Impact of Mycobacterium tuberculosis H37Rv Infection on Extracellular Vesicle Cargo in Macrophages: Implications for Host–Pathogen Interaction
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Biological Samples
2.2.1. Study Population
2.2.2. Peripheral Blood Mononuclear Cell Isolation
2.2.3. Monocyte Isolation, Culture, and Differentiation
2.2.4. Human Acute Monocytic Leukemia Cells (THP-1)
2.3. Strains and Culture Conditions
2.4. Human Monocyte-Derived Macrophage (MDM/Macrophage) Infection
2.5. Isolation of Extracellular Vesicles (EVs)
2.6. Quantitation of EV Proteins
2.7. Expression of CD9, CD63, CD81, and Syntenin in EVs Using Flow Cytometry
2.8. Internalization of EVs by THP1 Cells
2.9. Identification of Mtb Protein Cargo in EVs Using Western Blotting
2.10. Identification of Mtb RNA Cargo in EVs Using PCR Amplification
2.11. Statistical Analyses
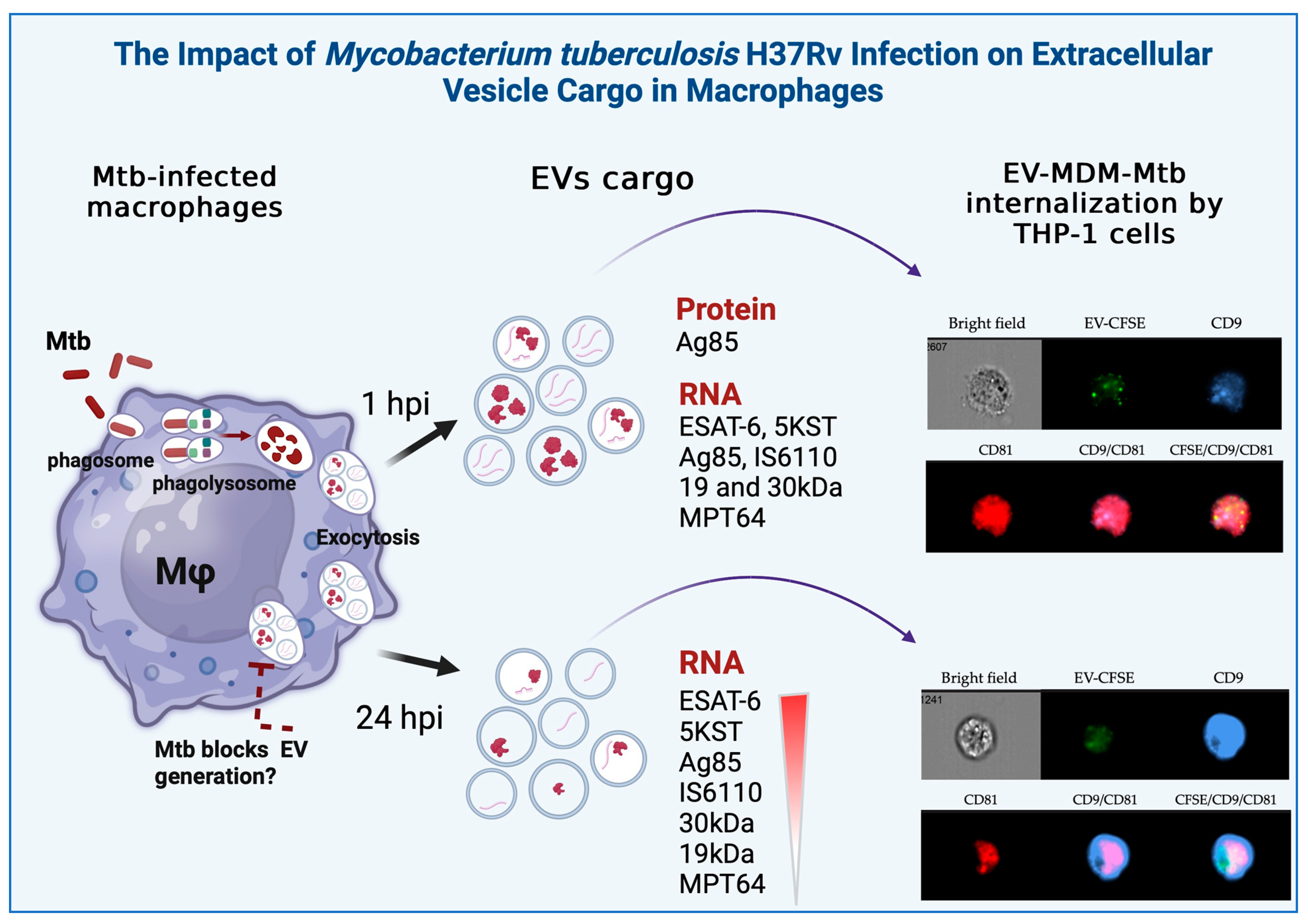
