Overview of Diagnostic Methods, Disease Prevalence and Transmission of Mpox (Formerly Monkeypox) in Humans and Animal Reservoirs
Abstract
1. Introduction
2. Methodology
2.1. Article Search Criteria
2.2. Article Inclusion and Exclusion Criteria
3. Results
3.1. Trends of Mpox Disease Investigations
3.2. Methods Used for Mpox Detection in Clinical and Tissue Samples of Humans and Animals over the Period and Their Significance
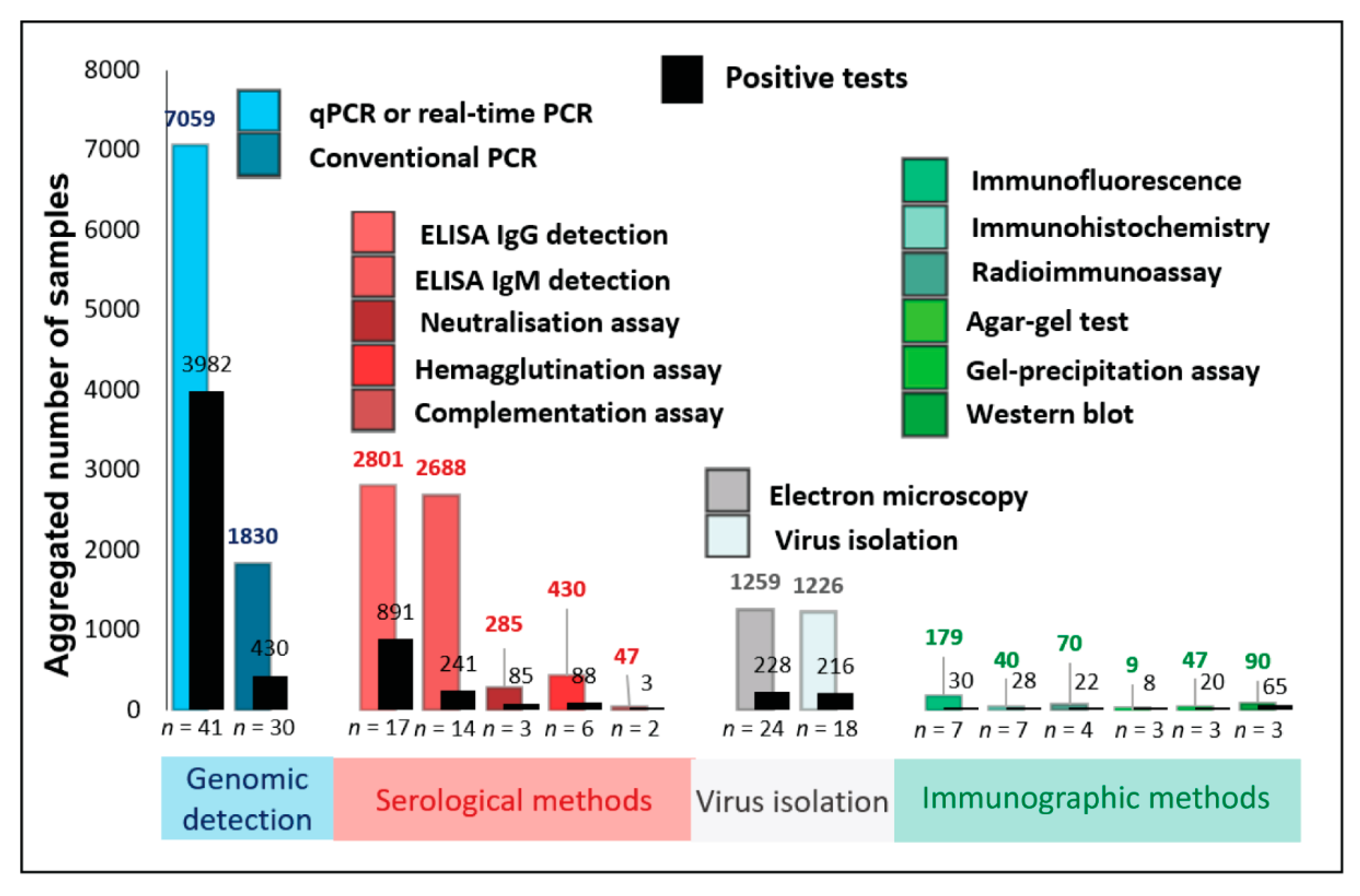
3.3. Comparison of Detection Methods by Prevalence of Use in Human Mpox Diagnosis
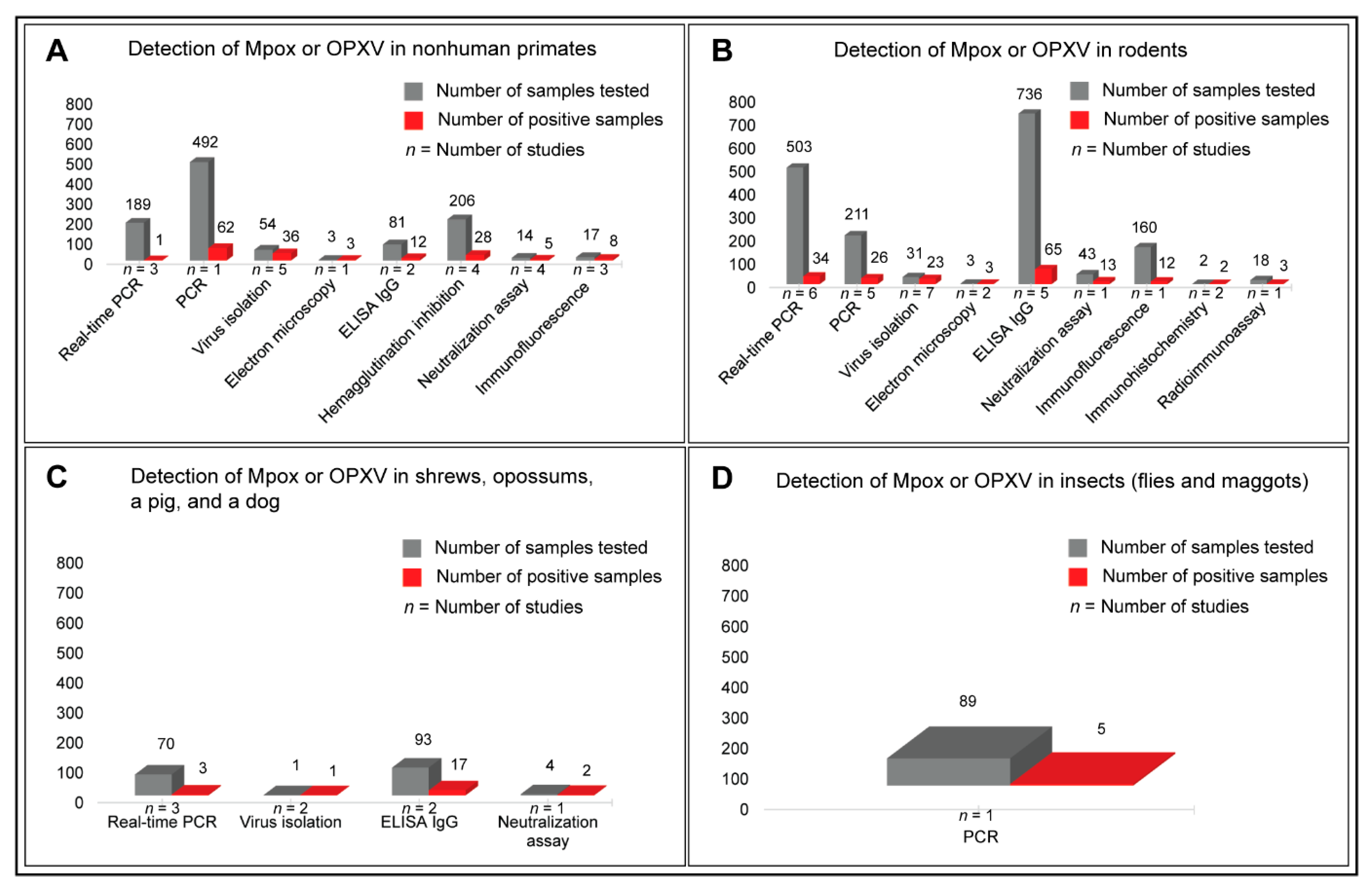
3.4. Prevalence of Mpox Disease in Animals
3.5. Host Ranges of Mpox
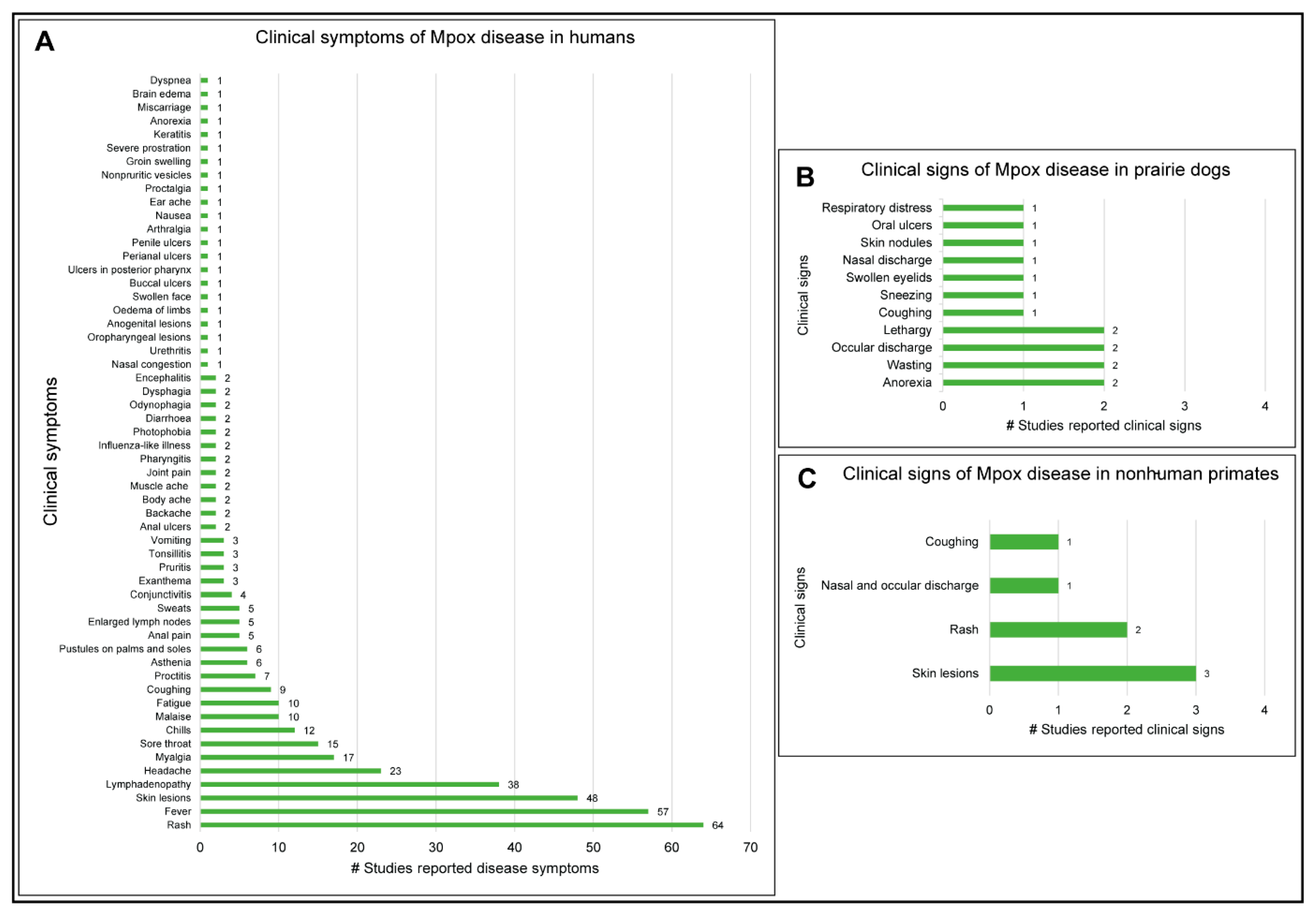
3.6. Clinical Manifestations of Mpox Disease in Humans and Animals
3.7. Oligonucleotide Primers Used for Mpox Detection in PCR and Real-Time PCR Assays
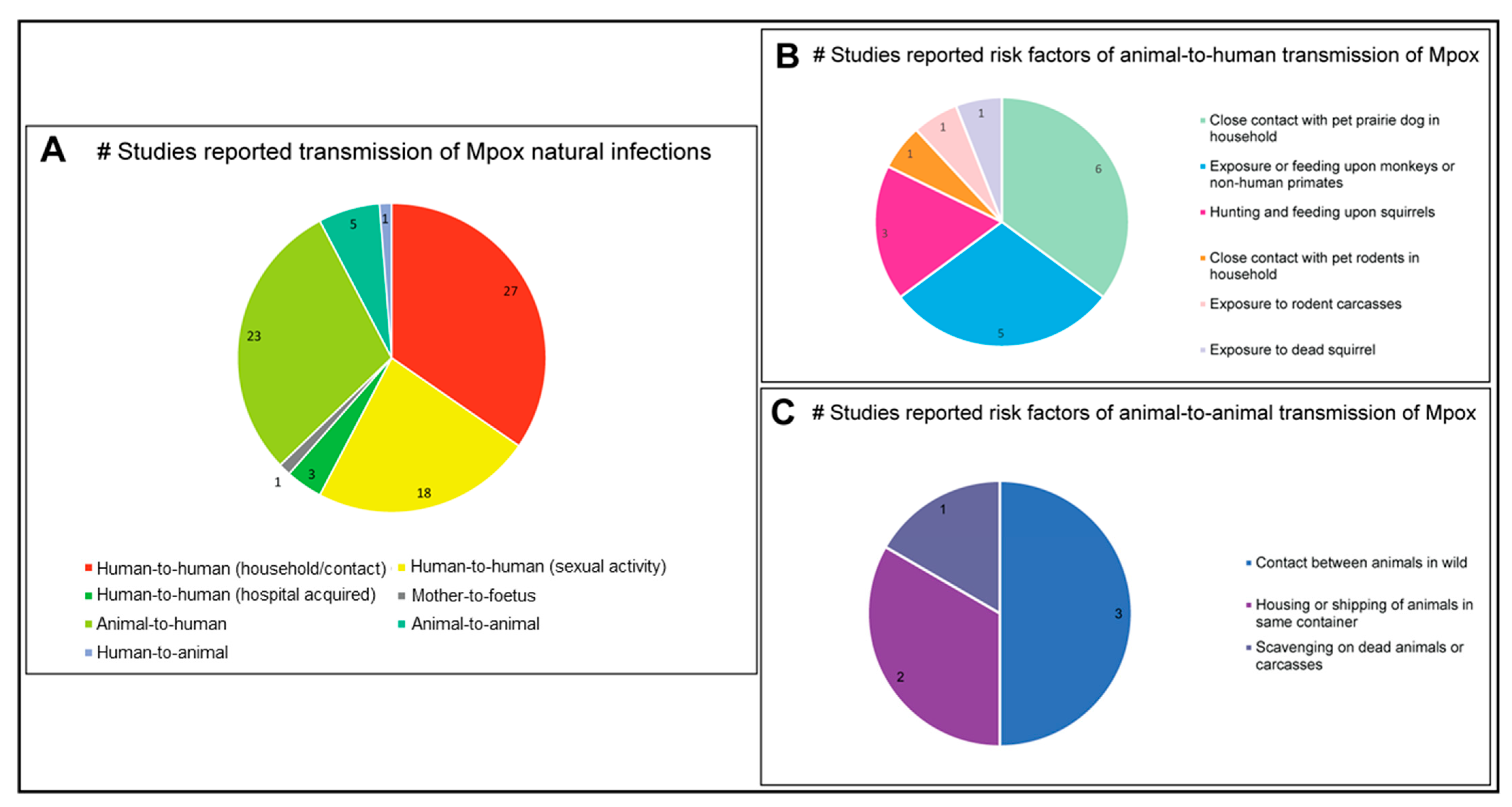
3.8. Transmission Dynamics of Mpox Disease
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Silva, N.I.O.; de Oliveira, J.S.; Kroon, E.G.; Trindade, G.S.; Drumond, B.P. Here, There, and Everywhere: The Wide Host Range and Geographic Distribution of Zoonotic Orthopoxviruses. Viruses 2020, 13, 43. [Google Scholar] [CrossRef] [PubMed]
- Kipkorir, V.; Dhali, A.; Srichawla, B.; Kutikuppala, S.; Cox, M.; Ochieng, D.; Nyaanga, F.; Găman, M.A. The re-emerging monkeypox disease. Trop. Med. Int. Health 2022, 27, 961–969. [Google Scholar] [CrossRef] [PubMed]
- Besombes, C.; Gonofio, E.; Konamna, X.; Selekon, B.; Grant, R.; Gessain, A.; Berthet, N.; Manuguerra, J.C.; Fontanet, A.; Nakouné, E. Intrafamily Transmission of Monkeypox Virus, Central African Republic, 2018. Emerg. Infect. Dis. 2019, 25, 1602–1604. [Google Scholar] [CrossRef] [PubMed]
- Cohen-Gihon, I.; Israeli, O.; Shifman, O.; Erez, N.; Melamed, S.; Paran, N.; Beth-Din, A.; Zvi, A. Identification and Whole-Genome Sequencing of a Monkeypox Virus Strain Isolated in Israel. Microbiol. Resour. Announc. 2020, 9, e01524-19. [Google Scholar] [CrossRef] [PubMed]
