Metagenome-Assembled Genomes of Pig Fecal Samples in Nine European Countries: Insights into Antibiotic Resistance Genes and Viruses
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. MAG Collection Construction
2.3. ARG Analysis
2.4. Virus Analysis
3. Results
3.1. Metagenome-Assembled Genomes
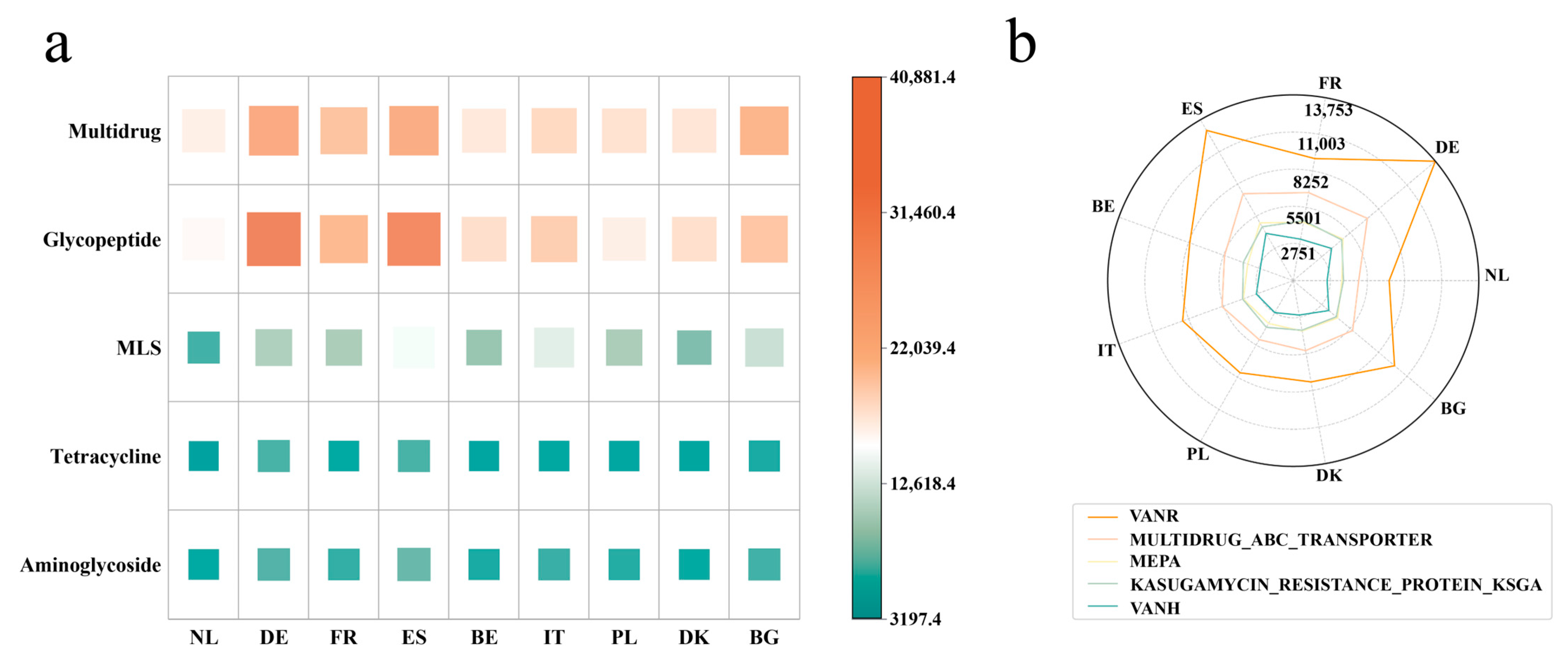
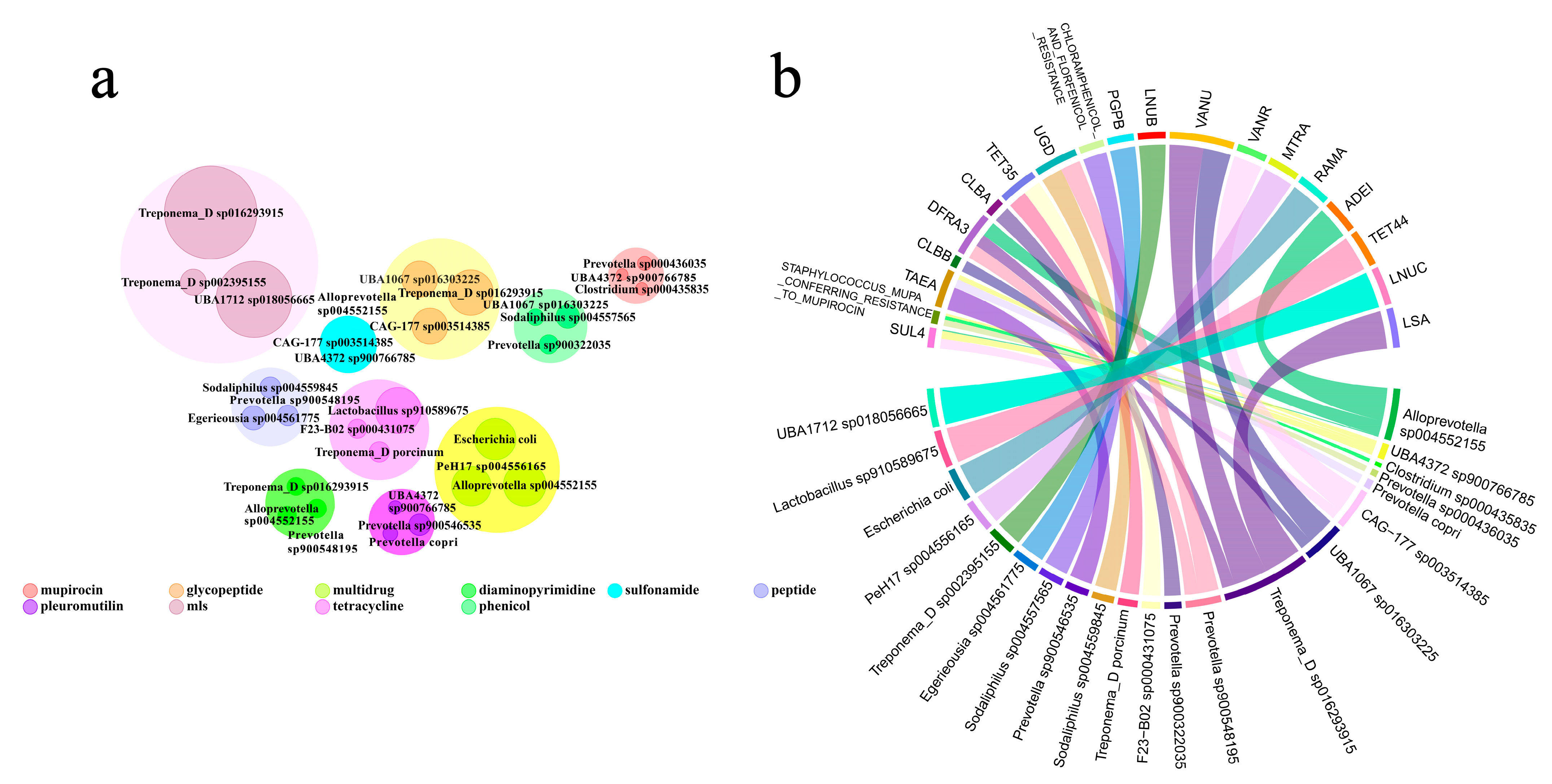
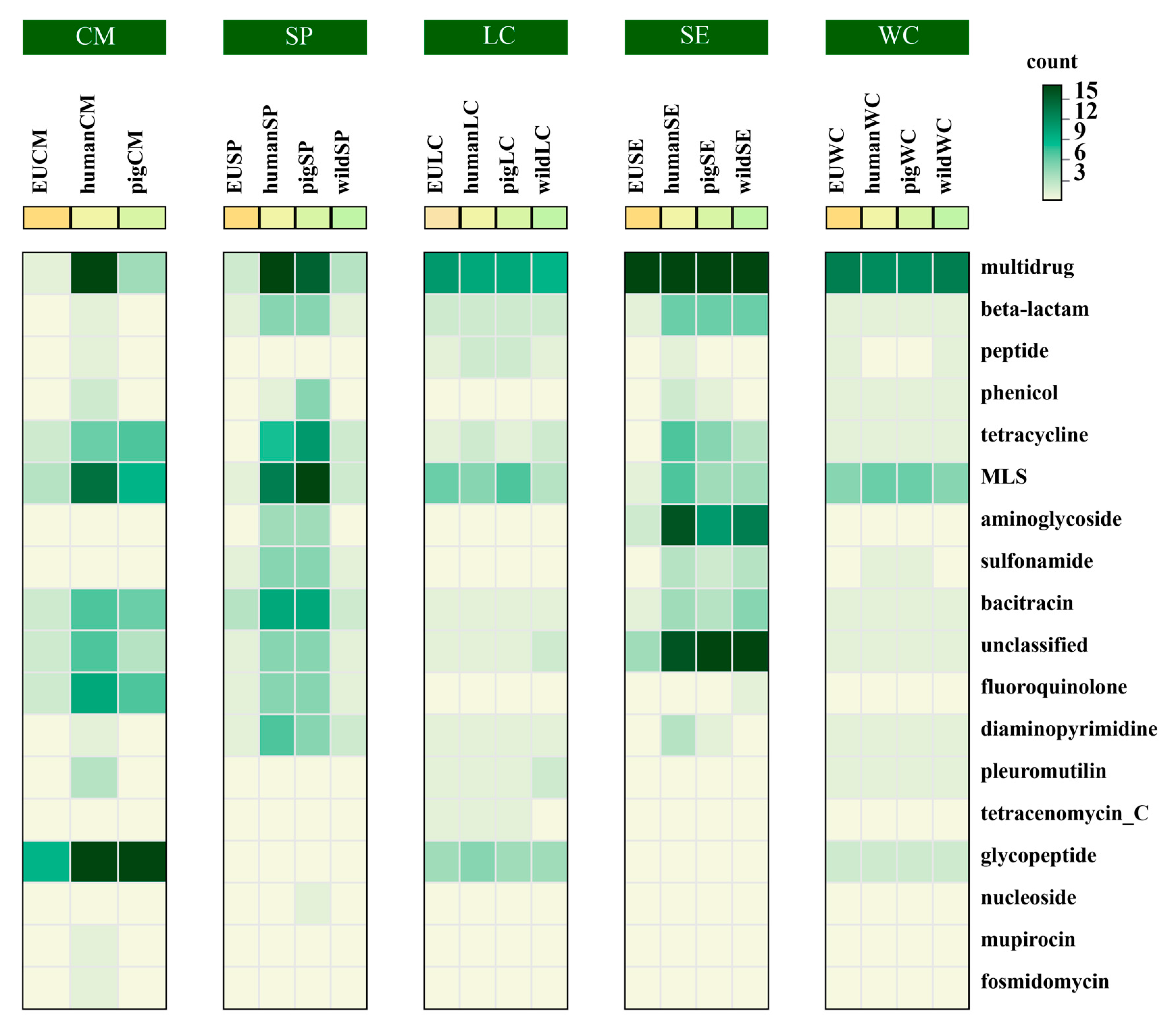
3.2. Antibiotic Resistance Gene Host Prediction
