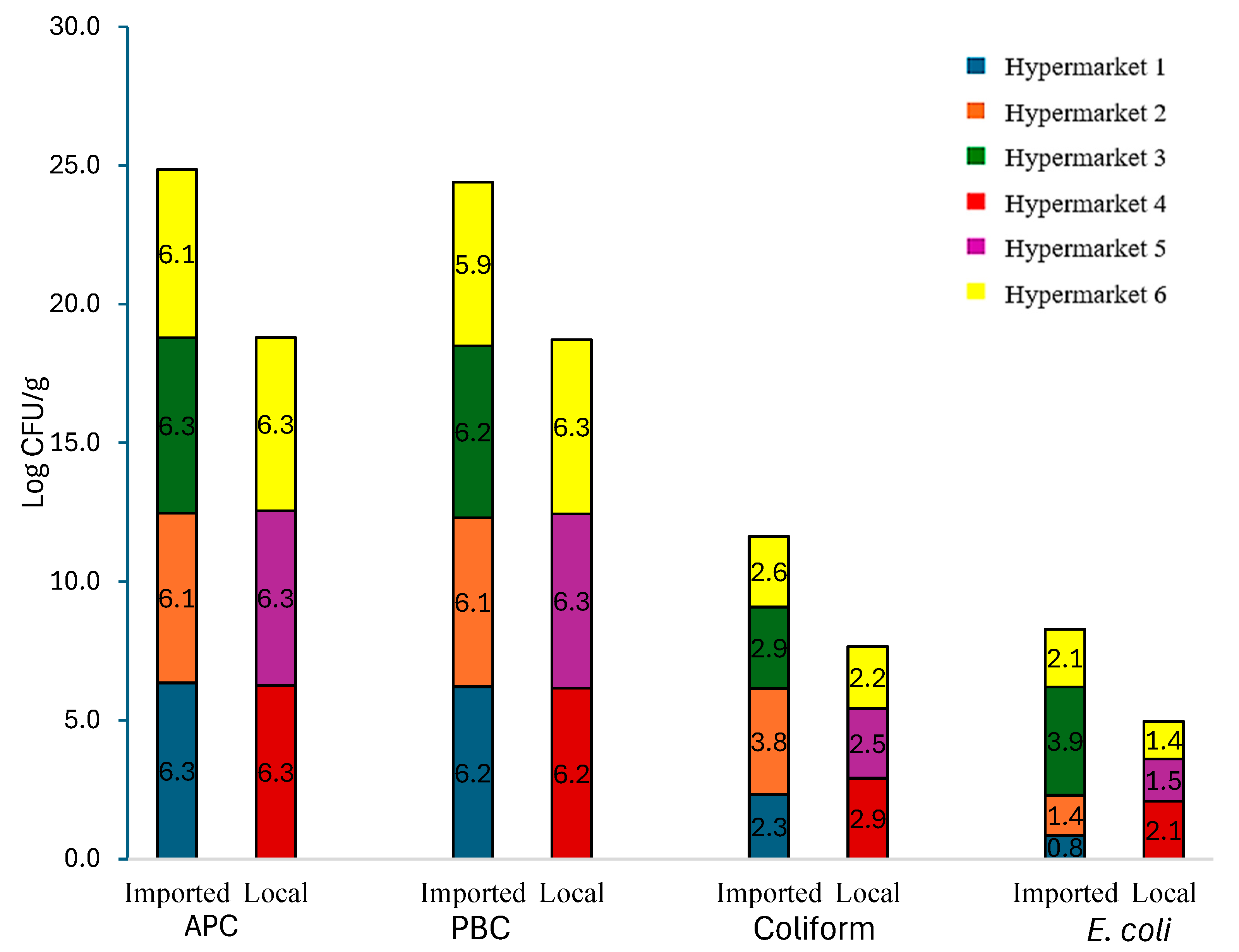
3.1. Microbial Counts
3.2. Bacterial Identification
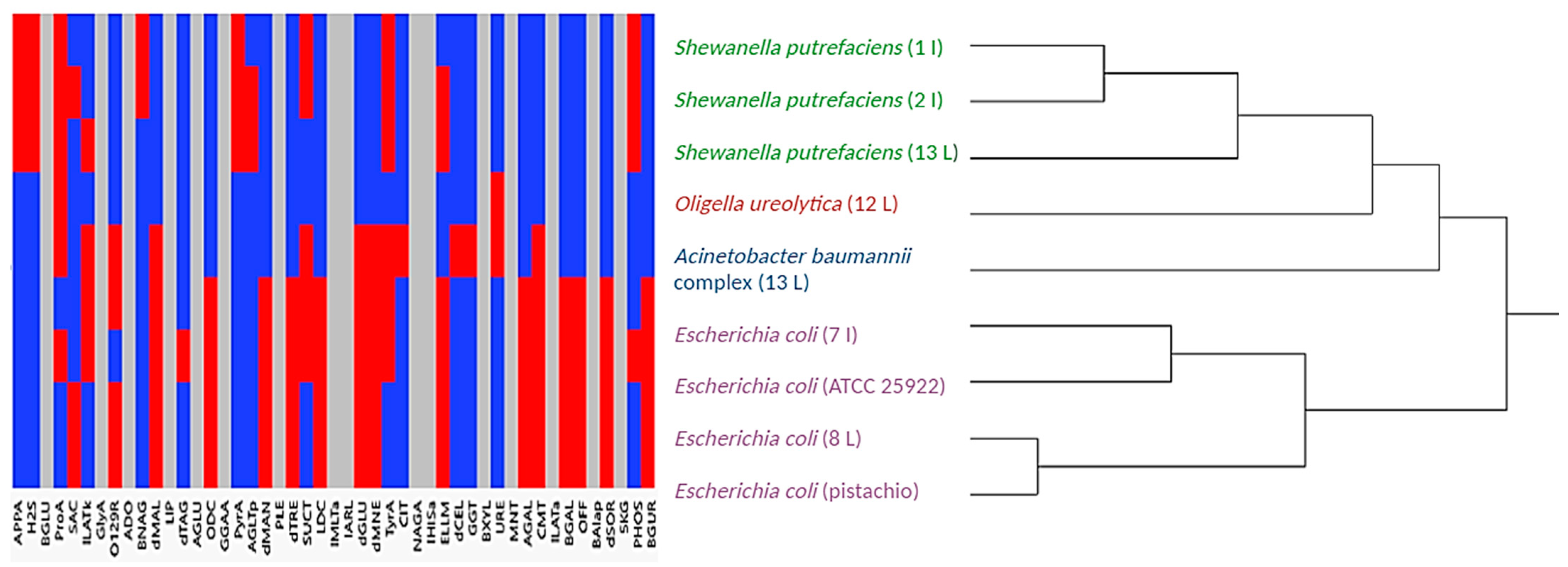
3.3. Cluster Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Al-Amri, I.; Kadim, I.T.; AlKindi, A.; Hamaed, A.; Al-Magbali, R.; Khalaf, S.; Al-Hosni, K.; Mabood, F. Determination of residues of pesticides, anabolic steroids, antibiotics, and antibacterial compounds in meat products in Oman by liquid chromatography/mass spectrometry and enzyme-linked immunosorbent assay. Vet. World 2021, 14, 709–720. [Google Scholar] [CrossRef] [PubMed]
- National Center for Statistics and Information (NCSI). Agriculture and Livestock Portal Data. 2023. Available online: https://data.gov.om/uvzbyhb/agriculture-livestock (accessed on 9 October 2024).