3. Results
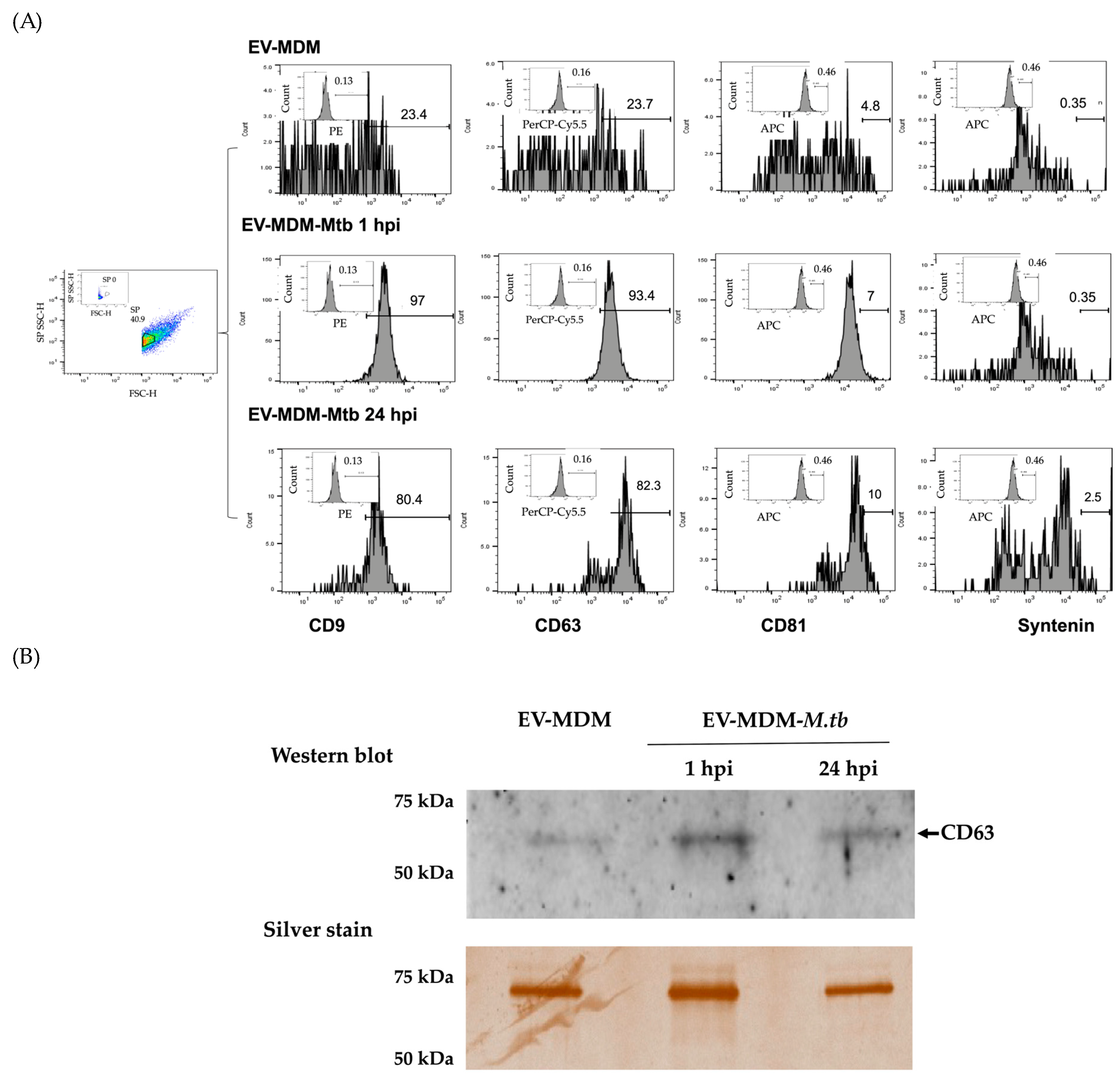
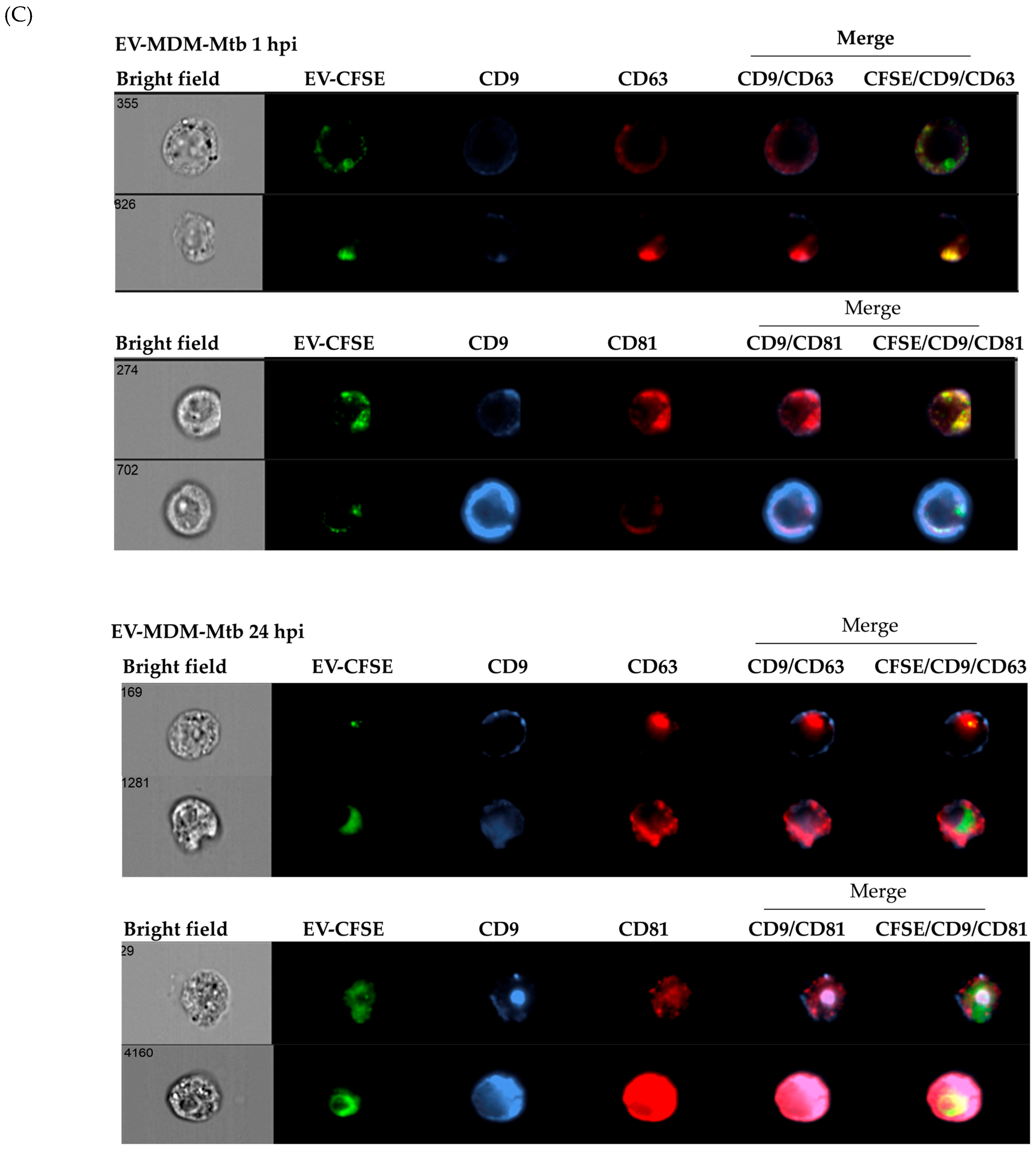
3.1. Characterization of EVs
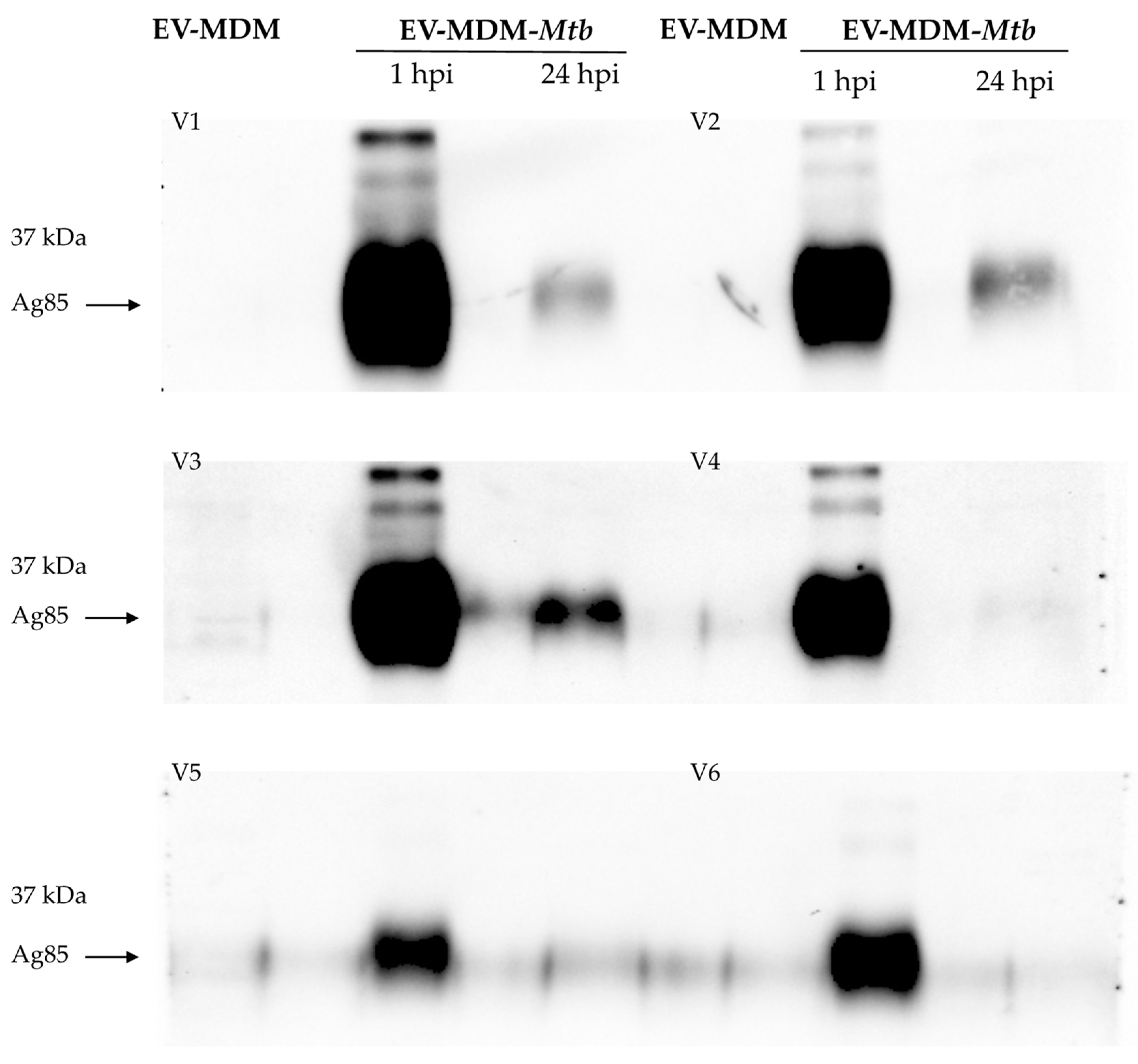
3.2. Detection of Mycobacterial Proteins in EVs
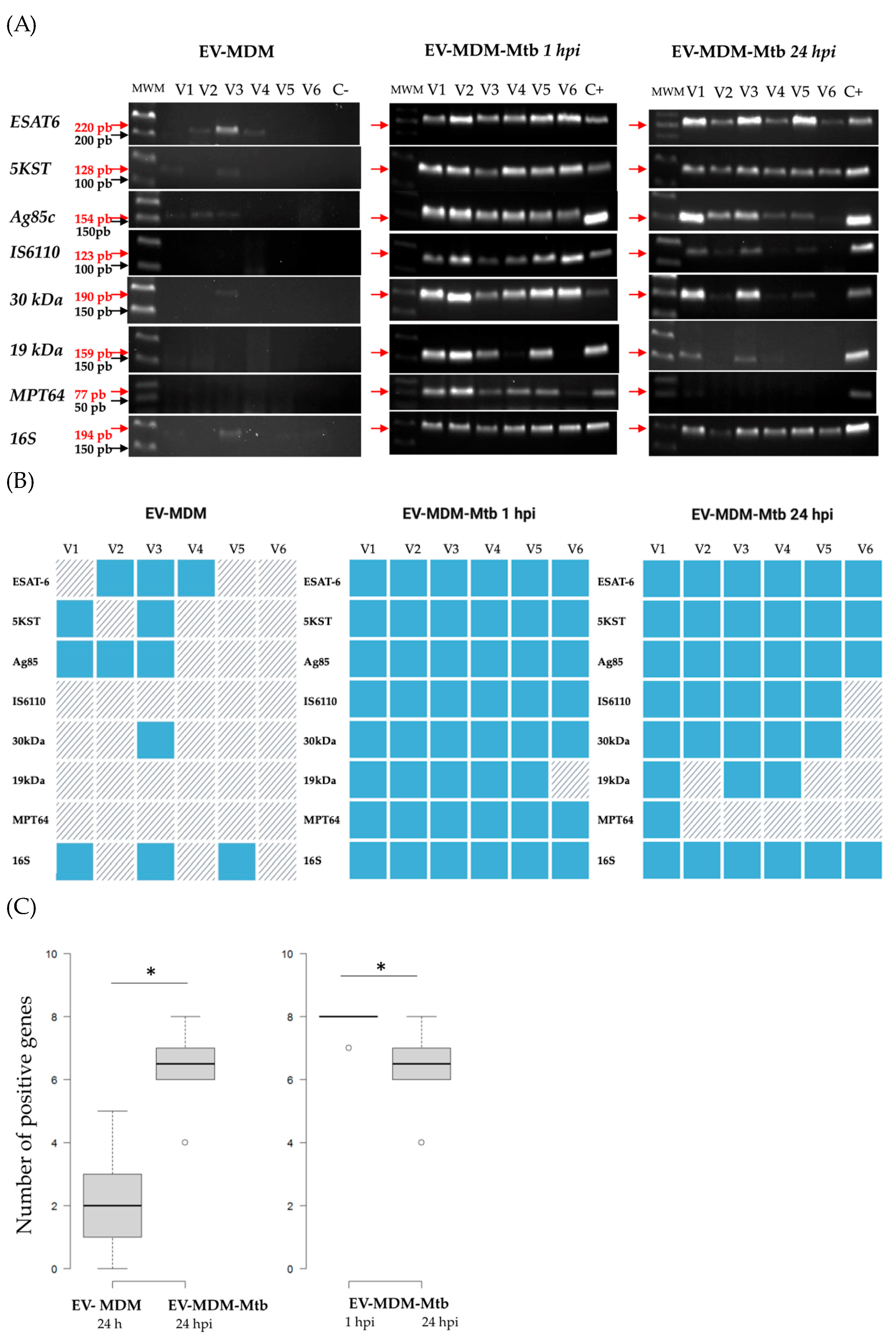
3.3. Detection of Mycobacterial RNAs in EVs
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Global Tuberculosis Report 2024; World Health Organization: Geneva, Switzerland, 2024. Available online: https://www.who.int/teams/global-tuberculosis-programme/tb-reports/global-tuberculosis-report-2024 (accessed on 31 October 2024).