- Fleischauer, A.T.; Kile, J.C.; Davidson, M.; Fischer, M.; Karem, K.L.; Teclaw, R.; Messersmith, H.; Pontones, P.; Beard, B.A.; Braden, Z.H.; et al. Evaluation of human-to-human transmission of monkeypox from infected patients to health care workers. Clin. Infect. Dis. 2005, 40, 689–694. [Google Scholar] [CrossRef]
- Froeschl, G.; Kayembe, P.K. Pox-like lesions and haemorrhagic fever in two concurrent cases in the Central African Republic: Case investigation and management in difficult circumstances. Pan Afr. Med. J. 2015, 22, 23. [Google Scholar] [CrossRef]
- Hobson, G.; Adamson, J.; Adler, H.; Firth, R.; Gould, S.; Houlihan, C.; Johnson, C.; Porter, D.; Rampling, T.; Ratcliffe, L.; et al. Family cluster of three cases of monkeypox imported from Nigeria to the United Kingdom, May 2021. Eurosurveillance 2021, 26, 2100745. [Google Scholar] [CrossRef] [PubMed]
- Hughes, C.M.; Liu, L.; Davidson, W.B.; Radford, K.W.; Wilkins, K.; Monroe, B.; Metcalfe, M.G.; Likafi, T.; Lushima, R.S.; Kabamba, J.; et al. A Tale of Two Viruses: Coinfections of Monkeypox and Varicella Zoster Virus in the Democratic Republic of Congo. Am. J. Trop. Med. Hyg. 2020, 104, 604–611. [Google Scholar] [CrossRef]
- Kugelman, J.R.; Johnston, S.C.; Mulembakani, P.M.; Kisalu, N.; Lee, M.S.; Koroleva, G.; McCarthy, S.E.; Gestole, M.C.; Wolfe, N.D.; Fair, J.N.; et al. Genomic variability of monkeypox virus among humans, Democratic Republic of the Congo. Emerg. Infect. Dis. 2014, 20, 232–239. [Google Scholar] [CrossRef]
- Costello, V.; Sowash, M.; Gaur, A.; Cardis, M.; Pasieka, H.; Wortmann, G.; Ramdeen, S. Imported Monkeypox from International Traveler, Maryland, USA, 2021. Emerg. Infect. Dis. 2022, 28, 1002–1005. [Google Scholar] [CrossRef]
- Croft, D.R.; Sotir, M.J.; Williams, C.J.; Kazmierczak, J.J.; Wegner, M.V.; Rausch, D.; Graham, M.B.; Foldy, S.L.; Wolters, M.; Damon, I.K.; et al. Occupational risks during a monkeypox outbreak, Wisconsin, 2003. Emerg. Infect. Dis. 2007, 13, 1150–1157. [Google Scholar] [CrossRef]
- Yinka-Ogunleye, A.; Aruna, O.; Dalhat, M.; Ogoina, D.; McCollum, A.; Disu, Y.; Mamadu, I.; Akinpelu, A.; Ahmad, A.; Burga, J.; et al. Outbreak of human monkeypox in Nigeria in 2017-18: A clinical and epidemiological report. Lancet Infect. Dis. 2019, 19, 872–879. [Google Scholar] [CrossRef] [PubMed]
