Gut Microbiota Comparison in Rectal Swabs Versus Stool Samples in Cats with Kidney Stones
Abstract
1. Introduction
2. Materials and Methods
2.1. Studied Population
2.1.1. Recruitment of Cats
2.1.2. Living Conditions
2.1.3. Inclusion Criteria
2.2. Collection of Fecal Samples
2.2.1. Rectal Swab
2.2.2. Fresh and Non-Fresh Stool
2.3. Stool Analysis
2.4. DNA Isolation and 16S rRNA Amplicon Sequencing
2.4.1. High-Throughput Bacterial DNA Sequencing Techniques Used
2.4.2. DNA Extraction
2.4.3. 16S rRNA Gene Amplification and Sequencing
2.4.4. Data Processing
2.5. Statistical Analyzes
3. Results
3.1. Studied Population
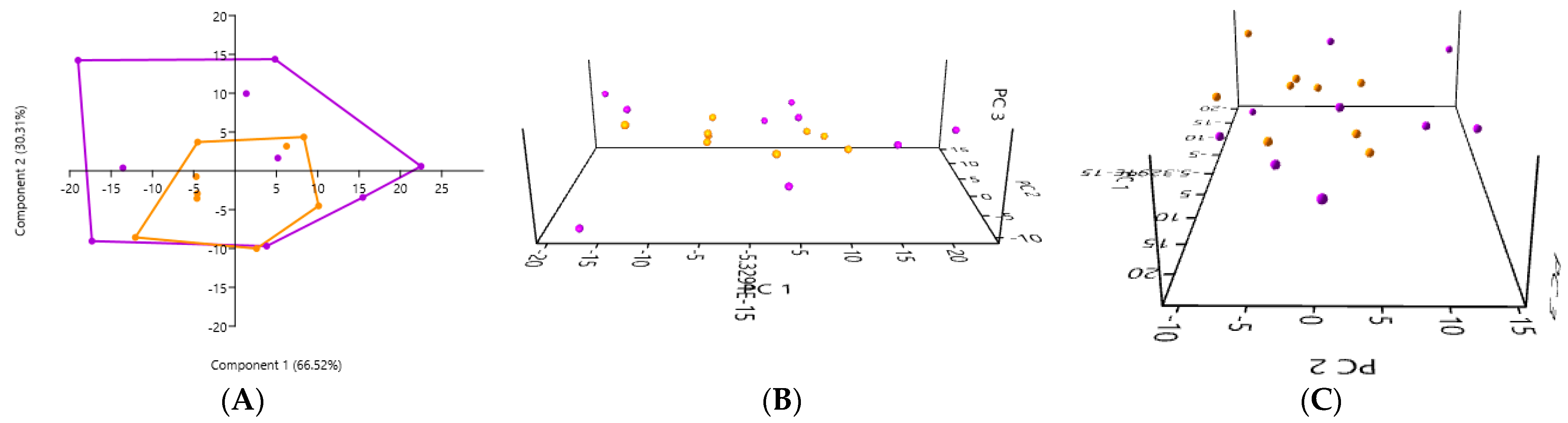
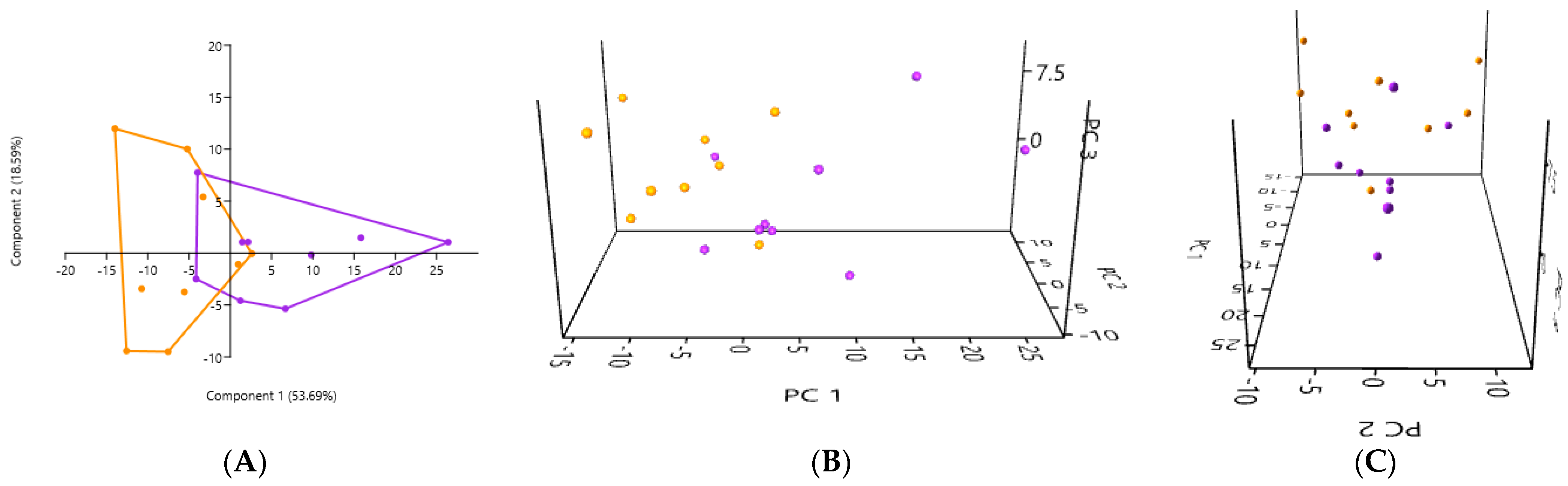
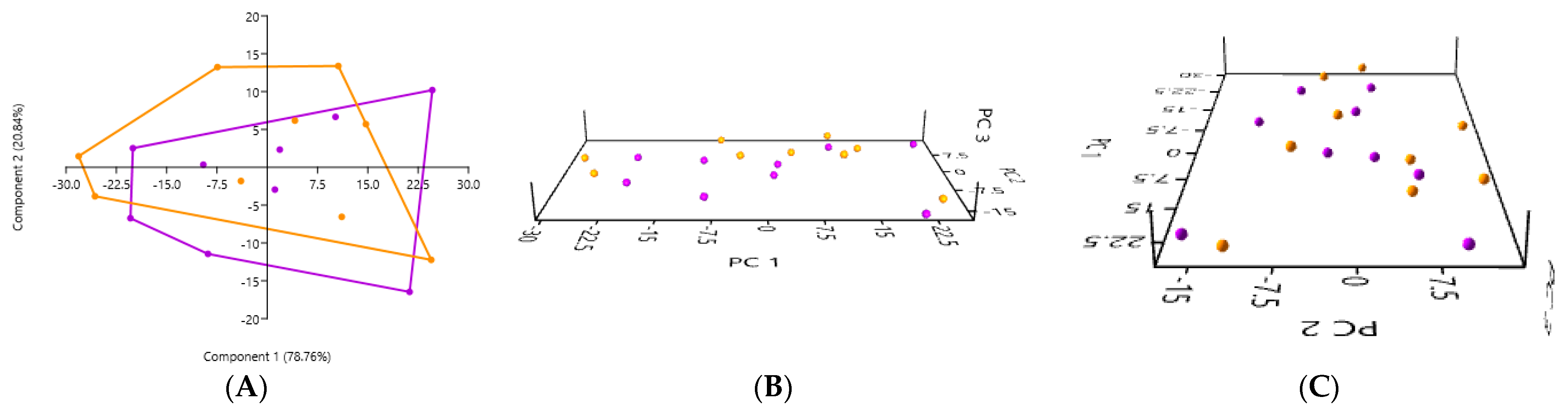
3.2. PCA According to the Collection Method and the Cat Population Studied
3.2.1. A Comparison of the Three Stool Collection Methods in the Healthy Cats
3.2.2. A Comparison of the Three Stool Collection Methods in the Kidney Stone-Diseased Cats
3.2.3. A Comparison of the Three Stool Collection Methods Among All Cats
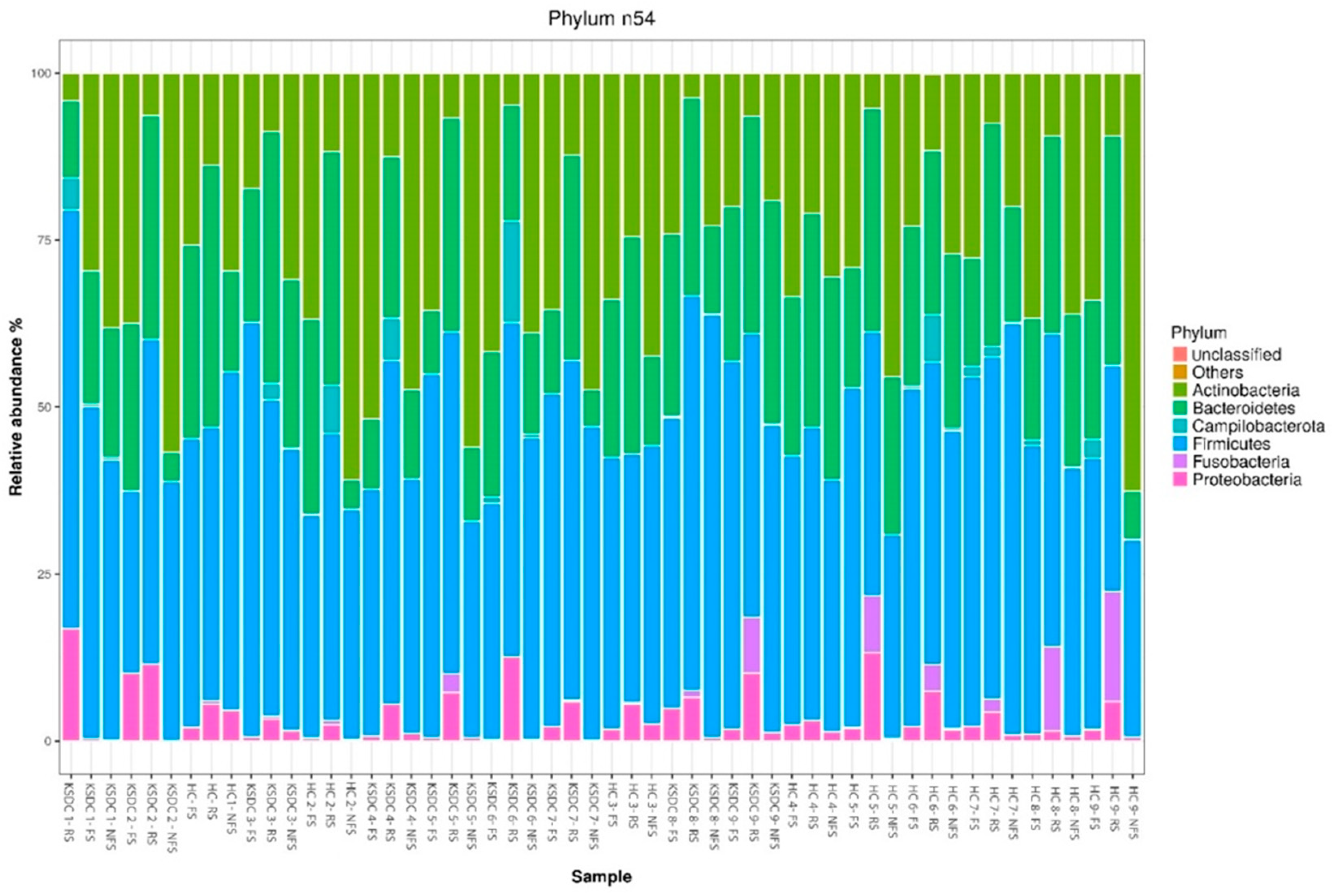
3.3. A Comparison of Main Bacterial Phyla According to the Method of Collection and the Studied Cat Population
3.3.1. Comparison of Bacterial Phyla Between Rectal Swab and Fresh Stool in Healthy Cats
| Phylum | Mean ± SEM for RS–HC | Mean ± SEM for FS–HC | p-Value | Significant |
|---|---|---|---|---|
| Actinobacteria | 12.697 ± 6.276 | 31.161 ± 4.983 | 0.0039 | Yes ** |
| Bacteroidetes | 32.681 ± 3.985 | 22.530 ± 4.657 | 0.0039 | Yes ** |
| Campylobacterota | 1.757 ± 3.046 | 0.653 ± 0.958 | 0.8125 | No |
| Candidatus Saccharibacteria | 0 | 0 | / | / |
| Chlamydiae | 0 | 0 | / | / |
| Firmicutes | 42,400 ± 5257 | 43.877 ± 6.238 | 0.4961 | No |
| Fusobacteria | 5.003 ± 6.097 | 0.063 ± 0.046 | 0.0039 | Yes ** |
| Proteobacteria | 5.440 ± 3.487 | 1.715 ± 0.65 | 0.0039 | Yes ** |
| Synergistetes | 0.022 ± 0.051 | 0 | 0.5 | No |
| Verrucomicrobia | 0 | 0 | / | / |
| Unclassified | 0 | 0.0003 ± 0.001 | >0.999 | No |
3.3.2. Comparison of Bacterial Phyla Between Rectal Swabs and Fresh Stool in Kidney Stone-Diseased Cats
| Phylum | Mean ± SEM for RS–KSDC | Mean ± SEM for FS–KSDC | p-Value | Significant |
|---|---|---|---|---|
| Actinobacteria | 7.314 ± 3.277 | 32.540 ± 10.987 | 0.0039 | Yes ** |
| Bacteroidetes | 27.702 ± 8.415 | 18.887 ± 6.470 | 0.0547 | No |
| Campylobacterota | 3.194 ± 5.089 | 0.179 ± 0.289 | 0.0938 | No |
| Candidatus Saccharibacteria | 0 | 0 | / | / |
| Chlamydiae | 0 | 0 | / | / |
| Firmicutes | 51.528 ± 6.069 | 46.040 ± 11.116 | 0.25 | No |
| Fusobacteria | 1.431 ± 2.769 | 0.015 ± 0.030 | 0.0156 | Yes * |
| Proteobacteria | 8.830 ± 4.255 | 2.338 ± 3.258 | 0.0039 | Yes ** |
| Synergistetes | 0 | 0 | / | / |
| Verrucomicrobia | 0.0002 ± 0.001 | 0 | >0.999 | No |
| Unclassified | 0 | 0.001 ± 0.001 | 0.25 | No |
3.3.3. Comparison of Bacterial Phyla Between Rectal Swab and Non-Fresh Stool in Healthy Cats
| Phylum | Mean ± SEM for RS–HC | Mean ± SEM for NFS–HC | p-Value | Significant |
|---|---|---|---|---|
| Actinobacteria | 12.697 ± 6.276 | 39.430 ± 14.836 | 0.0078 | Yes ** |
| Bacteroidetes | 32.681 ± 3.985 | 17.821 ± 8.717 | 0.0078 | Yes ** |
| Campylobacterota | 1.757 ± 3.046 | 0.082 ± 0.110 | 0.25 | No |
| Candidatus Saccharibacteria | 0 | 0.001 ± 0.002 | >0.999 | No |
| Chlamydiae | 0 | 0.002 ± 0.003 | 0.5 | No |
| Firmicutes | 42,400 ± 5257 | 41.180 ± 10.196 | 0.8203 | No |
| Fusobacteria | 5.003 ± 6.097 | 0.065 ± 0.078 | 0.0039 | Yes ** |
| Proteobacteria | 5.440 ± 3.487 | 1.419 ± 1.399 | 0.0039 | Yes ** |