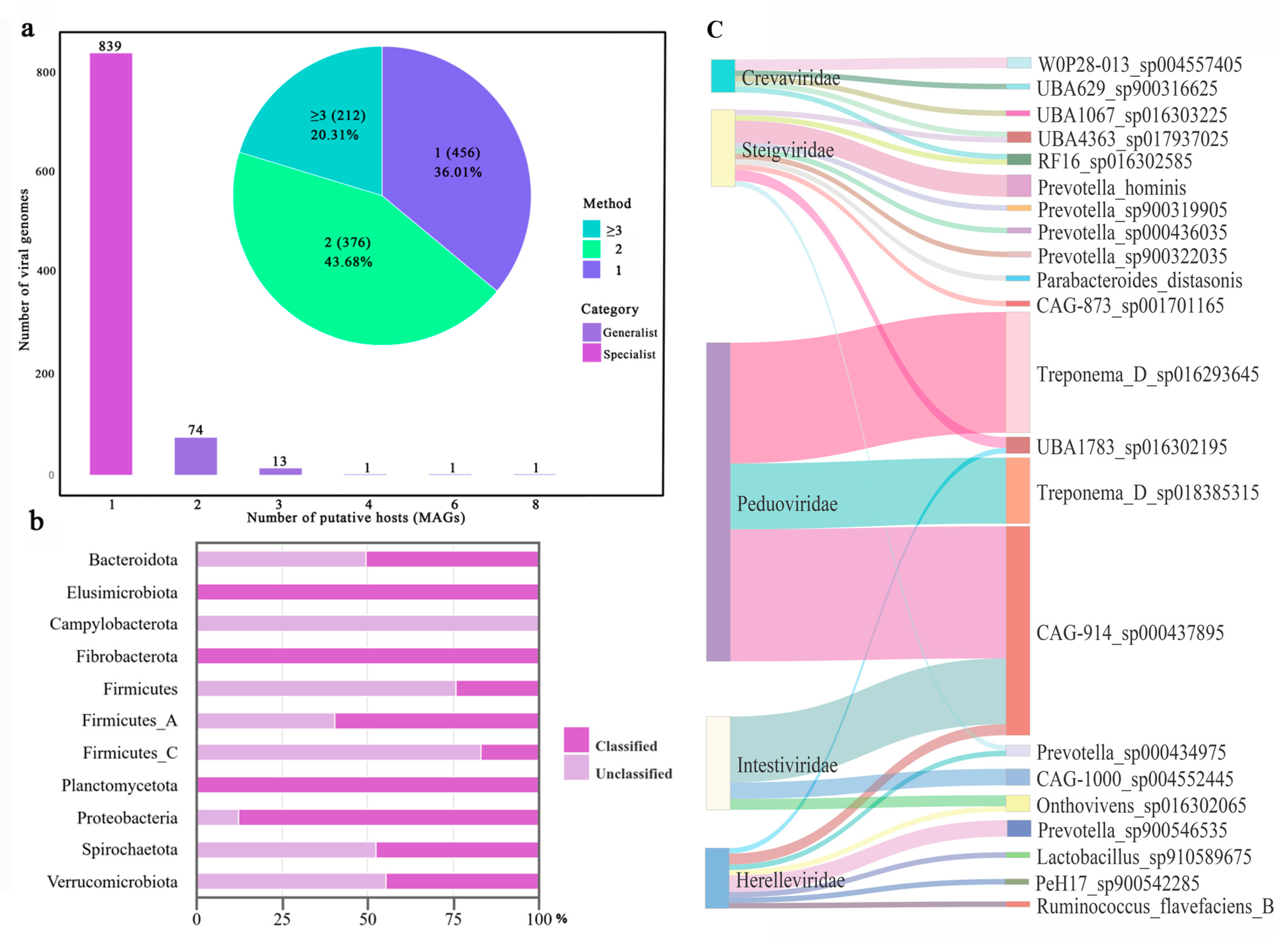
3.3. Viral Host Prediction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, Y.; Yang, Y.; Ma, L.; Liu, J.; An, Q.; Zhang, C.; Yin, G.; Cao, Z.; Pan, H. Comparative Analyses of Antibiotic Resistance Genes in Jejunum Microbiota of Pigs in Different Areas. Front. Cell. Infect. Microbiol. 2022, 12, 887428. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Song, Y.; Lyu, W.; Chen, Q.; Xiao, X.; Jin, Y.; Yang, H.; Wang, W.; Xiao, Y. Longitudinal metagenomic study reveals the dynamics of fecal antibiotic resistome in pigs throughout the lifetime. Anim. Microbiome 2023, 5, 55. [Google Scholar] [CrossRef] [PubMed]
- Ricker, N.; Trachsel, J.; Colgan, P.; Jones, J.; Choi, J.; Lee, J.; Coetzee, J.F.; Howe, A.; Brockmeier, S.L.; Loving, C.L. Toward antibiotic stewardship: Route of antibiotic administration impacts the microbiota and resistance gene diversity in swine feces. Front. Vet. Sci. 2020, 7, 255. [Google Scholar] [CrossRef] [PubMed]
- Authority, E.F.S. The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2019–2020. EFSA J. 2020, 20, e07209. [Google Scholar]
- European Centre for Disease Prevention and Control. Antimicrobial Resistance Surveillance in Europe 2015; Annual Report of the European Antimicrobial Resistance Surveillance Network (EARS-Net); ECDC: Stockholm, Sweden, 2015.