- Ng, C.Y.; Kee, L.T.; Al-Masawa, M.E.; Lee, Q.H.; Subramaniam, T.; Kok, D.; Ng, M.H.; Law, J.X. Scalable Production of Extracellular Vesicles and Its Therapeutic Values: A Review. Int. J. Mol. Sci. 2022, 23, 7986. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Kang, M.H.; Qasim, M.; Khan, K.; Kim, J.H. Biogenesis, Membrane Trafficking, Functions, and next Generation Nanotherapeutics Medicine of Extracellular Vesicles. Int. J. Nanomed. 2021, 16, 3357–3383. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, M.; Liu, S.; Guo, J.; Lu, Y.; Cheng, J.; Liu, J. Macrophage-Derived Extracellular Vesicles: Diverse Mediators of Pathology and Therapeutics in Multiple Diseases. Cell Death Dis. 2020, 11, 924. [Google Scholar] [CrossRef] [PubMed]
- Prados-Rosales, R.; Baena, A.; Martinez, L.R.; Luque-Garcia, J.; Kalscheuer, R.; Veeraraghavan, U.; Camara, C.; Nosanchuk, J.D.; Besra, G.S.; Chen, B.; et al. Mycobacteria Release Active Membrane Vesicles That Modulate Immune Responses in a TLR2-Dependent Manner in Mice. J. Clin. Investig. 2011, 121, 1471–1483. [Google Scholar] [CrossRef]
- Singh, P.P.; Li, L.; Schorey, J.S. Exosomal RNA from Mycobacterium Tuberculosis-Infected Cells Is Functional in Recipient Macrophages. Traffic 2015, 16, 555–571. [Google Scholar] [CrossRef]
- Lyu, L.; Zhang, X.; Li, C.; Yang, T.; Wang, J.; Pan, L.; Jia, H.; Li, Z.; Sun, Q.; Yue, L.; et al. Small RNA Profiles of Serum Exosomes Derived from Individuals with Latent and Active Tuberculosis. Front. Microbiol. 2019, 10, 1174. [Google Scholar] [CrossRef]
- Carranza, C.; Herrera, M.T.; Guzmán-Beltrán, S.; Salgado-Cantú, M.G.; Salido-Guadarrama, I.; Santiago, E.; Chávez-Galán, L.; Gutiérrez-González, L.H.; González, Y. A Dual Marker for Monitoring MDR-TB Treatment: Host-Derived MiRNAs and M. Tuberculosis-Derived RNA Sequences in Serum. Front. Immunol. 2021, 12, 760468. [Google Scholar] [CrossRef]
- Zheng, W.; LaCourse, S.M.; Song, B.; Singh, D.K.; Khanna, M.; Olivo, J.; Stern, J.; Escudero, J.N.; Vergara, C.; Zhang, F.; et al. Diagnosis of Paediatric Tuberculosis by Optically Detecting Two Virulence Factors on Extracellular Vesicles in Blood Samples. Nat. Biomed. Eng. 2022, 6, 979–991. [Google Scholar] [CrossRef]
- Mehaffy, C.; Dobos, K.M.; Nahid, P.; Kruh-Garcia, N.A. Second Generation Multiple Reaction Monitoring Assays for Enhanced Detection of Ultra-Low Abundance Mycobacterium Tuberculosis Peptides in Human Serum. Clin. Proteom. 2017, 14, 21. [Google Scholar] [CrossRef]
- Ramachandra, L.; Qu, Y.; Wang, Y.; Lewis, C.J.; Cobb, B.A.; Takatsu, K.; Boom, W.H.; Dubyak, G.R.; Harding, C.V. Mycobacterium Tuberculosis Synergizes with ATP to Induce Release of Microvesicles and Exosomes Containing Major Histocompatibility Complex Class II Molecules Capable of Antigen Presentation. Infect. Immun. 2010, 78, 5116–5125. [Google Scholar] [CrossRef]
- Bhatnagar, S.; Schorey, J.S. Exosomes Released from Infected Macrophages Contain Mycobacterium Avium Glycopeptidolipids and Are Proinflammatory. J. Biol. Chem. 2007, 282, 25779–25789. [Google Scholar] [CrossRef] [PubMed]
- García-Martínez, M.; Vázquez-Flores, L.; Álvarez-Jiménez, V.D.; Castañeda-Casimiro, J.; Ibáñez-Hernández, M.; Sánchez-Torres, L.E.; Barrios-Payán, J.; Mata-Espinosa, D.; Estrada-Parra, S.; Chacón-Salinas, R.; et al. Extracellular Vesicles Released by J774A.1 Macrophages Reduce the Bacterial Load in Macrophages and in an Experimental Mouse Model of Tuberculosis. Int. J. Nanomed. 2019, 14, 6707–6719. [Google Scholar] [CrossRef] [PubMed]