| Synergistetes | 0.022 ± 0.051 | 0 | 0.5 | No |
| Verrucomicrobia | 0 | 0.0003 ± 0.001 | >0.999 | No |
| Unclassified | 0 | 0.0003 ± 0.001 | >0.999 | No |
3.3.4. Comparison of Bacterial Phyla Between Rectal Swab and Non-Fresh Stool in Kidney Stone-Diseased Cats
| Phylum | Mean ± SEM for RS–KSDC | Mean ± SEM for NFS–KSDC | p-Value | Significant |
|---|---|---|---|---|
| Actinobacteria | 7.314 ± 3.277 | 39.741 ± 13.530 | 0.0039 | Yes ** |
| Bacteroidetes | 27.702 ± 8.415 | 15.643 ± 9.253 | 0.0273 | Yes * |
| Campylobacterota | 3.194 ± 5.089 | 0.139 ± 0.188 | 0.1563 | No |
| Candidatus Saccharibacteria | 0 | 0.001 ± 0.003 | >0.999 | No |
| Chlamydiae | 0 | 0.0004 ± 0.001 | >0.999 | No |
| Firmicutes | 51.528 ± 6.069 | 43.873 ± 8.599 | 0.0273 | Yes * |
| Fusobacteria | 1.431 ± 2.769 | 0.015 ± 0.029 | 0.0156 | Yes * |
| Proteobacteria | 8.830 ± 4.255 | 0.588 ± 0.570 | 0.0039 | Yes ** |
| Synergistetes | 0 | 0 | / | / |
| Verrucomicrobia | 0.0002 ± 0.001 | 0 | >0.999 | No |
| Unclassified | 0 | 0 | / | / |
3.3.5. Comparison of Bacterial Phyla Between Fresh Stool and Non-Fresh Stool in Healthy Cats
| Phylum | Mean ± SEM for FS–HC | Mean ± SEM for NFS–HC | p-Value | Significant |
|---|---|---|---|---|
| Actinobacteria | 31.161 ± 4.983 | 39.430 ± 14.836 | 0.0977 | No |
| Bacteroidetes | 22.530 ± 4.657 | 17.821 ± 8.717 | 0.4258 | No |
| Campylobacterota | 0.653 ± 0.958 | 0.082 ± 0.110 | 0.0234 | Yes * |
| Candidatus Saccharibacteria | 0 | 0.001 ± 0.002 | >0.999 | No |
| Chlamydiae | 0 | 0.002 ± 0.003 | 0.5 | No |
| Firmicutes | 43.877 ± 6.238 | 41.180 ± 10.196 | 0.4961 | No |
| Fusobacteria | 0.063 ± 0.046 | 0.065 ± 0.078 | >0.999 | No |
| Proteobacteria | 1.715 ± 0.65 | 1.419 ± 1.399 | 0.3008 | No |
| Synergistetes | 0 | 0 | / | / |
| Verrucomicrobia | 0 | 0.0003 ± 0.001 | >0.999 | No |
| Unclassified | 0.0003 ± 0.001 | 0.0003 ± 0.001 | >0.999 | No |
3.3.6. Comparison of Bacterial Phyla Between Fresh Stool and Non-Fresh Stool in Kidney Stone-Diseased Cats
| Phylum | Mean ± SEM for FS–KSDC | Mean ± SEM for NFS–KSDC | p-Value | Significant |
|---|---|---|---|---|
| Actinobacteria | 32.540 ± 10.987 | 39.741 ± 13.530 | 0.1641 | No |
| Bacteroidetes | 18.887 ± 6.470 | 15.643 ± 9.253 | 0.4961 | No |
| Campylobacterota | 0.179 ± 0.289 | 0.139 ± 0.188 | 0.8438 | No |
| Candidatus Saccharibacteria | 0 | 0.001 ± 0.003 | >0.999 | No |
| Chlamydiae | 0 | 0.0004 ± 0.001 | >0.999 | No |
| Firmicutes | 46.040 ± 11.116 | 43.873 ± 8.599 | 0.7344 | No |
| Fusobacteria | 0.015 ± 0.030 | 0.015 ± 0.029 | >0.999 | No |
| Proteobacteria | 2.338 ± 3.258 | 0.588 ± 0.570 | 0.3008 | No |
| Synergistetes | 0 | 0 | / | / |
| Verrucomicrobia | 0 | 0 | / | / |
| Unclassified | 0.001 ± 0.001 | 0 | 0.25 | No |
3.4. Comparison of Relative Abundance by Bacterial Phylum and by Bacterial Genus According to Collection Method and Studied Cat Population
3.5. Comparison of Main Alpha Diversity Indices According to Collection Method and Cat Population Studied
3.6. PCA of Samples Based on Cat Population Studied
3.6.1. A Comparison of the PCA of the Gut Microbiota Obtained by the Rectal Swab in the Healthy Cats and the Kidney Stone-Diseased Cats
3.6.2. A Comparison of the PCA of the Gut Microbiota in Fresh Stool Samples in the Healthy Cats and in the Kidney Stone-Diseased Cats
3.6.3. A Comparison of the PCA of the Gut Microbiota in Non-Fresh Stool Samples in the Healthy Cats and in the Kidney Stone-Diseased Cats
3.7. A Comparison of the Mean Proportion of Bacterial Phyla According to the Population Studied
4. Discussion
4.1. Existing Sampling Techniques