- Formenty, P.; Muntasir, M.O.; Damon, I.; Chowdhary, V.; Opoka, M.L.; Monimart, C.; Mutasim, E.M.; Manuguerra, J.C.; Davidson, W.B.; Karem, K.L.; et al. Human monkeypox outbreak caused by novel virus belonging to Congo Basin clade, Sudan, 2005. Emerg. Infect. Dis. 2010, 16, 1539–1545. [Google Scholar] [CrossRef] [PubMed]
- Hammerschlag, Y.; MacLeod, G.; Papadakis, G.; Adan Sanchez, A.; Druce, J.; Taiaroa, G.; Savic, I.; Mumford, J.; Roberts, J.; Caly, L.; et al. Monkeypox infection presenting as genital rash, Australia, May 2022. Eurosurveillance 2022, 27, 2200411. [Google Scholar] [CrossRef] [PubMed]
- Parrino, J.; Graham, B.S. Smallpox vaccines: Past, present, and future. J. Allergy Clin. Immunol. 2006, 118, 1320–1326. [Google Scholar] [CrossRef] [PubMed]
- Melamed, S.; Israely, T.; Paran, N. Challenges and Achievements in Prevention and Treatment of Smallpox. Vaccines 2018, 6, 8. [Google Scholar] [CrossRef]
- Li, Y.; Carroll, D.S.; Gardner, S.N.; Walsh, M.C.; Vitalis, E.A.; Damon, I.K. On the origin of smallpox: Correlating variola phylogenics with historical smallpox records. Proc. Natl. Acad. Sci. USA 2007, 104, 15787–15792. [Google Scholar] [CrossRef]
- Berche, P. Life and death of smallpox. La Presse Médicale 2022, 51, 104117. [Google Scholar] [CrossRef]
- Olson, V.A.; Shchelkunov, S.N. Are We Prepared in Case of a Possible Smallpox-Like Disease Emergence? Viruses 2017, 9, 242. [Google Scholar] [CrossRef]
- Nitsche, A.; Kurth, A.; Pauli, G. Viremia in human Cowpox virus infection. J. Clin. Virol. 2007, 40, 160–162. [Google Scholar] [CrossRef]
- Baxby, D.; Bennett, M.; Getty, B. Human cowpox 1969–93: A review based on 54 cases. Br. J. Dermatol. 1994, 131, 598–607. [Google Scholar] [CrossRef]
- Pether, J.V.S.; Trevains, P.H.; Harrison, S.R.B.; Baxby, D.; Bennett, M.; Gibb, A.P.R. Cowpox from cat to man. Lancet 1986, 1, 38–39. [Google Scholar] [CrossRef] [PubMed]
- Ninove, L.; Domart, Y.; Vervel, C.; Voinot, C.; Salez, N.; Raoult, D.; Meyer, H.; Capek, I.; Zandotti, C.; Charrel, R.N. Cowpox virus transmission from pet rats to humans, France. Emerg. Infect. Dis. 2009, 15, 781–784. [Google Scholar] [CrossRef]