- Raj, A.; Singh, N.; Gupta, K.B.; Chaudhary, D.; Yadav, A.; Chaudhary, A.; Agarwal, K.; Varma-Basil, M.; Prasad, R.; Khuller, G.K.; et al. Comparative Evaluation of Several Gene Targets for Designing a Multiplex-PCR for an Early Diagnosis of Extrapulmonary Tuberculosis. Yonsei Med. J. 2016, 57, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Neshani, A.; Kakhki, R.K.; Sankian, M.; Zare, H.; Chichaklu, A.H.; Sayyadi, M.; Ghazvini, K. Modified Genome Comparison Method: A New Approach for Identification of Specific Targets in Molecular Diagnostic Tests Using Mycobacterium Tuberculosis Complex as an Example. BMC Infect. Dis. 2018, 18, 517. [Google Scholar] [CrossRef]
- Parandaman, V.; Narayanan, P.R.; Venkatesan, P.; Girish, C.; Mahadevan, S.; Rajajee, S. Evaluation of PCR Using TRC(4) and IS6110 Primers in Detection of Tuberculous Meningitis. J. Clin. Microbiol. 2001, 39, 2006–2008. [Google Scholar] [CrossRef]
- Team, R.C. R Core Team 2023 R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2023. [Google Scholar]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV2018): A Position Statement of the International Society for Extracellular Vesicles and Update of the MISEV2014 Guidelines. J. Extracell Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Erwin, N.; Serafim, M.F.; He, M. Enhancing the Cellular Production of Extracellular Vesicles for Developing Therapeutic Applications. Pharm. Res. 2023, 40, 833–853. [Google Scholar] [CrossRef]
- Karam, J.; Blanchet, F.P.; Vivès, É.; Boisguérin, P.; Boudehen, Y.M.; Kremer, L.; Daher, W. Mycobacterium Abscessus Alkyl Hydroperoxide Reductase C Promotes Cell Invasion by Binding to Tetraspanin CD81. iScience 2023, 26, 106042. [Google Scholar] [CrossRef]
- Schumann, G.; Schleier, S.; Rosenkrands, I.; Nehmann, N.; Hälbich, S.; Zipfel, P.F.; De Jonge, M.I.; Cole, S.T.; Munder, T.; Möllmann, U. Mycobacterium Tuberculosis Secreted Protein ESAT-6 Interacts with the Human Protein Syntenin-1. Cent. Eur. J. Biol. 2006, 1, 183–202. [Google Scholar] [CrossRef]
- Palacios, A.; Gupta, S.; Rodriguez, G.M.; Prados-Rosales, R. Extracellular Vesicles in the Context of Mycobacterium Tuberculosis Infection. Mol. Immunol. 2021, 133, 175–181. [Google Scholar] [CrossRef]
- Jankovičová, J.; Sečová, P.; Michalková, K.; Antalíková, J. Tetraspanins, More than Markers of Extracellular Vesicles in Reproduction. Int. J. Mol. Sci. 2020, 21, 7568. [Google Scholar] [CrossRef] [PubMed]
- Seto, S.; Matsumoto, S.; Tsujimura, K.; Koide, Y. Differential Recruitment of CD63 and Rab7-Interacting-Lysosomal-Protein to Phagosomes Containing Mycobacterium Tuberculosis in Macrophages. Microbiol. Immunol. 2009, 54, 170–174. [Google Scholar] [CrossRef]
- Giri, P.K.; Kruh, N.A.; Dobos, K.M.; Schorey, J.S. Proteomic Analysis Identifies Highly Antigenic Proteins in Exosomes from M. Tuberculosis-Infected and Culture Filtrate Protein-Treated Macrophages. Proteomics 2010, 10, 3190–3202. [Google Scholar] [CrossRef] [PubMed]
- Bhatnagar, S.; Shinagawa, K.; Castellino, F.J.; Schorey, J.S. Exosomes Released from Macrophages Infected with Intracellular Pathogens Stimulate a Proinflammatory Response in Vitro and in Vivo. Blood 2007, 110, 3234–3244. [Google Scholar] [CrossRef]
- Wiker, H.G.; Harboe, M. The Antigen 85 Complex: A Major Secretion Product of Mycobacterium Tuberculosis. Microbiol. Rev. 1992, 56, 648–661. [Google Scholar] [CrossRef]