- Heredia, N.; García, S. Animals as sources of food-borne pathogens: A review. Anim. Nutr. 2018, 4, 250–255. [Google Scholar] [CrossRef] [PubMed]
- Al-Kharousi, Z.S.; Guizani, N.; Al-Sadi, A.M.; Al-Bulushi, I.M. Tetracycline resistance in enterococci and Escherichia coli isolated from fresh produce and why it matters. Int. J. Food Stud. 2021, 10, 359–370. [Google Scholar] [CrossRef]
- Pahalagedara, A.S.; Gkogka, E.; Hammershøj, M. A review on spore-forming bacteria and moulds implicated in the quality and safety of thermally processed acid foods: Focusing on their heat resistance. Food Control 2024, 166, 110716. [Google Scholar] [CrossRef]
- Ercolini, D.; Russo, F.; Nasi, A.; Ferranti, P.; Villani, F. Mesophilic and psychrotrophic bacteria from meat and their spoilage potential in vitro and in beef. Appl. Environ. Microbiol. 2009, 75, 1990–2001. [Google Scholar] [CrossRef]
- Hamza, M.A.; Lachisa, L.; Woyessa, D. Bacteriological quality of raw meat and dairy products and antibiogram profile of bacterial pathogens in Jimma town, southwest Ethiopia. Res. Sq. 2024, 1–19. [Google Scholar] [CrossRef]
- Afzaal, M.; Shehzadi, U.; Ali, R.; Ahmad, M.; Raza, M.A.; Shah, Y.A.; Mustafa, J. Assessing the microbiological safety of raw meat sold on different butcher’s shop in Faisalabad, Pakistan. Acta Sci. Nutr. Health 2019, 3, 110–113. [Google Scholar] [CrossRef]
- Serhan, M.; Hourieh, H.; El Deghel, M.; Serhan, C. Hygienic sanitary risk and microbiological quality of meat and meat-contact surfaces in traditional butcher shops and retail establishments—Lessons from a developing country. Int. J. Environ. Health Res. 2022, 34, 600–610. [Google Scholar] [CrossRef]
- Al-Kharousi, Z.S.; Guizani, N.; Al-Sadi, A.M.; Al-Bulushi, I.M.; Shaharoona, B. Hiding in fresh fruits and vegetables: Opportunistic pathogens may cross geographical barriers. Int. J. Microbiol. 2016, 2016, 4292417. [Google Scholar] [CrossRef]
- Yao, L.; Champagne, C.P.; Deschênes, L.; Raymond, Y.; Lemay, M.-J.; Ismail, A. Effect of the homogenization technique on the enumeration of psychrotrophic bacteria in food absorbent pads. J. Microbiol. Methods 2021, 187, 106275. [Google Scholar] [CrossRef]
- Ahmad, M.U.D.; Sarwar, A.; Najeeb, M.I.; Nawaz, M.; Anjum, A.A.; Ali, M.A.; Mansur, N. Assessment of microbial load of raw meat at abattoirs and retail outlets. J. Anim. Plant Sci. 2013, 23, 745–748. [Google Scholar]
- Al-Zaid, R.M.K.; Al-Attar, E.J.; Hadi, M.T. Detection of mineral and microbial contaminants in some types of imported meat. IOP Conf. Ser. Earth Environ. Sci. 2023, 1158, 112025. [Google Scholar] [CrossRef]
- Wakabayashi, R.; Aoyanagi, A.; Tominaga, T. Rapid counting of coliforms and Escherichia coli by deep learning-based classifier. J. Food Saf. 2024, 44, e13158. [Google Scholar] [CrossRef]
- Kang, J.Y.; Lee, S.H.; Jo, A.H.; Park, E.J.; Bak, Y.S.; Kim, J.B. Improving the accuracy of coliform detection in meat products using modified dry rehydratable film method. Food Sci. Biotechnol. 2020, 29, 1289–1294. [Google Scholar] [CrossRef]
- Manges, A.R. Escherichia coli and urinary tract infections: The role of poultry-meat. Clin. Microbiol. Infect. 2016, 22, 122–129. [Google Scholar] [CrossRef]
- Karasu-Yalcin, S.; Soylemez-Milli, N.; Eren, O.; Eryasar-Orer, K. Reducing Time in Detection of Listeria monocytogenes from Food by MALDI-TOF Mass Spectrometry. J. Food Sci. Technol. 2021, 58, 4102–4109. [Google Scholar] [CrossRef]
- Al Bulushi, I.M.; Al Kharousi, Z.S.; Rahman, M.S. Vitek: A Platform for a Better Understanding of Microbes. In Techniques to Measure Food Safety and Quality: Microbial, Chemical, and Sensory; Springer International Publishing: Cham, Switzerland, 2021; pp. 117–136. [Google Scholar] [CrossRef]
- Oxoid. Available online: http://www.oxoid.com/UK/blue/index.asp?c=UK&lang=EN (accessed on 12 November 2024).