- Campe, H.; Zimmermann, P.; Glos, K.; Bayer, M.; Bergemann, H.; Dreweck, C.; Graf, P.; Weber, B.K.; Meyer, H.; Büttner, M.; et al. Cowpox virus transmission from pet rats to humans, Germany. Emerg. Infect. Dis. 2009, 15, 777–780. [Google Scholar] [CrossRef] [PubMed]
- Mueller, N.H.; Gilden, D.H.; Cohrs, R.J.; Mahalingam, R.; Nagel, M.A. Varicella zoster virus infection: Clinical features, molecular pathogenesis of disease, and latency. Neurol. Clin. 2008, 26, 675–697. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.G.; Beecker, J. Chickenpox in an elderly man. Can. Fam. Physician 2020, 66, e213–e215. [Google Scholar]
- Vairo, F.; Di Bari, V.; Panella, V.; Quintavalle, G.; Torchia, S.; Serra, M.C.; Sinopoli, M.T.; Lopalco, M.; Ceccarelli, G.; Ferraro, F.; et al. An outbreak of chickenpox in an asylum seeker centre in Italy: Outbreak investigation and validity of reported chickenpox history, December 2015–May 2016. Eurosurveillance 2017, 22, 17-00020. [Google Scholar] [CrossRef]
- Egawa, G.; Egawa, K.; Kabashima, K. Case of chickenpox in which varicella zoster virus genotype E was identified for the first time in Japan. J. Dermatol. 2020, 47, 54–57. [Google Scholar] [CrossRef]
- Andrade, S.M.C.; Haslett, M.I.C.; Malta, J.; Renoiner, E.I.M.; Lucena, A.R.F.; Fantinato, F.F.S.; Cruz, V.O.; Costa, C.S.D.; Santos, E.D.D. Chickenpox outbreak among Venezuelan immigrants housed in shelters and occupancies in the state of Roraima, Brazil, 2019: A descriptive study. Epidemiol. Serv. Saude 2021, 30, e2021156. [Google Scholar] [CrossRef]
- WHO. Monkeypox. 2022. Available online: https://www.who.int/news-room/fact-sheets/detail/monkeypox (accessed on 23 April 2023).
- Minhaj, F.S.; Ogale, Y.P.; Whitehill, F.; Schultz, J.; Foote, M.; Davidson, W.; Hughes, C.M.; Wilkins, K.; Bachmann, L.; Chatelain, R.; et al. Monkeypox Outbreak—Nine States, May 2022. MMWR Morb. Mortal. Wkly. Rep. 2022, 71, 764–769. [Google Scholar] [CrossRef]
- WHO. Laboratory Testing for the Monkeypox Virus: Interim Guidance. 23 May 2022. Available online: https://apps.who.int/iris/bitstream/handle/10665/354488/WHO-MPX-Laboratory-2022.1-eng.pdf?sequence=1&isAllowed=y (accessed on 1 September 2022).