- Zhang, N.; Liu, E.; Tang, A.; Ye, M.C.; Wang, K.; Jia, Q.; Huang, Z. Data-driven analysis of antimicrobial resistance in foodborne pathogens from six states within the US. Int. J. Environ. Res. Public Health 2019, 16, 1811. [Google Scholar] [CrossRef]
- Chai, J.; Zhuang, Y.; Cui, K.; Bi, Y.; Zhang, N. Metagenomics reveals the temporal dynamics of the rumen resistome and microbiome in goat kids. Microbiome 2024, 12, 14. [Google Scholar] [CrossRef]
- Yang, D.; Heederik, D.J.; Mevius, D.J.; Scherpenisse, P.; Luiken, R.E.; Van Gompel, L.; Skarżyńska, M.; Wadepohl, K.; Chauvin, C.; Van Heijnsbergen, E. Risk factors for the abundance of antimicrobial resistance genes aph (3′)-III, erm (B), sul2 and tet (W) in pig and broiler faeces in nine European countries. J. Antimicrob. Chemother. 2022, 77, 969–978. [Google Scholar] [CrossRef]
- Van Gompel, L.; Luiken, R.E.C.; Sarrazin, S.; Munk, P.; Knudsen, B.E.; Hansen, R.B.; Bossers, A.; Aarestrup, F.M.; Dewulf, J.; Wagenaar, J.A.; et al. The antimicrobial resistome in relation to antimicrobial use and biosecurity in pig farming, a metagenome-wide association study in nine European countries. J. Antimicrob. Chemother. 2019, 74, 865–876. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Sui, L.; Kong, D.; Liu, D.; Gao, Y.; Jiang, Y.; Cui, W.; Li, J.; Li, Y.; Wang, L. Porcine epidemic diarrhea virus strain CH/HLJ/18 isolated in China: Characterization and phylogenetic analysis. Virol. J. 2024, 21, 28. [Google Scholar] [CrossRef]
- Li, C.; Wu, Q.; Song, H.; Lu, H.; Yang, K.; Liu, Z.; Liu, W.; Gao, T.; Yuan, F.; Zhu, J. Elucidating the biological characteristics and pathogenicity of the highly virulent G2a porcine epidemic diarrhea virus. J. Gen. Virol. 2024, 105, 001953. [Google Scholar] [CrossRef]
- Shkoporov, A.N.; Stockdale, S.R.; Lavelle, A.; Kondova, I.; Heuston, C.; Upadrasta, A.; Khokhlova, E.V.; Van Der Kamp, I.; Ouwerling, B.; Draper, L.A. Viral biogeography of the mammalian gut and parenchymal organs. Nat. Microbiol. 2022, 7, 1301–1311. [Google Scholar] [CrossRef] [PubMed]
- Munk, P.; Knudsen, B.E.; Lukjancenko, O.; Duarte, A.S.R.; Van Gompel, L.; Luiken, R.E.; Smit, L.A.; Schmitt, H.; Garcia, A.D.; Hansen, R.B. Abundance and diversity of the faecal resistome in slaughter pigs and broilers in nine European countries. Nat. Microbiol. 2018, 3, 898–908. [Google Scholar] [CrossRef] [PubMed]
- Deng, F.; Han, Y.; Huang, Y.; Li, D.; Chai, J.; Deng, L.; Wei, M.; Wu, K.; Zhao, H.; Yang, G. A comprehensive analysis of antibiotic resistance genes in the giant panda gut. iMeta 2024, 3, e171. [Google Scholar] [CrossRef] [PubMed]
- Antipov, D.; Korobeynikov, A.; McLean, J.S.; Pevzner, P.A. hybridSPAdes: An algorithm for hybrid assembly of short and long reads. Bioinformatics 2016, 32, 1009–1015. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.D.; Li, F.; Kirton, E.; Thomas, A.; Egan, R.; An, H.; Wang, Z. MetaBAT 2: An adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 2019, 7, e7359. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Olm, M.R.; Brown, C.T.; Brooks, B.; Banfield, J.F. dRep: A tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J. 2017, 11, 2864–2868. [Google Scholar] [CrossRef]
- Chaumeil, P.-A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: A Toolkit to Classify Genomes with the Genome Taxonomy Database; Oxford University Press: Oxford, UK, 2020. [Google Scholar]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.-L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed]
- Arango-Argoty, G.; Garner, E.; Pruden, A.; Heath, L.S.; Vikesland, P.; Zhang, L. DeepARG: A deep learning approach for predicting antibiotic resistance genes from metagenomic data. Microbiome 2018, 6, 23. [Google Scholar] [CrossRef] [PubMed]