- Nielsen, F. Hierarchical Clustering. In Introduction to HPC with MPI for Data Science; Springer: Cham, Switzerland, 2016. [Google Scholar] [CrossRef]
- Allen, M.J.; Edberg, S.C.; Reasoner, D.J. Heterotrophic plate count bacteria-what is their significance in drinking water? Int. J. Food Microbiol. 2004, 92, 265–274. [Google Scholar] [CrossRef]
- Siragusa, G.R.; Dorsa, W.J.; Cutter, C.N.; Bennett, G.L.; Keen, J.E.; Koohmaraie, M. The Incidence of Escherichia coli on Beef Carcasses and Its Association with Aerobic Mesophilic Plate Count Categories during the Slaughter Process. J. Food Prot. 1998, 61, 1269–1274. [Google Scholar] [CrossRef]
- Gounot, A.M. Psychrophilic and psychrotrophic microorganisms. Experientia 1986, 42, 1192–1197. [Google Scholar] [CrossRef]
- Saha, S.; Majumder, R.; Rout, P.; Hossain, S. Unveiling the significance of psychrotrophic bacteria in milk and milk product spoilage—A review. Microbe 2024, 2, 100034. [Google Scholar] [CrossRef]
- Yu, Z.; Joossens, M.; Kerkhof, P.; Houf, K. Bacterial shifts on broiler carcasses at retail upon frozen storage. Int. J. Food Microbiol. 2021, 340, 109051. [Google Scholar] [CrossRef] [PubMed]
- Riswandi, R.; Malaka, R.; Ali, H.M. Analysis of meat microbial contamination on the beef supply chain in Makassar City. BIO Web Conf. 2024, 96, 01034. [Google Scholar] [CrossRef]
- GSO/FDS 1016/2014; Microbiological Criteria for Foodstuffs. Standarization Organization for GCC (GSO): Riyadh, Saudi Arabia, 2014.
- Khalalfalla, F.A.; Fatma, H.M.; Ali, S.-A. Microbiological quality of retail meats. J. Vet. Med. Res. 2017, 24, 311–321. [Google Scholar] [CrossRef]
- Oh, H.; Lee, J. Psychrotrophic bacteria threatening the safety of animal-derived foods: Characteristics, Contamination, and control strategies. Food Sci. Anim. Resour. 2024, 44, 1011–1027. [Google Scholar] [CrossRef] [PubMed]
- Shoaib, S.; Wakil, W.; Zabihullah, N.; Nazir, T. Coliform contamination of raw beef at the slaughterhouse and butchery levels in herat city, Afghanistan. Int. J. Biosci. 2023, 2, 137–144. [Google Scholar] [CrossRef]
- Sudip, S.; Rittick, M.; Debasis, M.; Divya, J.; Devvret, V.; Samanwita, D. Microbial pollution of water with special reference to coliform bacteria and their nexus with environment. Energy Nexus 2021, 1, 100008. [Google Scholar] [CrossRef]
- Hamilton-Miller, J.M.T.; Shah, S. Identity and antibiotic susceptibility of enterobacterial flora of salad vegetables. Int. J. Antimicrob. Agents 2001, 18, 81–83. [Google Scholar] [CrossRef]
- Adzitey, F.; Assoah-Peprah, P.; Teye, G.A.; Somboro, A.M.; Kumalo, H.M.; Amoako, D.G. Prevalence and antimicrobial resistance of Escherichia coli isolated from various meat types in the tamale metropolis of Ghana. Int. J. Food Sci. 2020, 2020, 8877196. [Google Scholar] [CrossRef]
- Nguz, K.; Shindano, J.; Samapundo, S.; Huyghebaert, A. Microbiological Evaluation of Fresh-Cut Organic Vegetables Produced in Zambia. Food Control 2005, 16, 623–628. [Google Scholar] [CrossRef]
- Al-Chalaby, A.Y.H. Detection of Escherichia coli from imported and local beef meat in Mosul city. J. Pure Appl. Microbiol. 2020, 14, 383–388. [Google Scholar] [CrossRef]