- Reynolds, M.G.; Guagliardo, S.A.J.; Nakazawa, Y.J.; Doty, J.B.; Mauldin, M.R. Understanding orthopoxvirus host range and evolution: From the enigmatic to the usual suspects. Curr. Opin. Virol. 2018, 28, 108–115. [Google Scholar] [CrossRef] [PubMed]
- Balamurugan, V.; Venkatesan, G.; Bhanuprakash, V.; Singh, R.K. Camelpox, an emerging orthopox viral disease. Indian J. Virol. 2013, 24, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Kinnunen, P.M.; Henttonen, H.; Hoffmann, B.; Kallio, E.R.; Korthase, C.; Laakkonen, J.; Niemimaa, J.; Palva, A.; Schlegel, M.; Ali, H.S.; et al. Orthopox virus infections in Eurasian wild rodents. Vector Borne Zoonotic Dis. 2011, 11, 1133–1140. [Google Scholar] [CrossRef] [PubMed]
- Pauli, G.; Blümel, J.; Burger, R.; Drosten, C.; Gröner, A.; Gürtler, L.; Heiden, M.; Hildebrandt, M.; Jansen, B.; Montag-Lessing, T.; et al. Orthopox Viruses: Infections in Humans. Transfus. Med. Hemother. 2010, 37, 351–364. [Google Scholar] [CrossRef] [PubMed]
- Beer, E.M.; Rao, V.B. A systematic review of the epidemiology of human monkeypox outbreaks and implications for outbreak strategy. PLoS Negl. Trop. Dis. 2019, 13, e0007791. [Google Scholar] [CrossRef] [PubMed]
- Benites-Zapata, V.A.; Ulloque-Badaracco, J.R.; Alarcon-Braga, E.A.; Hernandez-Bustamante, E.A.; Mosquera-Rojas, M.D.; Bonilla-Aldana, D.K.; Rodriguez-Morales, A.J. Clinical features, hospitalisation and deaths associated with monkeypox: A systematic review and meta-analysis. Ann. Clin. Microbiol. Antimicrob. 2022, 21, 36. [Google Scholar] [CrossRef] [PubMed]
- Besombes, C.; Mbrenga, F.; Schaeffer, L.; Malaka, C.; Gonofio, E.; Landier, J.; Vickos, U.; Konamna, X.; Selekon, B.; Dankpea, J.N.; et al. National Monkeypox Surveillance, Central African Republic, 2001–2021. Emerg. Infect. Dis. 2022, 28, 2435–2445. [Google Scholar] [CrossRef]
- Mbala, P.K.; Huggins, J.W.; Riu-Rovira, T.; Ahuka, S.M.; Mulembakani, P.; Rimoin, A.W.; Martin, J.W.; Muyembe, J.T. Maternal and Fetal Outcomes among Pregnant Women with Human Monkeypox Infection in the Democratic Republic of Congo. J. Infect. Dis. 2017, 216, 824–828. [Google Scholar] [CrossRef]
- D'Antonio, F.; Pagani, G.; Buca, D.; Khalil, A. Monkeypox infection in pregnancy: A systematic review and metaanalysis. Am. J. Obstet. Gynecol. MFM 2023, 5, 100747. [Google Scholar] [CrossRef]
- Adler, H.; Gould, S.; Hine, P.; Snell, L.B.; Wong, W.; Houlihan, C.F.; Osborne, J.C.; Rampling, T.; Beadsworth, M.B.; Duncan, C.J.; et al. Clinical features and management of human monkeypox: A retrospective observational study in the UK. Lancet Infect. Dis. 2022, 22, 1153–1162. [Google Scholar] [CrossRef]
- Ogoina, D.; Iroezindu, M.; James, H.I.; Oladokun, R.; Yinka-Ogunleye, A.; Wakama, P.; Otike-Odibi, B.; Usman, L.M.; Obazee, E.; Aruna, O.; et al. Clinical Course and Outcome of Human Monkeypox in Nigeria. Clin. Infect. Dis. 2020, 71, e210–e214. [Google Scholar] [CrossRef] [PubMed]
- Sejvar, J.J.; Chowdary, Y.; Schomogyi, M.; Stevens, J.; Patel, J.; Karem, K.; Fischer, M.; Kuehnert, M.J.; Zaki, S.R.; Paddock, C.D.; et al. Human monkeypox infection: A family cluster in the midwestern United States. J. Infect. Dis. 2004, 190, 1833–1840. [Google Scholar] [CrossRef] [PubMed]
- Shafaati, M.; Zandi, M. Monkeypox virus neurological manifestations in comparison to other orthopoxviruses. Travel. Med. Infect. Dis. 2022, 49, 102414. [Google Scholar] [CrossRef] [PubMed]
- Forni, D.; Molteni, C.; Cagliani, R.; Sironi, M. Geographic structuring and divergence time frame of monkeypox virus in the endemic region. J. Infect. Dis. 2022, 227, 742–751. [Google Scholar] [CrossRef]
- Yeh, T.Y.; Hsieh, Z.Y.; Feehley, M.C.; Feehley, P.J.; Contreras, G.P.; Su, Y.C.; Hsieh, S.L.; Lewis, D.A. Recombination shapes the 2022 monkeypox (mpox) outbreak. Med (N. Y.) 2022, 3, 824–826. [Google Scholar] [CrossRef]
- Thornhill, J.P.; Barkati, S.; Walmsley, S.; Rockstroh, J.; Antinori, A.; Harrison, L.B.; Palich, R.; Nori, A.; Reeves, I.; Habibi, M.S.; et al. Monkeypox Virus Infection in Humans across 16 Countries—April–June 2022. N. Engl. J. Med. 2022, 387, 679–691. [Google Scholar] [CrossRef]
- Kohli, R.M.; Isaacs, S.N. Mpox Evolution: Has the Current Outbreak Revealed a Pox on “U”? J. Infect. Dis. 2023, 227, 828–830. [Google Scholar] [CrossRef]
- Isidro, J.; Borges, V.; Pinto, M.; Sobral, D.; Santos, J.D.; Nunes, A.; Mixão, V.; Ferreira, R.; Santos, D.; Duarte, S.; et al. Phylogenomic characterization and signs of microevolution in the 2022 multi-country outbreak of monkeypox virus. Nat. Med. 2022, 28, 1569–1572. [Google Scholar] [CrossRef]
- Magnus, P.v.; Andersen, E.K.; Petersen, K.B.; Birch-Andersen, A. A pox-like disease in Cynomolgus monkeys. Acta Pathol. Microbiol. Scand. 1959, 46, 156–176. [Google Scholar] [CrossRef]
- Tiee, M.S.; Harrigan, R.J.; Thomassen, H.A.; Smith, T.B. Ghosts of infections past: Using archival samples to understand a century of monkeypox virus prevalence among host communities across space and time. R. Soc. Open. Sci. 2018, 5, 171089. [Google Scholar] [CrossRef]
- Babkin, I.V.; Babkina, I.N.; Tikunova, N.V. An Update of Orthopoxvirus Molecular Evolution. Viruses 2022, 14, 388. [Google Scholar] [CrossRef] [PubMed]
- Babkin, I.V.; Babkina, I.N. A retrospective study of the orthopoxvirus molecular evolution. Infect. Genet. Evol. 2012, 12, 1597–1604. [Google Scholar] [CrossRef] [PubMed]
- WHO. WHO Recommends New Name for Monkeypox Disease; WHO: Geneva, Switzerland, 28 November 2022.