4.1.1. Rectal Swab: A Preferred Method in Certain Hospital Settings
4.1.2. Collection of Spontaneously Produced Stool: The Standard Method
4.1.3. Intestinal Biopsy: A More Invasive Method
4.2. Comparison of Bacterial Microbiota Based on Sampling Method
4.2.1. Rectal Swab
4.2.2. Fresh Stool
4.2.3. Non-Fresh Stool
4.3. Rarity of Certain Phyla
4.4. Comparison of Bacterial Microbiota Based on Studied Population
4.4.1. Comparison of PCA Graphs
4.4.2. Comparison of Bacterial Phyla
Firmicutes
Bacteroidetes
Proteobacteria
Fusobacteria
Actinobacteria
4.4.3. Summary of Bacterial Microbiota Comparison
4.5. Advantages of This Animal Experimental Model
4.6. Study Limitations and Potential Research Directions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huang, Z.; Huang, Z.; Liu, K.; Ma, W.; Li, D.; Mo, T.; Liu, Q. The gut microbiome in human health and disease—Where are we and where are we going? A bibliometric analysis. Front. Microbiol. 2022, 13, 1018594. [Google Scholar] [CrossRef] [PubMed]
- Mehta, M.; Goldfarb, D.S.; Nazzal, L. The role of the microbiome in kidney stone formation. Int. J. Surg. 2016, 36, 607–612. [Google Scholar] [CrossRef] [PubMed]
- Lin, E.; Xu, J.; Liu, M.; Nazzal, L.; Katz, S. Enteric Hyperoxaluria and Kidney Stone Management in Inflammatory Bowel Disease. Curr. Treat. Options Gastroenterol. 2020, 18, 384–393. [Google Scholar] [CrossRef]
- Kachroo, N.; Lange, D.; Penniston, K.L.; Stern, J.; Tasian, G.; Bajic, P.; Wolfe, A.J.; Suryavanshi, M.; Ticinesi, A.; Meschi, T.; et al. Meta-analysis of Clinical Microbiome Studies in Urolithiasis Reveal Age, Stone Composition, and Study Location as the Predominant Factors in Urolithiasis-Associated Microbiome Composition. mBio 2021, 12, e0200721. [Google Scholar] [CrossRef]
- Miller, A.W.; Penniston, K.L.; Fitzpatrick, K.; Agudelo, J.; Tasian, G.; Lange, D. Mechanisms of the intestinal and urinary microbiome in kidney stone disease. Nat. Rev. Urol. 2022, 19, 695–707. [Google Scholar] [CrossRef]
- Galan-Llopis, J.A.; Sanchez-Pellicer, P.; Navarro-Lopez, V. Role of microbiome in kidney stone disease. Curr. Opin. Urol. 2023, 33, 84–89. [Google Scholar] [CrossRef]
- Jung, H.D.; Cho, S.; Lee, J.Y. Update on the Effect of the Urinary Microbiome on Urolithiasis. Diagnostics 2023, 13, 951. [Google Scholar] [CrossRef] [PubMed]
- Yuan, T.; Xia, Y.; Li, B.; Yu, W.; Rao, T.; Ye, Z.; Yan, X.; Song, B.; Li, L.; Lin, F.; et al. Gut microbiota in patients with kidney stones: A systematic review and meta-analysis. BMC Microbiol. 2023, 23, 143. [Google Scholar] [CrossRef]
- Kyles, A.E.; Hardie, E.M.; Wooden, B.G.; Adin, C.A.; Stone, E.A.; Gregory, C.R.; Mathews, K.G.; Cowgill, L.D.; Vaden, S.; Nyland, T.G.; et al. Clinical, clinicopathologic, radiographic, and ultrasonographic abnormalities in cats with ureteral calculi: 163 cases (1984–2002). J. Am. Vet. Med. Assoc. 2005, 226, 932–936. [Google Scholar] [CrossRef]
- Kopecny, L.; Palm, C.A.; Segev, G.; Larsen, J.A.; Westropp, J.L. Urolithiasis in cats: Evaluation of trends in urolith composition and risk factors (2005–2018). J. Vet. Intern. Med. 2021, 35, 1397–1405. [Google Scholar] [CrossRef]
- Lekcharoensuk, C.; Lulich, J.P.; Osborne, C.A.; Koehler, L.A.; Urlich, L.K.; Carpenter, K.A.; Swanson, L.L. Association between patient-related factors and risk of calcium oxalate and magnesium ammonium phosphate urolithiasis in cats. J. Am. Vet. Med. Assoc. 2000, 217, 520–525. [Google Scholar] [CrossRef] [PubMed]