- Marcelli, V.; Osimani, A.; Aquilanti, L. Research progress in the use of lactic acid bacteria as natural biopreservatives against Pseudomonas spp. in meat and meat products: A review. Food Res. Int. 2024, 196, 115129. [Google Scholar] [CrossRef] [PubMed]
- Palevich, N.; Palevich, F.P.; Gardner, A.; Brightwell, G.; Mills, J. Genome collection of Shewanella spp. isolated from spoiled lamb. Front. Microbiol. 2022, 13, 976152. [Google Scholar] [CrossRef] [PubMed]
- Yi, Z.; Xie, J. Prediction in the dynamics and spoilage of Shewanella putrefaciens in bigeye tuna (Thunnus obesus) by gas sensors stored at different refrigeration temperatures. Foods 2021, 10, 2132. [Google Scholar] [CrossRef] [PubMed]
- Fukatsu, A.; Tsuzukibashi, O.; Yamamoto, H.; Takahashi, Y.; Usuda, K.; Fuchigami, M.; Komine, C.; Uchibori, S.; Umezawa, K.; Hayashi, S.; et al. Study on Distribution of Acinetobacter baumannii complex in dental hospital using multiplex PCR. Open J. Stomatol. 2023, 13, 212–221. [Google Scholar] [CrossRef]
- Ahuatzin-Flores, O.E.; Torres, E.; Chávez-Bravo, E. Acinetobacter baumannii, a multidrug-resistant opportunistic pathogen in new habitats: A systematic review. Microorganisms 2024, 12, 644. [Google Scholar] [CrossRef]
- Elbehiry, A.; Marzouk, E.; Moussa, I.M.; Dawoud, T.M.; Mubarak, A.S.; Al-Sarar, D.; Alsubki, R.A.; Alhaji, J.H.; Hamada, M.; Abalkhail, A.; et al. Acinetobacter baumannii as a community foodborne pathogen: Peptide mass fingerprinting analysis, genotypic of biofilm formation and phenotypic pattern of antimicrobial resistance. Saudi J. Biol. Sci. 2021, 28, 1158–1166. [Google Scholar] [CrossRef]
- Serandour, P.; Plouzeau, C.; Michaud, A.; Broutin, L.; Cremniter, J.; Burucoa, C.; Pichon, M. The first lethal infection by Oligella ureolytica: A case report and review of the literature. Antibiotics 2023, 12, 1470. [Google Scholar] [CrossRef]
- Xi, H.; Ji, Y.; Fu, Y.; Chen, C.; Han, W.; Gu, J. Biological Characterization of the Phage Lysin AVPL and Its Efficiency against Aerococcus viridans-Induced Mastitis in a Murine Model. Appl. Environ. Microbiol. 2024, 90, e00461-24. [Google Scholar] [CrossRef]
- Sahu, K.K.; Lal, A.; Mishra, A.K.; Abraham, G.M. Aerococcus-related infections and their significance: A 9-Year Retrospective Study. J. Microsc. Ultrastruct. 2021, 9, 18–25. [Google Scholar] [CrossRef]
- Cebeci, T.; Tanrıverdi, E.S.; Otlu, B. A First Study of Meat-Borne Enterococci from Butcher Shops: Prevalence, Virulence Characteristics, Antibiotic Resistance and Clonal Relationship. Vet. Res. Commun. 2024, 48, 3669–3682. [Google Scholar] [CrossRef]
- Nüesch-Inderbinen, M.; Heyvaert, L.; Treier, A.; Zurfluh, K.; Cernela, N.; Biggel, M.; Stephan, R. High Occurrence of Enterococcus faecalis, Enterococcus faecium, and Vagococcus lutrae Harbouring Oxazolidinone Resistance Genes in Raw Meat-Based Diets for Companion Animals—A Public Health Issue, Switzerland, September 2018 to May 2020. Eurosurveillance 2023, 28, 2200496. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Chon, J.W.; Jeong, H.W.; Song, K.Y.; Kim, D.H.; Bae, D.; Kim, H.; Seo, K.H. Identification and phylogenetic analysis of Enterococcus isolates using MALDI-TOF MS and VITEK 2. AMB Express 2023, 13, 21. [Google Scholar] [CrossRef] [PubMed]