- Chandra, P.; Grigsby, S.J.; Philips, J.A. Immune Evasion and Provocation by Mycobacterium Tuberculosis. Nat. Rev. Microbiol. 2022, 20, 750–766. [Google Scholar] [CrossRef]
- Cheng, Y.; Schorey, J.S. Extracellular Vesicles Deliver Mycobacterium RNA to Promote Host Immunity and Bacterial Killing. EMBO Rep. 2019, 20, e46613. [Google Scholar] [CrossRef]
- Mehaffy, C.; Kruh-Garcia, N.A.; Graham, B.; Jarlsberg, L.G.; Willyerd, C.E.; Borisov, A.; Sterling, T.R.; Nahid, P.; Dobos, K.M. Identification of Mycobacterium Tuberculosis Peptides in Serum Extracellular Vesicles from Persons with Latent Tuberculosis Infection. J. Clin. Microbiol. 2020, 58, e00393-20. [Google Scholar] [CrossRef]
- Kruh-Garcia, N.A.; Wolfe, L.M.; Chaisson, L.H.; Worodria, W.O.; Nahid, P.; Schorey, J.S.; Davis, J.L.; Dobos, K.M. Detection of Mycobacterium Tuberculosis Peptides in the Exosomes of Patients with Active and Latent M. Tuberculosis Infection Using MRM-MS. PLoS ONE 2014, 9, e103811. [Google Scholar] [CrossRef]
- Lv, L.; Li, C.; Zhang, X.; Ding, N.; Cao, T.; Jia, X.; Wang, J.; Pan, L.; Jia, H.; Li, Z.; et al. RNA Profiling Analysis of the Serum Exosomes Derived from Patients with Active and Latent Mycobacterium Tuberculosis Infection. Front. Microbiol. 2017, 8, 1051. [Google Scholar] [CrossRef]

| Target | Primer Sequences | Size (bp) | Reference |
|---|---|---|---|
| ESAT-6 | F: GTCCATTCATTCCCTCCT R. CTATGCGAACATCCCAGT | 220 | [14] |
| 5KST | F: TTGCTGAACTTGACCTGCCCGTA R: GCGTCTCTGCCTTCCTCCGAT | 128 | [15] |
| Ag85 | F: AAGGTCCAGTTCCAGGGCG R: ATTGGCCGCCCACGGGCATGAT | 154 | [8] |
| IS6110 | F: CCTGCGAGCGTAGGCGTCGG R: CTCGTCCAGCGCCGCTFCGG | 123 | [16] |
| 30 kDa | F: TGTACCAGTCGCTGTAGAAG R: GACATCAAGGTTCAGTTCC | 190 | [14] |
| 19 kDa | F: GAGACCACGACCGCGGCAGG R: AATGCCGGTCGCCGCCCCGCCGAT | 159 | [8] |
| MPT64 | F: GTGAACTGAGCAAGCAGACCG R: GTTCTGATAATTCACCGGGTCC | 77 | [8] |
| rRNA16S | F: GCCGTAAACGGTGGGTACTA R: TGCATGTCAAACCCAGGTAA | 194 | [8] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salgado-Cantú, M.G.; Gutiérrez-González, L.H.; Guzmán-Beltrán, S.; Herrera, M.T.; Sarabia, C.; González, Y. Impact of Mycobacterium tuberculosis H37Rv Infection on Extracellular Vesicle Cargo in Macrophages: Implications for Host–Pathogen Interaction. Microorganisms 2024, 12, 2405. https://doi.org/10.3390/microorganisms12122405
Salgado-Cantú MG, Gutiérrez-González LH, Guzmán-Beltrán S, Herrera MT, Sarabia C, González Y. Impact of Mycobacterium tuberculosis H37Rv Infection on Extracellular Vesicle Cargo in Macrophages: Implications for Host–Pathogen Interaction. Microorganisms. 2024; 12(12):2405. https://doi.org/10.3390/microorganisms12122405
Chicago/Turabian StyleSalgado-Cantú, Manuel G., Luis Horacio Gutiérrez-González, Silvia Guzmán-Beltrán, María Teresa Herrera, Carmen Sarabia, and Yolanda González. 2024. "Impact of Mycobacterium tuberculosis H37Rv Infection on Extracellular Vesicle Cargo in Macrophages: Implications for Host–Pathogen Interaction" Microorganisms 12, no. 12: 2405. https://doi.org/10.3390/microorganisms12122405
APA StyleSalgado-Cantú, M. G., Gutiérrez-González, L. H., Guzmán-Beltrán, S., Herrera, M. T., Sarabia, C., & González, Y. (2024). Impact of Mycobacterium tuberculosis H37Rv Infection on Extracellular Vesicle Cargo in Macrophages: Implications for Host–Pathogen Interaction. Microorganisms, 12(12), 2405. https://doi.org/10.3390/microorganisms12122405