- Mi, J.; Jing, X.; Ma, C.; Yang, Y.; Li, Y.; Zhang, Y.; Long, R.; Zheng, H. Massive expansion of the pig gut virome based on global metagenomic mining. Npj Biofilms Microbiomes 2024, 10, 76. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Feng, T.; Wu, Y.; Xu, Y.; Du, L.; Wang, T.; Luo, Y.; Wang, Y.; Li, Z.; Xuan, Z. The multi-kingdom microbiome of the goat gastrointestinal tract. Microbiome 2023, 11, 219. [Google Scholar] [CrossRef]
- Ryu, W.-S. Molecular Virology of Human Pathogenic Viruses; Academic Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Nayfach, S.; Roux, S.; Seshadri, R.; Udwary, D.; Varghese, N.; Schulz, F.; Wu, D.; Paez-Espino, D.; Chen, I.-M.; Huntemann, M. A genomic catalog of Earth’s microbiomes. Nat. Biotechnol. 2021, 39, 499–509. [Google Scholar] [CrossRef]
- Deng, F.; Wang, C.; Li, D.; Peng, Y.; Deng, L.; Zhao, Y.; Zhang, Z.; Wei, M.; Wu, K.; Zhao, J. The unique gut microbiome of giant pandas involved in protein metabolism contributes to the host’s dietary adaption to bamboo. Microbiome 2023, 11, 180. [Google Scholar] [CrossRef]
- Jiminez, J.A.; Uwiera, T.C.; Abbott, D.W.; Uwiera, R.R.; Inglis, G.D. Impacts of resistant starch and wheat bran consumption on enteric inflammation in relation to colonic bacterial community structures and short-chain fatty acid concentrations in mice. Gut Pathog. 2016, 8, 67. [Google Scholar] [CrossRef]
- Wielgosz-Grochowska, J.P.; Domanski, N.; Drywień, M.E. Efficacy of an irritable bowel syndrome diet in the treatment of small intestinal bacterial overgrowth: A narrative review. Nutrients 2022, 14, 3382. [Google Scholar] [CrossRef]
- Bevins, C.L.; Salzman, N.H. The potter’s wheel: The host’s role in sculpting its microbiota. Cell. Mol. Life Sci. 2011, 68, 3675–3685. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Li, A.; Shi, C.; Chen, L.; Zhao, Z.; Yin, X.; Zhang, Q.; Huang, Y.; Pan, H. Mulberry branch fiber improved lipid metabolism and egg yolk fatty acid composition of laying hens via the enterohepatic axis. Microbiome 2024, 12, 73. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Tsai, T.; Deng, F.; Wei, X.; Chai, J.; Knapp, J.; Apple, J.; Maxwell, C.V.; Lee, J.A.; Li, Y.; et al. Longitudinal investigation of the swine gut microbiome from birth to market reveals stage and growth performance associated bacteria. Microbiome 2019, 7, 109. [Google Scholar] [CrossRef] [PubMed]
- Holman, D.B.; Kommadath, A.; Tingley, J.P.; Abbott, D.W. Novel insights into the pig gut microbiome using metagenome-assembled genomes. Microbiol. Spectr. 2022, 10, e0238022. [Google Scholar] [CrossRef] [PubMed]
- Lapidus, A.L.; Korobeynikov, A.I. Metagenomic data assembly–the way of decoding unknown microorganisms. Front. Microbiol. 2021, 12, 613791. [Google Scholar] [CrossRef]
- Ma, L.; Lyu, W.; Zeng, T.; Wang, W.; Chen, Q.; Zhao, J.; Zhang, G.; Lu, L.; Yang, H.; Xiao, Y. Duck gut metagenome reveals the microbiome signatures linked to intestinal regional, temporal development, and rearing condition. iMeta 2024, 3, e198. [Google Scholar] [CrossRef]
- Sun, J.; Chen, C.; Cui, C.-Y.; Zhang, Y.; Liu, X.; Cui, Z.-H.; Ma, X.-Y.; Feng, Y.; Fang, L.-X.; Lian, X.-L. Plasmid-encoded tet (X) genes that confer high-level tigecycline resistance in Escherichia coli. Nat. Microbiol. 2019, 4, 1457–1464. [Google Scholar] [CrossRef]
- Deckert, A.; Gow, S.; Rosengren, L.; Leger, D.; Avery, B.; Daignault, D.; Dutil, L.; Reid-Smith, R.; Irwin, R. Canadian integrated program for antimicrobial resistance surveillance (CIPARS) farm program: Results from finisher pig surveillance. Zoonoses Public Health 2010, 57, 71–84. [Google Scholar] [CrossRef]
- Part IV, USDA. Health and Health Management on US Feedlots with a Capacity of 1000 or More Head; APHIS, Ed.; National Animal Health Monitoring System: Fort Collins, CO, USA, 2011.