- Al-Kharousi, Z.S.; Al-Ramadhani, Z.; Al-Malki, F.A.; Al-Habsi, N. Date Vinegar: First Isolation of Acetobacter and Formulation of a Starter Culture. Foods 2024, 13, 1389. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Ma, F.; Wu, Y.; Tan, S.; Niu, A.; Qiu, W.; Wang, G. Biosurfactant from Pseudomonas fragi Enhances the Competitive Advantage of Pseudomonas but Reduces the Overall Spoilage Ability of the Microbial Community in Chilled Meat. Food Microbiol. 2023, 115, 104311. [Google Scholar] [CrossRef] [PubMed]
- Sequino, G.; Cobo-Diaz, J.F.; Valentino, V.; Tassou, C.; Volpe, S.; Torrieri, E.; De Filippis, F. Microbiome Mapping in Beef Processing Reveals Safety-Relevant Variations in Microbial Diversity and Genomic Features. Food Res. Int. 2024, 186, 114318. [Google Scholar] [CrossRef]

| Well | Test | Abbreviation | Well | Test | Abbreviation |
|---|---|---|---|---|---|
| 2 | Ala-Phe-Pro-arylamidase | APPA | 33 | Saccharose/sucrose | SAC |
| 3 | Adonitol | ADO | 34 | D-Tagatose | dTAG |
| 4 | L-Pyrrolydonyl-arylamidase | PyrA | 35 | D-Trehalose | dTRE |
| 5 | L-Arabitol | IARL | 36 | Citrate (sodium) | CIT |
| 7 | D-Cellobiose | dCEL | 37 | Malonate | MNT |
| 9 | Beta-galactosidase | BGAL | 39 | 5-Keto-D-gluconate | 5KG |
| 10 | H2S Production | H2S | 40 | L-lactate alkalinization | ILATk |
| 11 | Beta-N-acetyl-glucosaminidase | BNAG | 41 | Alpha-glucosidase | AGLU |
| 12 | Glutamyl Arylamidase pNA | AGLTp | 42 | Succinate alkalinization | SUCT |
| 13 | D-glucose | dGLU | 43 | Beta-N-acetyl-galactosaminidase | NAGA |
| 14 | gamma-glutamyl-transferase | GGT | 44 | Alpha-galactosidase | AGAL |
| 15 | Fermentation/glucose | OFF | 45 | Phosphatase | PHOS |
| 17 | Beta-glucosidase | BGLU | 46 | Glycine arylamidase | GlyA |
| 18 | D-Maltose | dMAL | 47 | Ornithine decarboxylase | ODC |
| 19 | D-Mannitol | dMAN | 48 | Lysine decarboxylase | LDC |
| 20 | D-Mannose | dMNE | 53 | L-Histidine assimilation | IHISa |
| 21 | Beta-xylosidase | BXYL | 56 | coumarate | CMT |
| 22 | Beta-alanine arylamidase pNA | BAIap | 57 | Beta-glucuronidase | BGUR |
| 23 | L-Proline arylamidase | ProA | 58 | O/129 resistance | O129R |
| 26 | Lipase | LIP | 59 | Glu-Gly-Arg-Arylamidase | GGAA |
| 27 | Palatinose | PLE | 61 | L-Malate assimilation | IMLTa |
| 29 | Tyrosine arylamidase | TyrA | 62 | Ellman | ELLM |
| 31 | Urease | URE | 64 | L-Lactate assimilation | ILATa |
| 32 | D-Sorbitol | dSOR |
| Well | Test | Abbreviation | Well | Test | Abbreviation |
|---|---|---|---|---|---|
| 2 | D-amygdalin | AMY | 32 | Polymixin B resistance | POLYB |
| 4 | Phosphatidylinositol phospholipase C | PIPLC | 37 | D-galactose | dGAL |
| 5 | D-xylose | dXYL | 38 | D-ribose | dRIB |
| 8 | Arginine dihydrolase 1 | ADH1 | 39 | L-lactate alkalinization | ILATk |
| 9 | Beta-galactosidase | BGAL | 42 | Lactose | LAC |
| 11 | Alpha-glucosidase | AGLU | 44 | N-acetyl-D-glucosamine | NAG |
| 13 | Ala-phe-pro arylamidase | APPA | 45 | D-maltose | dMAL |
| 14 | Cyclodextrin | CDEX | 46 | Bacitracin resistance | BACI |
| 15 | L-aspartate arylamidase | AspA | 47 | Novobiocin resistance | NOVO |
| 16 | Beta galactopyranosidase | BGAR | 50 | Growth in 6.5% nacl | NC6.5 |
| 17 | Alpha-mannosidase | AMAN | 52 | D-mannitol | dMAN |
| 19 | Phosphatase | PHOS | 53 | D-mannose | dMNE |