- Lekcharoensuk, C.; Osborne, C.A.; Lulich, J.P.; Pusoonthornthum, R.; Kirk, C.A.; Ulrich, L.K.; Koehler, L.A.; Carpenter, K.A.; Swanson, L.L. Association between dietary factors and calcium oxalate and magnesium ammonium phosphate urolithiasis in cats. J. Am. Vet. Med. Assoc. 2001, 219, 1228–1237. [Google Scholar] [CrossRef] [PubMed]
- Lekcharoensuk, C.; Osborne, C.A.; Lulich, J.P.; Albasan, H.; Ulrich, L.K.; Koehler, L.A.; Carpenter, K.A.; Swanson, L.L.; Pederson, L.A. Trends in the frequency of calcium oxalate uroliths in the upper urinary tract of cats. J. Am. Anim. Hosp. Assoc. 2005, 41, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Cannon, A.B.; Westropp, J.L.; Ruby, A.L.; Kass, P.H. Evaluation of trends in urolith composition in cats: 5230 cases (1985–2004). J. Am. Vet. Med. Assoc. 2007, 231, 570–576. [Google Scholar] [CrossRef]
- Osborne, C.A.; Lulich, J.P.; Kruger, J.M.; Ulrich, L.K.; Koehler, L.A. Analysis of 451,891 canine uroliths, feline uroliths, and feline urethral plugs from 1981 to 2007: Perspectives from the Minnesota Urolith Center. Vet. Clin. N. Am. Small Anim. Pract. 2009, 39, 183–197. [Google Scholar] [CrossRef]
- Joubran, P.; Roux, F.A.; Serino, M.; Deschamps, J.Y. Gut and Urinary Microbiota in Cats with Kidney Stones. Microorganisms 2024, 12, 1098. [Google Scholar] [CrossRef]
- Cammarota, G.; Ianiro, G.; Bibbo, S.; Gasbarrini, A. Gut microbiota modulation: Probiotics, antibiotics or fecal microbiota transplantation? Intern. Emerg. Med. 2014, 9, 365–373. [Google Scholar] [CrossRef]
- Vemuri, R.C.; Gundamaraju, R.; Shinde, T.; Eri, R. Therapeutic interventions for gut dysbiosis and related disorders in the elderly: Antibiotics, probiotics or faecal microbiota transplantation? Benef. Microbes 2017, 8, 179–192. [Google Scholar] [CrossRef]
- Araújo-Pérez, F.; McCoy, A.N.; Okechukwu, C.; Carroll, I.M.; Smith, K.M.; Jeremiah, K.; Sandler, R.S.; Asher, G.N.; Keku, T.O. Differences in microbial signatures between rectal mucosal biopsies and rectal swabs. Gut Microbes 2012, 3, 530–535. [Google Scholar] [CrossRef]
- Espey, M.G. Role of oxygen gradients in shaping redox relationships between the human intestine and its microbiota. Free Radic. Biol. Med. 2013, 55, 130–140. [Google Scholar] [CrossRef]
- Albenberg, L.; Esipova, T.V.; Judge, C.P.; Bittinger, K.; Chen, J.; Laughlin, A.; Grunberg, S.; Baldassano, R.N.; Lewis, J.D.; Li, H.; et al. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology 2014, 147, 1055–1063.e8. [Google Scholar] [CrossRef] [PubMed]
- Stearns, J.C.; Lynch, M.D.; Senadheera, D.B.; Tenenbaum, H.C.; Goldberg, M.B.; Cvitkovitch, D.G.; Croitoru, K.; Moreno-Hagelsieb, G.; Neufeld, J.D. Bacterial biogeography of the human digestive tract. Sci. Rep. 2011, 1, 170. [Google Scholar] [CrossRef] [PubMed]
- Nowicki, C.; Ray, L.; Engen, P.; Madrigrano, A.; Witt, T.; Lad, T.; Cobleigh, M.; Mutlu, E.A. Comparison of gut microbiome composition in colonic biopsies, endoscopically-collected and at-home-collected stool samples. Front. Microbiol. 2023, 14, 1148097. [Google Scholar] [CrossRef]
- Jones, R.B.; Zhu, X.; Moan, E.; Murff, H.J.; Ness, R.M.; Seidner, D.L.; Sun, S.; Yu, C.; Dai, Q.; Fodor, A.A.; et al. Inter-niche and inter-individual variation in gut microbial community assessment using stool, rectal swab, and mucosal samples. Sci. Rep. 2018, 8, 4139. [Google Scholar] [CrossRef]
- Biehl, L.M.; Garzetti, D.; Farowski, F.; Ring, D.; Koeppel, M.B.; Rohde, H.; Schafhausen, P.; Stecher, B.; Vehreschild, M.J.G.T. Usability of rectal swabs for microbiome sampling in a cohort study of hematological and oncological patients. PLoS ONE 2019, 14, e0215428. [Google Scholar] [CrossRef] [PubMed]