- Zaheer, R.; Cook, S.R.; Barbieri, R.; Goji, N.; Cameron, A.; Petkau, A.; Polo, R.O.; Tymensen, L.; Stamm, C.; Song, J. Surveillance of Enterococcus spp. reveals distinct species and antimicrobial resistance diversity across a One-Health continuum. Sci. Rep. 2020, 10, 3937. [Google Scholar]
- Tang, K.L.; Caffrey, N.P.; Nóbrega, D.B.; Cork, S.C.; Ronksley, P.E.; Barkema, H.W.; Polachek, A.J.; Ganshorn, H.; Sharma, N.; Kellner, J.D. Restricting the use of antibiotics in food-producing animals and its associations with antibiotic resistance in food-producing animals and human beings: A systematic review and meta-analysis. Lancet Planet. Health 2017, 1, e316–e327. [Google Scholar] [CrossRef]
- Wang, Z.; Yun, H.; Li, S.; Ji, J.; Khan, A.; Fu, X.; Zhang, P.; Li, X. Factors influencing the transfer and abundance of antibiotic resistance genes in livestock environments in China. Int. J. Environ. Sci. Technol. 2023, 20, 2197–2208. [Google Scholar] [CrossRef]
- Andrade-Martínez, J.S.; Camelo Valera, L.C.; Chica Cardenas, L.A.; Forero-Junco, L.; López-Leal, G.; Moreno-Gallego, J.L.; Rangel-Pineros, G.; Reyes, A. Computational tools for the analysis of uncultivated phage genomes. Microbiol. Mol. Biol. Rev. 2022, 86, e0000421. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, B.; Yang, J.; Chen, R.; Chai, J.; Wei, X.; Zhao, J.; Zhao, Y.; Deng, F.; Li, Y. Metagenome-Assembled Genomes of Pig Fecal Samples in Nine European Countries: Insights into Antibiotic Resistance Genes and Viruses. Microorganisms 2024, 12, 2409. https://doi.org/10.3390/microorganisms12122409
Yang B, Yang J, Chen R, Chai J, Wei X, Zhao J, Zhao Y, Deng F, Li Y. Metagenome-Assembled Genomes of Pig Fecal Samples in Nine European Countries: Insights into Antibiotic Resistance Genes and Viruses. Microorganisms. 2024; 12(12):2409. https://doi.org/10.3390/microorganisms12122409
Chicago/Turabian StyleYang, Boxuan, Jianbo Yang, Routing Chen, Jianmin Chai, Xiaoyuan Wei, Jiangchao Zhao, Yunxiang Zhao, Feilong Deng, and Ying Li. 2024. "Metagenome-Assembled Genomes of Pig Fecal Samples in Nine European Countries: Insights into Antibiotic Resistance Genes and Viruses" Microorganisms 12, no. 12: 2409. https://doi.org/10.3390/microorganisms12122409
APA StyleYang, B., Yang, J., Chen, R., Chai, J., Wei, X., Zhao, J., Zhao, Y., Deng, F., & Li, Y. (2024). Metagenome-Assembled Genomes of Pig Fecal Samples in Nine European Countries: Insights into Antibiotic Resistance Genes and Viruses. Microorganisms, 12(12), 2409. https://doi.org/10.3390/microorganisms12122409