- Graham, F. Daily briefing: Mpox-a new name for monkeypox. Nature 2022. [Google Scholar] [CrossRef]
- Ladnyj, I.D.; Ziegler, P.; Kima, E. A human infection caused by monkeypox virus in Basankusu Territory, Democratic Republic of the Congo. Bull. World Health Organ. 1972, 46, 593–597. [Google Scholar] [PubMed]
- Marennikova, S.S.; Seluhina, E.M.; Mal’ceva, N.N.; Cimiskjan, K.L.; Macevic, G.R. Isolation and properties of the causal agent of a new variola-like disease (monkeypox) in man. Bull. World Health Organ. 1972, 46, 599–611. [Google Scholar]
- Lourie, B.; Bingham, P.G.; Evans, H.H.; Foster, S.O.; Nakano, J.H.; Herrmann, K.L. Human infection with monkeypox virus: Laboratory investigation of six cases in West Africa. Bull. World Health Organ. 1972, 46, 633–639. [Google Scholar]
- Breman, J.G.; Kalisa, R.; Steniowski, M.V.; Zanotto, E.; Gromyko, A.I.; Arita, I. Human monkeypox, 1970–1979. Bull. World Health Organ. 1980, 58, 165–182. [Google Scholar]
- Foster, S.O.; Brink, E.W.; Hutchins, D.L.; Pifer, J.M.; Lourie, B.; Moser, C.R.; Cummings, E.C.; Kuteyi, O.E.; Eke, R.E.; Titus, J.B.; et al. Human monkeypox. Bull. World Health Organ. 1972, 46, 569–576. [Google Scholar]
- Jezek, Z.; Marennikova, S.S.; Mutumbo, M.; Nakano, J.H.; Paluku, K.M.; Szczeniowski, M. Human monkeypox: A study of 2,510 contacts of 214 patients. J. Infect. Dis. 1986, 154, 551–555. [Google Scholar] [CrossRef]
- Levine, R.S.; Peterson, A.T.; Yorita, K.L.; Carroll, D.; Damon, I.K.; Reynolds, M.G. Ecological niche and geographic distribution of human monkeypox in Africa. PLoS ONE 2007, 2, e176. [Google Scholar] [CrossRef]
- Guarner, J.; Johnson, B.J.; Paddock, C.D.; Shieh, W.J.; Goldsmith, C.S.; Reynolds, M.G.; Damon, I.K.; Regnery, R.L.; Zaki, S.R. Monkeypox transmission and pathogenesis in prairie dogs. Emerg. Infect. Dis. 2004, 10, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Reed, K.D.; Melski, J.W.; Graham, M.B.; Regnery, R.L.; Sotir, M.J.; Wegner, M.V.; Kazmierczak, J.J.; Stratman, E.J.; Li, Y.; Fairley, J.A.; et al. The detection of monkeypox in humans in the Western Hemisphere. N. Engl. J. Med. 2004, 350, 342–350. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.G.; Frenkel, L.D.; Homann, S.; Guffey, J. A case of severe monkeypox virus disease in an American child: Emerging infections and changing professional values. Pediatr. Infect. Dis. J. 2003, 22, 1093–1096; discussion 1096–1098. [Google Scholar] [CrossRef] [PubMed]
- Cunha, B.E. Monkeypox in the United States: An occupational health look at the first cases. AAOHN J. 2004, 52, 164–168. [Google Scholar] [CrossRef]
- Mauldin, M.R.; McCollum, A.M.; Nakazawa, Y.J.; Mandra, A.; Whitehouse, E.R.; Davidson, W.; Zhao, H.; Gao, J.; Li, Y.; Doty, J.; et al. Exportation of Monkeypox Virus From the African Continent. J. Infect. Dis. 2022, 225, 1367–1376. [Google Scholar] [CrossRef]
- Rao, A.K.; Schulte, J.; Chen, T.H.; Hughes, C.M.; Davidson, W.; Neff, J.M.; Markarian, M.; Delea, K.C.; Wada, S.; Liddell, A.; et al. Monkeypox in a Traveler Returning from Nigeria—Dallas, Texas, July 2021. MMWR Morb. Mortal. Wkly. Rep. 2022, 71, 509–516. [Google Scholar] [CrossRef]
- Yong, S.E.F.; Ng, O.T.; Ho, Z.J.M.; Mak, T.M.; Marimuthu, K.; Vasoo, S.; Yeo, T.W.; Ng, Y.K.; Cui, L.; Ferdous, Z.; et al. Imported Monkeypox, Singapore. Emerg. Infect. Dis. 2020, 26, 1826–1830. [Google Scholar] [CrossRef]
- Vaughan, A.; Aarons, E.; Astbury, J.; Balasegaram, S.; Beadsworth, M.; Beck, C.R.; Chand, M.; O’Connor, C.; Dunning, J.; Ghebrehewet, S.; et al. Two cases of monkeypox imported to the United Kingdom, September 2018. Eurosurveillance 2018, 23, 1800509. [Google Scholar] [CrossRef]
- Laiton-Donato, K.; Álvarez-Díaz, D.A.; Franco-Muñoz, C.; Ruiz-Moreno, H.A.; Rojas-Estévez, P.; Prada, A.; Rosales, A.; Ospina, M.L.; Mercado-Reyes, M. Monkeypox virus genome sequence from an imported human case in Colombia. Biomedica 2022, 42, 541–545. [Google Scholar] [CrossRef]
- Alcamí, A. Pathogenesis of the circulating mpox virus and its adaptation to humans. Proc. Natl. Acad. Sci. USA 2023, 120, e2301662120. [Google Scholar] [CrossRef]
- Vusirikala, A.; Charles, H.; Balasegaram, S.; Macdonald, N.; Kumar, D.; Barker-Burnside, C.; Cumiskey, K.; Dickinson, M.; Watson, M.; Olufon, O.; et al. Epidemiology of Early Monkeypox Virus Transmission in Sexual Networks of Gay and Bisexual Men, England, 2022. Emerg. Infect. Dis. 2022, 28, 2082–2086. [Google Scholar] [CrossRef]
- Patel, A.; Bilinska, J.; Tam, J.C.H.; Da Silva Fontoura, D.; Mason, C.Y.; Daunt, A.; Snell, L.B.; Murphy, J.; Potter, J.; Tuudah, C.; et al. Clinical features and novel presentations of human monkeypox in a central London centre during the 2022 outbreak: Descriptive case series. BMJ 2022, 378, e072410. [Google Scholar] [CrossRef] [PubMed]
- Philpott, D.; Hughes, C.M.; Alroy, K.A.; Kerins, J.L.; Pavlick, J.; Asbel, L.; Crawley, A.; Newman, A.P.; Spencer, H.; Feldpausch, A.; et al. Epidemiologic and Clinical Characteristics of Monkeypox Cases—United States, May 17–July 22, 2022. MMWR Morb. Mortal. Wkly. Rep. 2022, 71, 1018–1022. [Google Scholar] [CrossRef]
- Vivancos, R.; Anderson, C.; Blomquist, P.; Balasegaram, S.; Bell, A.; Bishop, L.; Brown, C.S.; Chow, Y.; Edeghere, O.; Florence, I.; et al. Community transmission of monkeypox in the United Kingdom, April to May 2022. Eurosurveillance 2022, 27, 2200422. [Google Scholar] [CrossRef] [PubMed]
- Girometti, N.; Byrne, R.; Bracchi, M.; Heskin, J.; McOwan, A.; Tittle, V.; Gedela, K.; Scott, C.; Patel, S.; Gohil, J.; et al. Demographic and clinical characteristics of confirmed human monkeypox virus cases in individuals attending a sexual health centre in London, UK: An observational analysis. Lancet Infect. Dis. 2022, 22, 1321–1328. [Google Scholar] [CrossRef]
- Heskin, J.; Belfield, A.; Milne, C.; Brown, N.; Walters, Y.; Scott, C.; Bracchi, M.; Moore, L.S.; Mughal, N.; Rampling, T.; et al. Transmission of monkeypox virus through sexual contact—A novel route of infection. J. Infect. 2022, 85, 334–363. [Google Scholar] [CrossRef]
- WHO. 2022-23 Mpox (Monkeypox) Outbreak: Global Trends; WHO: Geneva, Switzerland, 28 February 2023.