- Suchodolski, J.S.; Foster, M.L.; Sohail, M.U.; Leutenegger, C.; Queen, E.V.; Steiner, J.M.; Marks, S.L. The fecal microbiome in cats with diarrhea. PLoS ONE 2015, 10, e0127378. [Google Scholar] [CrossRef]
- Suchodolski, J.S. Analysis of the gut microbiome in dogs and cats. Vet. Clin. Pathol. 2022, 50 (Suppl. S1), 6–17. [Google Scholar] [CrossRef]
- Fraher, M.H.; O’Toole, P.W.; Quigley, E.M. Techniques used to characterize the gut microbiota: A guide for the clinician. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 312–322. [Google Scholar] [CrossRef]
- Lemberger, U.; Pjevac, P.; Hausmann, B.; Berry, D.; Moser, D.; Jahrreis, V.; Ozsoy, M.; Shariat, S.F.; Veser, J. The microbiome of kidney stones and urine of patients with nephrolithiasis. Urolithiasis 2023, 51, 27. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glockner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Ansorge, R.; Birolo, G.; James, S.A.; Telatin, A. Dadaist2: A Toolkit to Automate and Simplify Statistical Analysis and Plotting of Metabarcoding Experiments. Int. J. Mol. Sci. 2021, 22, 5309. [Google Scholar] [CrossRef] [PubMed]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. Past: Paleontological statistics software package for educaton and data analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
- Ritchie, L.E.; Steiner, J.M.; Suchodolski, J.S. Assessment of microbial diversity along the feline intestinal tract using 16S rRNA gene analysis. FEMS Microbiol. Ecol. 2008, 66, 590–598. [Google Scholar] [CrossRef] [PubMed]
- Desai, A.R.; Musil, K.M.; Carr, A.P.; Hill, J.E. Characterization and quantification of feline fecal microbiota using cpn60 sequence-based methods and investigation of animal-to-animal variation in microbial population structure. Vet. Microbiol. 2009, 137, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Handl, S.; Dowd, S.E.; Garcia-Mazcorro, J.F.; Steiner, J.M.; Suchodolski, J.S. Massive parallel 16S rRNA gene pyrosequencing reveals highly diverse fecal bacterial and fungal communities in healthy dogs and cats. FEMS Microbiol. Ecol. 2011, 76, 301–310. [Google Scholar] [CrossRef]
- Barry, K.A.; Middelbos, I.S.; Vester Boler, B.M.; Dowd, S.E.; Suchodolski, J.S.; Henrissat, B.; Coutinho, P.M.; White, B.A.; Fahey, G.C., Jr.; Swanson, K.S. Effects of dietary fiber on the feline gastrointestinal metagenome. J. Proteome Res. 2012, 11, 5924–5933. [Google Scholar] [CrossRef]
- Tun, H.M.; Brar, M.S.; Khin, N.; Jun, L.; Hui, R.K.; Dowd, S.E.; Leung, F.C. Gene-centric metagenomics analysis of feline intestinal microbiome using 454 junior pyrosequencing. J. Microbiol. Methods 2012, 88, 369–376. [Google Scholar] [CrossRef]
- Tal, M.; Verbrugghe, A.; Gomez, D.E.; Chau, C.; Weese, J.S. The effect of storage at ambient temperature on the feline fecal microbiota. BMC Vet. Res. 2017, 13, 256. [Google Scholar] [CrossRef]
- Jha, A.R.; Shmalberg, J.; Tanprasertsuk, J.; Perry, L.; Massey, D.; Honaker, R.W. Characterization of gut microbiomes of household pets in the United States using a direct-to-consumer approach. PLoS ONE 2020, 15, e0227289. [Google Scholar] [CrossRef]
- Ganz, H.H.; Jospin, G.; Rojas, C.A.; Martin, A.L.; Dahlhausen, K.; Kingsbury, D.D.; Osborne, C.X.; Entrolezo, Z.; Redner, S.; Ramirez, B.; et al. The Kitty Microbiome Project: Defining the Healthy Fecal “Core Microbiome” in Pet Domestic Cats. Vet. Sci. 2022, 9, 635. [Google Scholar] [CrossRef]
- Sung, C.-H.; Marsilio, S.; Pilla, R.; Wu, Y.-A.; Cavasin, J.P.; Hong, M.-P.; Suchodolski, J.S. Temporal Variability of the Dominant Fecal Microbiota in Healthy Adult Cats. Vet. Sci. 2024, 11, 31. [Google Scholar] [CrossRef]