| 20 | Leucine arylamidase | LeuA | 54 | Methyl-B-D-glucopyranoside | MBdG |
| 23 | L-proline arylamidase | ProA | 56 | Pullulan | PUL |
| 24 | Beta glucuronidase | BGURr | 57 | D-raffinose | dRAF |
| 25 | Alpha-galactosidase | AGAL | 58 | O/129 resistance | O129R |
| 26 | L-pyrrolydonyl-arylamidase | PyrA | 59 | Salicin | SAL |
| 27 | Beta-glucuronidase | BGUR | 60 | Saccharose/sucrose | SAC |
| 28 | Alanine arylamidase | AlaA | 62 | D-trehalose | dTRE |
| 29 | Tyrosine arylamidase | TyrA | 63 | Arginine dihydrolase 2 | ADH2s |
| 30 | D-sorbitol | dSOR | 64 | Optochin resistance | OPTO |
| 31 | Urease | URE |
| Sample | Name | Bionumber | % Identification |
|---|---|---|---|
| Gram-negative | |||
| 13 (L) | Acinetobacter baumannii complex (A. nosocomialis, A. pittii, A. baumannii, A. calcoaceticus) | 0203211301500210 | 93% (very good) |
| 7 (I) | Escherichia coli | 0405610540526611 | 99% (excellent) |
| 8 (L) | Escherichia coli | 0405610450026611 | 99% (excellent) |
| ATCC | Escherichia coli ATCC 25922 | 0405611560566601 | 98% (excellent) |
| Pistachio (I) | Escherichia coli | 0405610450026611 | 99% (excellent) |
| 12 (L) | Oligella ureolytica | 0000001200000000 | 97% (excellent) |
| 14 (I) | Pseudomonas fluorescens | 4203251101500210 | 88% (acceptable) |
| 13 (L) | Pseudomonas fluorescens | 4203211101100210 | 89% (good) |
| 2 (I) | Pseudomonas luteola | 4207211301500250 | 85% (acceptable) |
| 1 (I) | Shewanella putrefaciens | 5030001100440000 | 96% (excellent) |
| 2 (I) | Shewanella putrefaciens | 5070001110440000 | 97% (excellent) |
| 13 (L) | Shewanella putrefaciens | 5050001100140001 | 98% (excellent) |
| Gram-positive | |||
| 12 (L) | Aerococcus viridans | 000000200363430 | 87% (acceptable) |
| 14 (I) | Enterococcus faecalis | 176012665773671 | 94% (very good) |
| ATCC | Staphylococcus aureus ATCC 25923 | 030402067763231 | 99% (excellent) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Mazrouei, M.A.; Al-Kharousi, Z.S.; Al-Kharousi, J.M.; Al-Barashdi, H.M. Microbiological Evaluation of Local and Imported Raw Beef Meat at Retail Sites in Oman with Emphasis on Spoilage and Pathogenic Psychrotrophic Bacteria. Microorganisms 2024, 12, 2545. https://doi.org/10.3390/microorganisms12122545
Al-Mazrouei MA, Al-Kharousi ZS, Al-Kharousi JM, Al-Barashdi HM. Microbiological Evaluation of Local and Imported Raw Beef Meat at Retail Sites in Oman with Emphasis on Spoilage and Pathogenic Psychrotrophic Bacteria. Microorganisms. 2024; 12(12):2545. https://doi.org/10.3390/microorganisms12122545
Chicago/Turabian StyleAl-Mazrouei, Musallam A., Zahra S. Al-Kharousi, Jamila M. Al-Kharousi, and Hajer M. Al-Barashdi. 2024. "Microbiological Evaluation of Local and Imported Raw Beef Meat at Retail Sites in Oman with Emphasis on Spoilage and Pathogenic Psychrotrophic Bacteria" Microorganisms 12, no. 12: 2545. https://doi.org/10.3390/microorganisms12122545
APA StyleAl-Mazrouei, M. A., Al-Kharousi, Z. S., Al-Kharousi, J. M., & Al-Barashdi, H. M. (2024). Microbiological Evaluation of Local and Imported Raw Beef Meat at Retail Sites in Oman with Emphasis on Spoilage and Pathogenic Psychrotrophic Bacteria. Microorganisms, 12(12), 2545. https://doi.org/10.3390/microorganisms12122545