- Altindis, M.; Puca, E.; Shapo, L. Diagnosis of monkeypox virus—An overview. Travel. Med. Infect. Dis. 2022, 50, 102459. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed]
- Yinka-Ogunleye, A.; Aruna, O.; Ogoina, D.; Aworabhi, N.; Eteng, W.; Badaru, S.; Mohammed, A.; Agenyi, J.; Etebu, E.N.; Numbere, T.W.; et al. Reemergence of Human Monkeypox in Nigeria, 2017. Emerg. Infect. Dis. 2018, 24, 1149–1151. [Google Scholar] [CrossRef] [PubMed]
- Ligon, B.L. Monkeypox: A review of the history and emergence in the Western hemisphere. Semin. Pediatr. Infect. Dis. 2004, 15, 280–287. [Google Scholar] [CrossRef]
- Whitehouse, E.R.; Bonwitt, J.; Hughes, C.M.; Lushima, R.S.; Likafi, T.; Nguete, B.; Kabamba, J.; Monroe, B.; Doty, J.B.; Nakazawa, Y.; et al. Clinical and Epidemiological Findings from Enhanced Monkeypox Surveillance in Tshuapa Province, Democratic Republic of the Congo During 2011–2015. J. Infect. Dis. 2021, 223, 1870–1878. [Google Scholar] [CrossRef] [PubMed]
- Iñigo Martínez, J.; Gil Montalbán, E.; Jiménez Bueno, S.; Martín Martínez, F.; Nieto Juliá, A.; Sánchez Díaz, J.; García Marín, N.; Córdoba Deorador, E.; Nunziata Forte, A.; Alonso García, M.; et al. Monkeypox outbreak predominantly affecting men who have sex with men, Madrid, Spain, 26 April to 16 June 2022. Eurosurveillance 2022, 27, 2200471. [Google Scholar] [CrossRef] [PubMed]
- Osadebe, L.; Hughes, C.M.; Shongo Lushima, R.; Kabamba, J.; Nguete, B.; Malekani, J.; Pukuta, E.; Karhemere, S.; Muyembe Tamfum, J.J.; Wemakoy Okitolonda, E.; et al. Enhancing case definitions for surveillance of human monkeypox in the Democratic Republic of Congo. PLoS Negl. Trop. Dis. 2017, 11, e0005857. [Google Scholar] [CrossRef]
- Hoff, N.A.; Morier, D.S.; Kisalu, N.K.; Johnston, S.C.; Doshi, R.H.; Hensley, L.E.; Okitolonda-Wemakoy, E.; Muyembe-Tamfum, J.J.; Lloyd-Smith, J.O.; Rimoin, A.W. Varicella Coinfection in Patients with Active Monkeypox in the Democratic Republic of the Congo. Ecohealth 2017, 14, 564–574. [Google Scholar] [CrossRef] [PubMed]
- Kulesh, D.A.; Loveless, B.M.; Norwood, D.; Garrison, J.; Whitehouse, C.A.; Hartmann, C.; Mucker, E.; Miller, D.; Wasieloski, L.P., Jr.; Huggins, J.; et al. Monkeypox virus detection in rodents using real-time 3′-minor groove binder TaqMan assays on the Roche LightCycler. Lab. Investig. 2004, 84, 1200–1208. [Google Scholar] [CrossRef]
- Bayer-Garner, I.B. Monkeypox virus: Histologic, immunohistochemical and electron-microscopic findings. J. Cutan. Pathol. 2005, 32, 28–34. [Google Scholar] [CrossRef]
- Langohr, I.M.; Stevenson, G.W.; Thacker, H.L.; Regnery, R.L. Extensive lesions of monkeypox in a prairie dog (Cynomys sp). Vet. Pathol. 2004, 41, 702–707. [Google Scholar] [CrossRef]
- Kile, J.C.; Fleischauer, A.T.; Beard, B.; Kuehnert, M.J.; Kanwal, R.S.; Pontones, P.; Messersmith, H.J.; Teclaw, R.; Karem, K.L.; Braden, Z.H.; et al. Transmission of monkeypox among persons exposed to infected prairie dogs in Indiana in 2003. Arch. Pediatr. Adolesc. Med. 2005, 159, 1022–1025. [Google Scholar] [CrossRef]
- Jang, Y.R.; Lee, M.; Shin, H.; Kim, J.W.; Choi, M.M.; Kim, Y.M.; Lee, M.J.; Kim, J.; Na, H.K.; Kim, J.Y. The First Case of Monkeypox in the Republic of Korea. J. Korean Med. Sci. 2022, 37, e224. [Google Scholar] [CrossRef]
- Patrocinio-Jesus, R.; Peruzzu, F. Monkeypox Genital Lesions. N. Engl. J. Med. 2022, 387, 66. [Google Scholar] [CrossRef]
- Moschese, D.; Pozza, G.; Mileto, D.; Giacomelli, A.; Cutrera, M.; Cossu, M.V.; Matone, M.; Beltrami, M.; Salari, F.; Antinori, S.; et al. Isolation of viable monkeypox virus from anal and urethral swabs, Italy, May to July 2022. Eurosurveillance 2022, 27, 2200675. [Google Scholar] [CrossRef] [PubMed]
- Orviz, E.; Negredo, A.; Ayerdi, O.; Vázquez, A.; Muñoz-Gomez, A.; Monzón, S.; Clavo, P.; Zaballos, A.; Vera, M.; Sánchez, P.; et al. Monkeypox outbreak in Madrid (Spain): Clinical and virological aspects. J. Infect. 2022, 85, 412–417. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, L.E.; Jadoo, A.; Kirsner, R.S. Human monkeypox virus infection in an immunocompromised man: Trial with tecovirimat. Lancet 2022, 400, e8. [Google Scholar] [CrossRef] [PubMed]
- De Baetselier, I.; Van Dijck, C.; Kenyon, C.; Coppens, J.; Michiels, J.; de Block, T.; Smet, H.; Coppens, S.; Vanroye, F.; Bugert, J.J.; et al. Retrospective detection of asymptomatic monkeypox virus infections among male sexual health clinic attendees in Belgium. Nat. Med. 2022, 28, 2288–2292. [Google Scholar] [CrossRef]
- Noe, S.; Zange, S.; Seilmaier, M.; Antwerpen, M.H.; Fenzl, T.; Schneider, J.; Spinner, C.D.; Bugert, J.J.; Wendtner, C.M.; Wölfel, R. Clinical and virological features of first human monkeypox cases in Germany. Infection 2022, 51, 265–270. [Google Scholar] [CrossRef]
- Meduri, E.; Malclès, A.; Kecik, M. Conjunctivitis with Monkeypox Virus Positive Conjunctival Swabs. Ophthalmology 2022, 129, 1095. [Google Scholar] [CrossRef]
- Likos, A.M.; Sammons, S.A.; Olson, V.A.; Frace, A.M.; Li, Y.; Olsen-Rasmussen, M.; Davidson, W.; Galloway, R.; Khristova, M.L.; Reynolds, M.G.; et al. A tale of two clades: Monkeypox viruses. J. Gen. Virol. 2005, 86, 2661–2672. [Google Scholar] [CrossRef]