- Minamoto, Y.; Hooda, S.; Swanson, K.S.; Suchodolski, J.S. Feline gastrointestinal microbiota. Anim. Health Res. Rev. 2012, 13, 64–77. [Google Scholar] [CrossRef] [PubMed]
- Barko, P.C.; McMichael, M.A.; Swanson, K.S.; Williams, D.A. The Gastrointestinal Microbiome: A Review. J. Vet. Intern. Med. 2018, 32, 9–25. [Google Scholar] [CrossRef] [PubMed]
- Lyu, Y.; Su, C.; Verbrugghe, A.; Van de Wiele, T.; Martos Martinez-Caja, A.; Hesta, M. Past, Present, and Future of Gastrointestinal Microbiota Research in Cats. Front. Microbiol. 2020, 11, 1661. [Google Scholar] [CrossRef] [PubMed]
- Pilla, R.; Suchodolski, J.S. The Gut Microbiome of Dogs and Cats, and the Influence of Diet. Vet. Clin. N. Am. Small Anim. Pract. 2021, 51, 605–621. [Google Scholar] [CrossRef]
- Drut, A.; Mkaouar, H.; Kriaa, A.; Mariaule, V.; Akermi, N.; Méric, T.; Sénécat, O.; Maguin, E.; Hernandez, J.; Rhimi, M. Gut microbiota in cats with inflammatory bowel disease and low-grade intestinal T-cell lymphoma. Front. Microbiol. 2024, 15, 1346639. [Google Scholar] [CrossRef]
- Thomas, V.; Clark, J.; Dore, J. Fecal microbiota analysis: An overview of sample collection methods and sequencing strategies. Future Microbiol. 2015, 10, 1485–1504. [Google Scholar] [CrossRef]
- Budding, A.E.; Grasman, M.E.; Eck, A.; Bogaards, J.A.; Vandenbroucke-Grauls, C.M.; van Bodegraven, A.A.; Savelkoul, P.H. Rectal swabs for analysis of the intestinal microbiota. PLoS ONE 2014, 9, e101344. [Google Scholar] [CrossRef] [PubMed]
- Bassis, C.M.; Moore, N.M.; Lolans, K.; Seekatz, A.M.; Weinstein, R.A.; Young, V.B.; Hayden, M.K.; CDC Prevention Epicenters Program. Comparison of stool versus rectal swab samples and storage conditions on bacterial community profiles. BMC Microbiol. 2017, 17, 78. [Google Scholar] [CrossRef]
- Salonen, A.; Nikkila, J.; Jalanka-Tuovinen, J.; Immonen, O.; Rajilic-Stojanovic, M.; Kekkonen, R.A.; Palva, A.; de Vos, W.M. Comparative analysis of fecal DNA extraction methods with phylogenetic microarray: Effective recovery of bacterial and archaeal DNA using mechanical cell lysis. J. Microbiol. Methods 2010, 81, 127–134. [Google Scholar] [CrossRef]
- Wu, W.K.; Chen, C.C.; Panyod, S.; Chen, R.A.; Wu, M.S.; Sheen, L.Y.; Chang, S.C. Optimization of fecal sample processing for microbiome study—The journey from bathroom to bench. J. Formos. Med. Assoc. 2019, 118, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Cardona, S.; Eck, A.; Cassellas, M.; Gallart, M.; Alastrue, C.; Dore, J.; Azpiroz, F.; Roca, J.; Guarner, F.; Manichanh, C. Storage conditions of intestinal microbiota matter in metagenomic analysis. BMC Microbiol. 2012, 12, 158. [Google Scholar] [CrossRef] [PubMed]
- Lieske, J.C. Probiotics for prevention of urinary stones. Ann. Transl. Med. 2017, 5, 29. [Google Scholar] [CrossRef] [PubMed]
- Wolff, B.J.; Price, T.K.; Joyce, C.J.; Wolfe, A.J.; Mueller, E.R. Oral probiotics and the female urinary microbiome: A double-blinded randomized placebo-controlled trial. Int. Urol. Nephrol. 2019, 51, 2149–2159. [Google Scholar] [CrossRef]
- Miller, A.W.; Oakeson, K.F.; Dale, C.; Dearing, M.D. Microbial Community Transplant Results in Increased and Long-Term Oxalate Degradation. Microb. Ecol. 2016, 72, 470–478. [Google Scholar] [CrossRef] [PubMed]
- Stern, J.M.; Urban-Maldonado, M.; Usyk, M.; Granja, I.; Schoenfeld, D.; Davies, K.P.; Agalliu, I.; Asplin, J.; Burk, R.; Suadicani, S.O. Fecal transplant modifies urine chemistry risk factors for urinary stone disease. Physiol. Rep. 2019, 7, e14012. [Google Scholar] [CrossRef]
- Hall, J.A.; Jackson, M.I.; Jewell, D.E.; Ephraim, E. Chronic kidney disease in cats alters response of the plasma metabolome and fecal microbiome to dietary fiber. PLoS ONE 2020, 15, e0235480. [Google Scholar] [CrossRef]
- Jewell, D.E.; Tavener, S.K.; Hollar, R.L.; Panickar, K.S. Metabolomic changes in cats with renal disease and calcium oxalate uroliths. Metabolomics 2022, 18, 68. [Google Scholar] [CrossRef]
- Kim, Y.S.; Unno, T.; Kim, B.Y.; Park, M.S. Sex Differences in Gut Microbiota. World J. Mens. Health 2020, 38, 48–60. [Google Scholar] [CrossRef]
- Stern, J.M.; Moazami, S.; Qiu, Y.; Kurland, I.; Chen, Z.; Agalliu, I.; Burk, R.; Davies, K.P. Evidence for a distinct gut microbiome in kidney stone formers compared to non-stone formers. Urolithiasis 2016, 44, 399–407. [Google Scholar] [CrossRef]
- Gnanandarajah, J.S.; Abrahante, J.E.; Lulich, J.P.; Murtaugh, M.P. Presence of Oxalobacter formigenes in the intestinal tract is associated with the absence of calcium oxalate urolith formation in dogs. Urol. Res. 2012, 40, 467–473. [Google Scholar] [CrossRef] [PubMed]
- Gnanandarajah, J.S.; Johnson, T.J.; Kim, H.B.; Abrahante, J.E.; Lulich, J.P.; Murtaugh, M.P. Comparative faecal microbiota of dogs with and without calcium oxalate stones. J. Appl. Microbiol. 2012, 113, 745–756. [Google Scholar] [CrossRef] [PubMed]
- Coffey, E.L.; Gomez, A.M.; Burton, E.N.; Granick, J.L.; Lulich, J.P.; Furrow, E. Characterization of the urogenital microbiome in Miniature Schnauzers with and without calcium oxalate urolithiasis. J. Vet. Intern. Med. 2022, 36, 1341–1352. [Google Scholar] [CrossRef] [PubMed]
- Zhernakova, A.; Kurilshikov, A.; Bonder, M.J.; Tigchelaar, E.F.; Schirmer, M.; Vatanen, T.; Mujagic, Z.; Vila, A.V.; Falony, G.; Vieira-Silva, S.; et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 2016, 352, 565–569. [Google Scholar] [CrossRef] [PubMed]
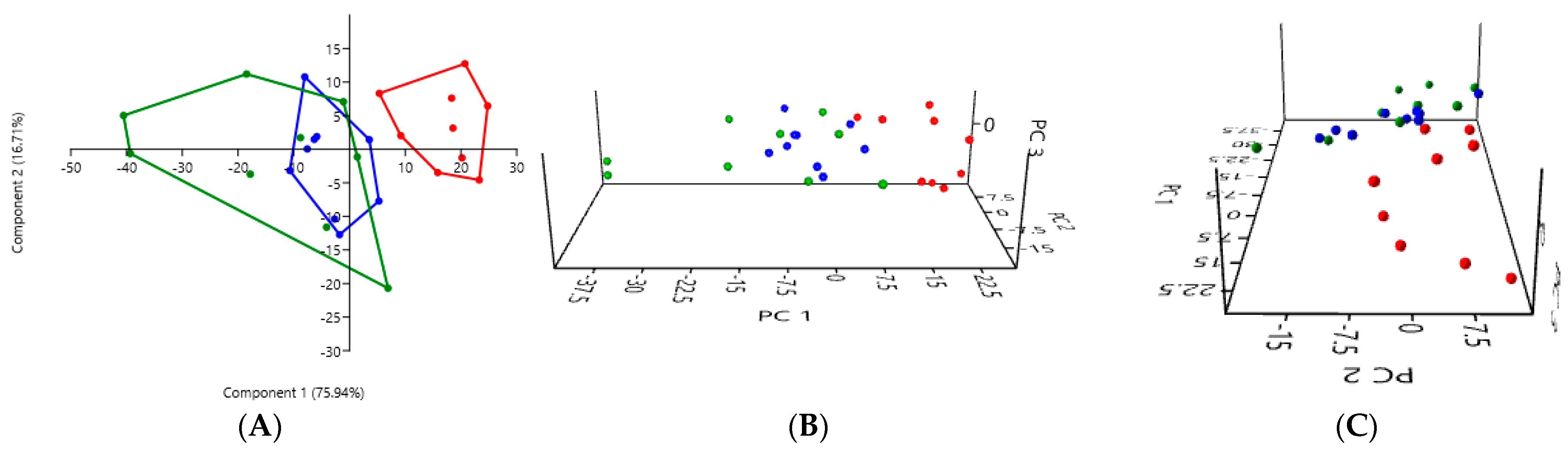
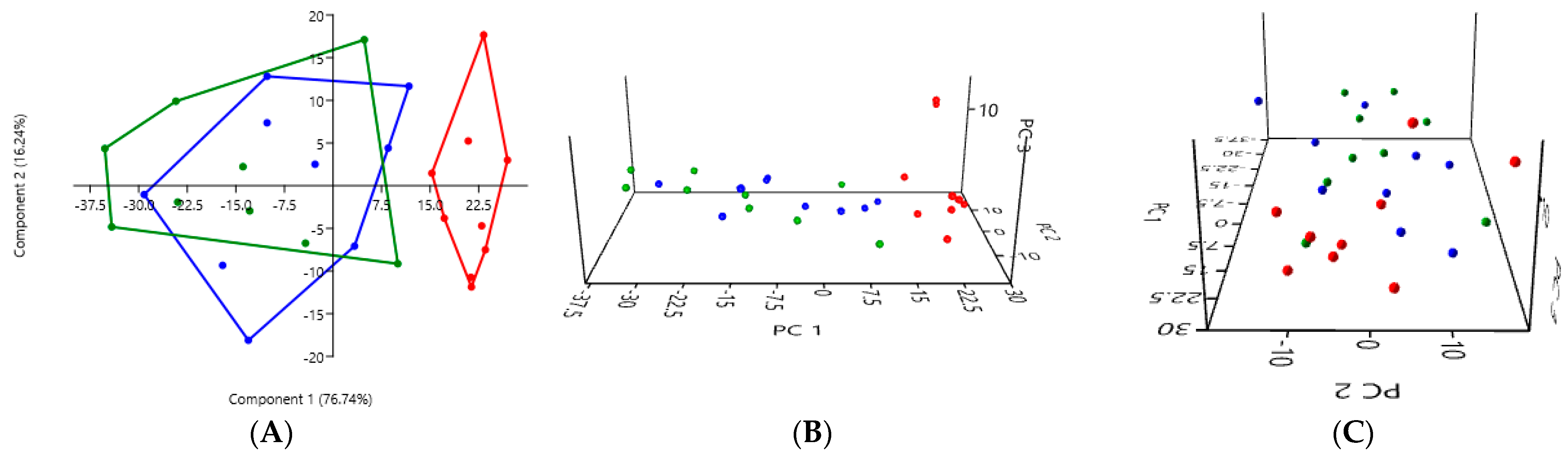
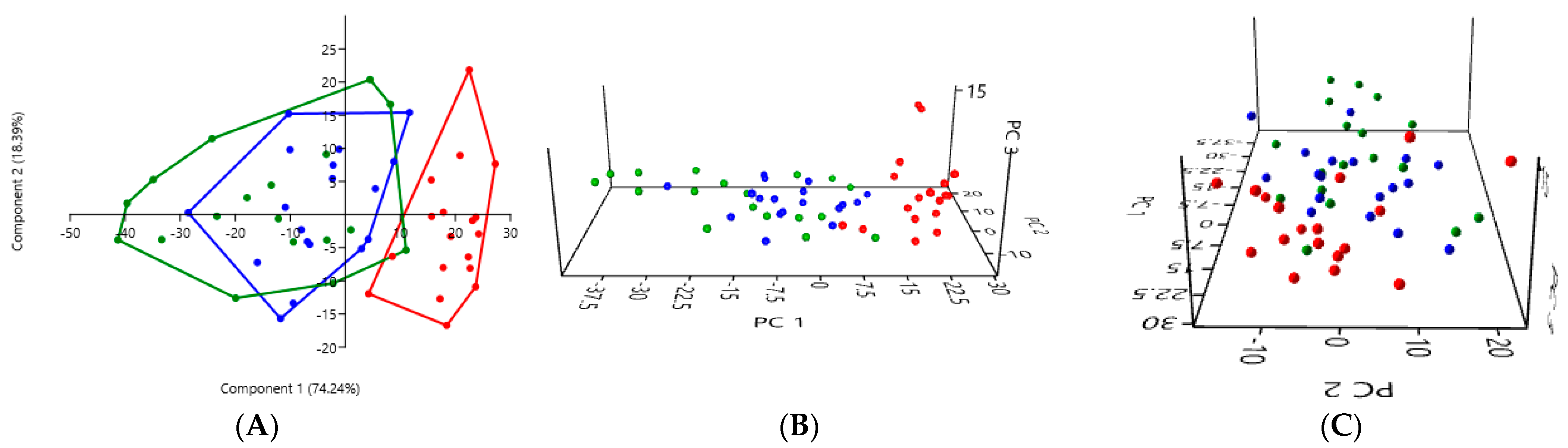
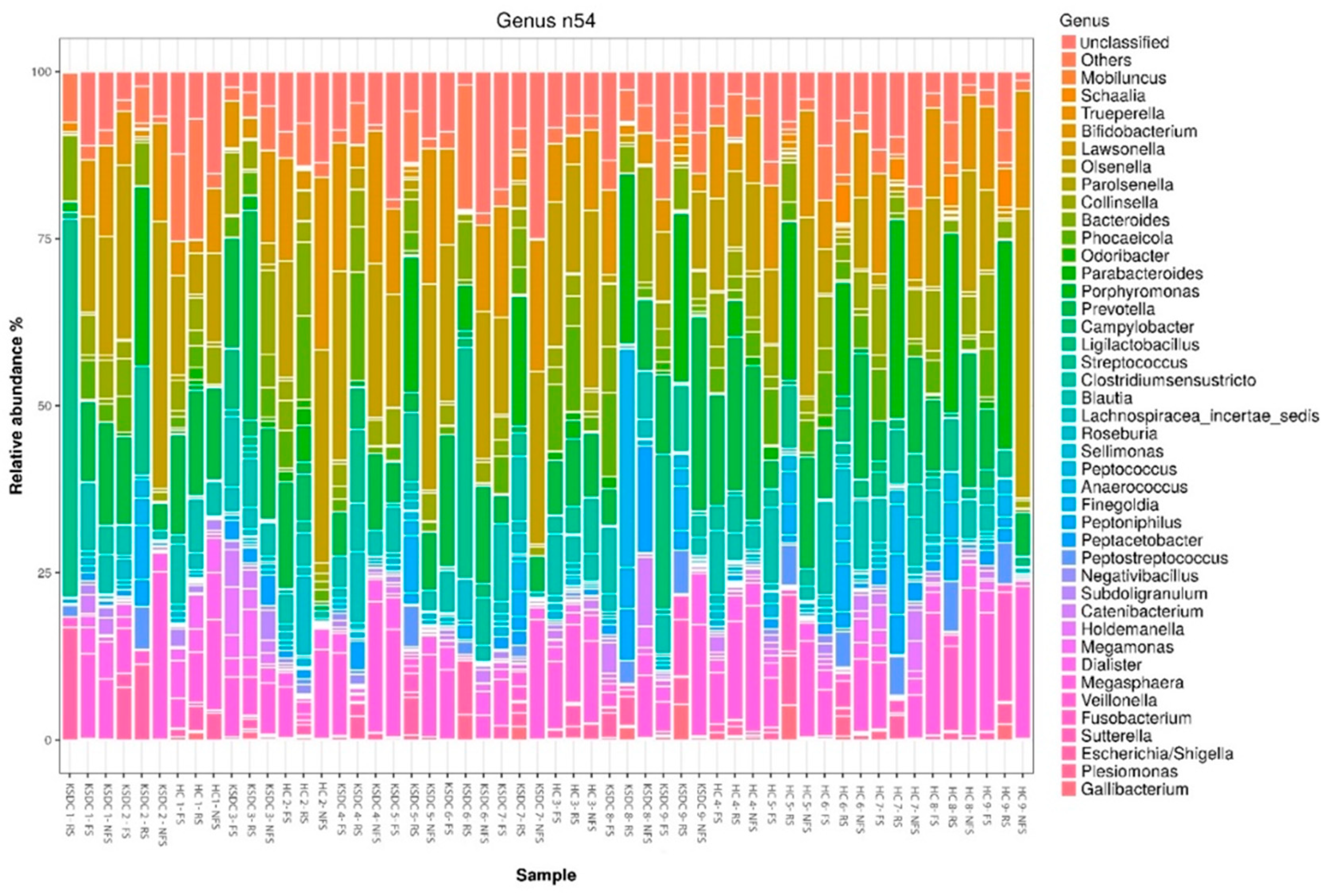
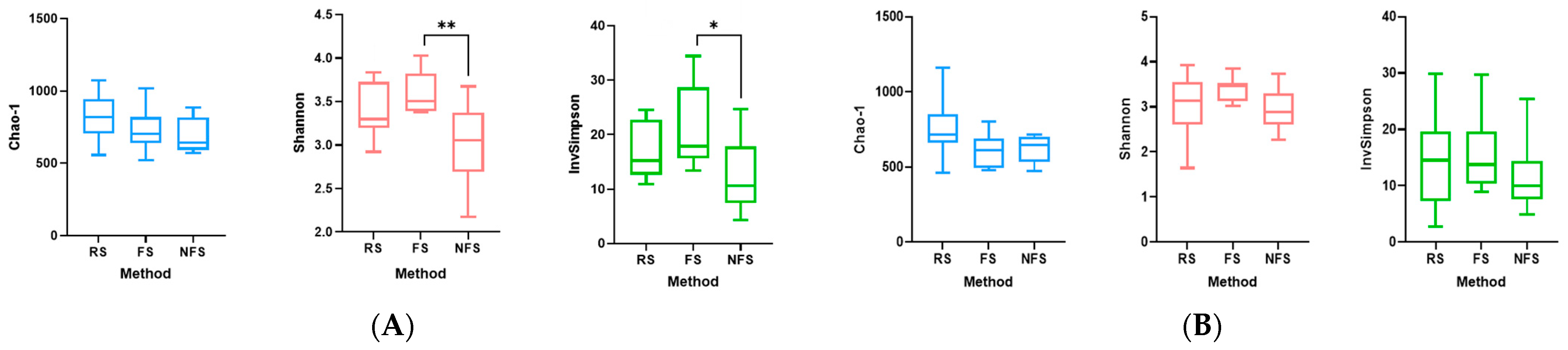
| Rectal Swab | Fresh Stool | Non-Fresh Stool | |
|---|---|---|---|
| Rectal swab | - | 0.0001 *** | 0.0003 *** |
| Fresh stool | 0.0001 *** | - | 0.1927 |
| Non-fresh stool | 0.0003 *** | 0.1927 | - |
| Rectal Swab | Fresh Stool | Non-Fresh Stool | |
|---|---|---|---|
| Rectal swab | - | 0.0001 *** | 0.0001 *** |
| Fresh stool | 0.0001 *** | - | 0.3634 |
| Non-fresh stool | 0.0001 *** | 0.3634 | - |
| Rectal Swab | Fresh Stool | Non-Fresh Stool | |
|---|---|---|---|
| Rectal swab | - | 0.0003 *** | 0.0003 *** |
| Fresh stool | 0.0003 *** | - | 0.0651 |
| Non-fresh stool | 0.0003 *** | 0.0651 | - |
| Phylum | Mean ± SEM for RS–HC | Mean ± SEM for RS–KSDC | p-Value | Significant |
|---|---|---|---|---|
| Actinobacteria | 12.697 ± 6.276 | 7.314 ± 3.277 | 0.0273 | Yes * |
| Bacteroidetes | 32.681 ± 3.985 | 27.702 ± 8.415 | 0.0742 | No |
| Campylobacterota | 1.757 ± 3.046 | 3.194 ± 5.089 | 0.6523 | No |
| Candidatus Saccharibacteria | 0 | 0 | / | / |
| Chlamydiae | 0 | 0 | / | / |
| Firmicutes | 42,400 ± 5257 | 51.528 ± 6.069 | 0.0078 | Yes ** |
| Fusobacteria | 5.003 ± 6.097 | 1.431 ± 2.769 | 0.0117 | Yes * |
| Proteobacteria | 5.440 ± 3.487 | 8.830 ± 4.255 | 0.1289 | No |
| Synergistetes | 0.022 ± 0.051 | 0 | 0.5 | No |
| Verrucomicrobia | 0 | 0.0002 ± 0.001 | 1 | No |
| Unclassified | 0 | 0 | / | / |
| Phylum | Mean ± SEM for FS–HC | Mean ± SEM for FS–KSDC | p-Value | Significant |
|---|---|---|---|---|
| Actinobacteria | 31.161 ± 4.983 | 32.540 ± 10.987 | 0.6523 | No |
| Bacteroidetes | 22.530 ± 4.657 | 18.887 ± 6.470 | 0.1641 | No |
| Campylobacterota | 0.653 ± 0.958 | 0.179 ± 0.289 | 0.4961 | No |
| Candidatus Saccharibacteria | 0 | 0 | / | / |
| Chlamydiae | 0 | 0 | / | / |
| Firmicutes | 43.877 ± 6.238 | 46.040 ± 11.116 | 0.6523 | No |
| Fusobacteria | 0.063 ± 0.046 | 0.015 ± 0.030 | 0.0078 | Yes ** |
| Proteobacteria | 1.715 ± 0.65 | 2.338 ± 3.258 | 0.7344 | No |
| Synergistetes | 0 | 0 | / | / |
| Verrucomicrobia | 0 | 0 | / | / |
| Unclassified | 0.0003 ± 0.001 | 0.001 ± 0.001 | 0.25 | No |
| Phylum | Mean ± SEM for NFS–HC | Mean ± SEM for NFS–KSDC | p-Value | Significant |
|---|---|---|---|---|
| Actinobacteria | 39.430 ± 14.836 | 39.741 ± 13.530 | 0.8203 | No |
| Bacteroidetes | 17.821 ± 8.717 | 15.643 ± 9.253 | 0.5703 | No |
| Campylobacterota | 0.082 ± 0.110 | 0.139 ± 0.188 | 0.7422 | No |
| Candidatus Saccharibacteria | 0.001 ± 0.002 | 0.001 ± 0.003 | 1 | No |
| Chlamydiae | 0.002 ± 0.003 | 0.0004 ± 0.001 | 0.5 | No |
| Firmicutes | 41.180 ± 10.196 | 43.873 ± 8.599 | 0.3008 | No |
| Fusobacteria | 0.065 ± 0.078 | 0.015 ± 0.029 | 0.2969 | No |
| Proteobacteria | 1.419 ± 1.399 | 0.588 ± 0.570 | 0.1289 | No |
| Synergistetes | 0 | 0 | / | / |
| Verrucomicrobia | 0.0003 ± 0.001 | 0 | 1 | No |
| Unclassified | 0.0003 ± 0.001 | 0 | 1 | No |
| Phylum | Ritchie et al. (2008) [33] (Luminal Intestinal Content) | Handl et al. (2011) [35] (Stool) | Barry et al. (2012) [36] (Stool) | Tun et al. (2012) [37] (Stool) | Tal et al. (2017) [38] (Fresh and Non-Fresh Stool) | Jha et al. (2020) [39] (Stool) | Present Study, Joubran et al. (2024) (Rectal Swab) | Present Study, Joubran et al. (2024) (Fresh Stool) | Present Study, Joubran et al. (2024) (Non-Fresh Stool) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 healthy cats | 12 healthy cats | 4 healthy cats | 5 healthy cats | 12 healthy cats | 46 healthy cats | 9 healthy cats | 9 cats with kidney stones | 9 healthy cats | 9 cats with kidney stones | 9 healthy cats | 9 cats with kidney stones | |
| Firmicutes | 68% | 92.1% | 36.3% | 12.98% | ~70–80% | 38.43% | 42.4% | 51.53% | 43.88% | 46.04% | 41.18% | 43.87% |
| Bacteroidetes | 10% | 0.45% | 36.1% | 67.54% | ~3–5% | 14.30% | 32.68% | 27.7% | 22.53% | 18.89% | 17.82% | 15.64% |
| Proteobacteria | 14% | unspecified | 12.4% | 5.85% | ~5–10% | 37.47% | 5.44% | 8.83% | 1.72% | 2.34% | 1.42% | 0.59% |
| Fusobacteria | 5% | 0.04% | - | 0.68% | <1% | 1.58% | 5% | 1.43% | 0.063% | 0.015% | 0.065% | 0.015% |
| Actinobacteria | 4% | 7.31% | 7.7% | 1.16% | ~10–15% | 8.06% | 12.7% | 7.31% | 31.16% | 32.54% | 39.43% | 39.74% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Joubran, P.; Roux, F.A.; Serino, M.; Deschamps, J.-Y. Gut Microbiota Comparison in Rectal Swabs Versus Stool Samples in Cats with Kidney Stones. Microorganisms 2024, 12, 2411. https://doi.org/10.3390/microorganisms12122411
Joubran P, Roux FA, Serino M, Deschamps J-Y. Gut Microbiota Comparison in Rectal Swabs Versus Stool Samples in Cats with Kidney Stones. Microorganisms. 2024; 12(12):2411. https://doi.org/10.3390/microorganisms12122411
Chicago/Turabian StyleJoubran, Patrick, Françoise A. Roux, Matteo Serino, and Jack-Yves Deschamps. 2024. "Gut Microbiota Comparison in Rectal Swabs Versus Stool Samples in Cats with Kidney Stones" Microorganisms 12, no. 12: 2411. https://doi.org/10.3390/microorganisms12122411
APA StyleJoubran, P., Roux, F. A., Serino, M., & Deschamps, J.-Y. (2024). Gut Microbiota Comparison in Rectal Swabs Versus Stool Samples in Cats with Kidney Stones. Microorganisms, 12(12), 2411. https://doi.org/10.3390/microorganisms12122411