- Nakazawa, Y.; Mauldin, M.R.; Emerson, G.L.; Reynolds, M.G.; Lash, R.R.; Gao, J.; Zhao, H.; Li, Y.; Muyembe, J.J.; Kingebeni, P.M.; et al. A phylogeographic investigation of African monkeypox. Viruses 2015, 7, 2168–2184. [Google Scholar] [CrossRef]
- Luna, N.; Ramírez, A.L.; Muñoz, M.; Ballesteros, N.; Patiño, L.H.; Castañeda, S.A.; Bonilla-Aldana, D.K.; Paniz-Mondolfi, A.; Ramírez, J.D. Phylogenomic analysis of the monkeypox virus (MPXV) 2022 outbreak: Emergence of a novel viral lineage? Travel. Med. Infect. Dis. 2022, 49, 102402. [Google Scholar] [CrossRef]
- Yadav, P.D.; Reghukumar, A.; Sahay, R.R.; Sudeep, K.; Shete, A.M.; Raman, A.; Vk, P.; Abraham, P.; Benson, R.; Sm, S.; et al. First two cases of Monkeypox virus infection in travellers returned from UAE to India, July 2022. J. Infect. 2022, 85, e145–e148. [Google Scholar] [CrossRef]
- Atkinson, B.; Burton, C.; Pottage, T.; Thompson, K.A.; Ngabo, D.; Crook, A.; Pitman, J.; Summers, S.; Lewandowski, K.; Furneaux, J.; et al. Infection-competent monkeypox virus contamination identified in domestic settings following an imported case of monkeypox into the UK. Environ. Microbiol. 2022, 24, 4561–4569. [Google Scholar] [CrossRef] [PubMed]
- Radonić, A.; Metzger, S.; Dabrowski, P.W.; Couacy-Hymann, E.; Schuenadel, L.; Kurth, A.; Mätz-Rensing, K.; Boesch, C.; Leendertz, F.H.; Nitsche, A. Fatal monkeypox in wild-living sooty mangabey, Côte d'Ivoire, 2012. Emerg. Infect. Dis. 2014, 20, 1009–1011. [Google Scholar] [CrossRef] [PubMed]
- Patrono, L.V.; Pléh, K.; Samuni, L.; Ulrich, M.; Röthemeier, C.; Sachse, A.; Muschter, S.; Nitsche, A.; Couacy-Hymann, E.; Boesch, C.; et al. Monkeypox virus emergence in wild chimpanzees reveals distinct clinical outcomes and viral diversity. Nat. Microbiol. 2020, 5, 955–965. [Google Scholar] [CrossRef]
- Sadeuh-Mba, S.A.; Yonga, M.G.; Els, M.; Batejat, C.; Eyangoh, S.; Caro, V.; Etoundi, A.; Carniel, E.; Njouom, R. Monkeypox virus phylogenetic similarities between a human case detected in Cameroon in 2018 and the 2017–2018 outbreak in Nigeria. Infect. Genet. Evol. 2019, 69, 8–11. [Google Scholar] [CrossRef] [PubMed]
- Ye, F.; Song, J.; Zhao, L.; Zhang, Y.; Xia, L.; Zhu, L.; Kamara, I.L.; Ren, J.; Wang, W.; Tian, H.; et al. Molecular Evidence of Human Monkeypox Virus Infection, Sierra Leone. Emerg. Infect. Dis. 2019, 25, 1220–1222. [Google Scholar] [CrossRef] [PubMed]
- Erez, N.; Achdout, H.; Milrot, E.; Schwartz, Y.; Wiener-Well, Y.; Paran, N.; Politi, B.; Tamir, H.; Israely, T.; Weiss, S.; et al. Diagnosis of Imported Monkeypox, Israel, 2018. Emerg. Infect. Dis. 2019, 25, 980–983. [Google Scholar] [CrossRef] [PubMed]
- Marien, J.; Laudisoit, A.; Patrono, L.; Baelo, P.; van Vredendaal, R.; Akawa, P.; Tungaluna, G.-C.; Mande, C.; Ngoy, S.; Mussaw, M.; et al. Monkeypox viruses circulate in distantly-related small mammal species in the Democratic Republic of the Congo. Biology 2021. [Google Scholar] [CrossRef]
- Berthet, N.; Descorps-Declère, S.; Besombes, C.; Curaudeau, M.; Nkili Meyong, A.A.; Selekon, B.; Labouba, I.; Gonofio, E.C.; Ouilibona, R.S.; Simo Tchetgna, H.D.; et al. Genomic history of human monkey pox infections in the Central African Republic between 2001 and 2018. Sci. Rep. 2021, 11, 13085. [Google Scholar] [CrossRef]
- Hutin, Y.J.; Williams, R.J.; Malfait, P.; Pebody, R.; Loparev, V.N.; Ropp, S.L.; Rodriguez, M.; Knight, J.C.; Tshioko, F.K.; Khan, A.S.; et al. Outbreak of human monkeypox, Democratic Republic of Congo, 1996 to 1997. Emerg. Infect. Dis. 2001, 7, 434–438. [Google Scholar] [CrossRef]
- McCollum, A.M.; Nakazawa, Y.; Ndongala, G.M.; Pukuta, E.; Karhemere, S.; Lushima, R.S.; Ilunga, B.K.; Kabamba, J.; Wilkins, K.; Gao, J.; et al. Human Monkeypox in the Kivus, a Conflict Region of the Democratic Republic of the Congo. Am. J. Trop. Med. Hyg. 2015, 93, 718–721. [Google Scholar] [CrossRef]
- Rimoin, A.W.; Kisalu, N.; Kebela-Ilunga, B.; Mukaba, T.; Wright, L.L.; Formenty, P.; Wolfe, N.D.; Shongo, R.L.; Tshioko, F.; Okitolonda, E.; et al. Endemic human monkeypox, Democratic Republic of Congo, 2001–2004. Emerg. Infect. Dis. 2007, 13, 934–937. [Google Scholar] [CrossRef] [PubMed]
- Antinori, A.; Mazzotta, V.; Vita, S.; Carletti, F.; Tacconi, D.; Lapini, L.E.; D’Abramo, A.; Cicalini, S.; Lapa, D.; Pittalis, S.; et al. Epidemiological, clinical and virological characteristics of four cases of monkeypox support transmission through sexual contact, Italy, May 2022. Eurosurveillance 2022, 27, 2200421. [Google Scholar] [CrossRef] [PubMed]
- Perez Duque, M.; Ribeiro, S.; Martins, J.V.; Casaca, P.; Leite, P.P.; Tavares, M.; Mansinho, K.; Duque, L.M.; Fernandes, C.; Cordeiro, R.; et al. Ongoing monkeypox virus outbreak, Portugal, 29 April to 23 May 2022. Eurosurveillance 2022, 27, 2200424. [Google Scholar] [CrossRef]
- Vandenbogaert, M.; Kwasiborski, A.; Gonofio, E.; Descorps-Declère, S.; Selekon, B.; Nkili Meyong, A.A.; Ouilibona, R.S.; Gessain, A.; Manuguerra, J.-C.; Caro, V.; et al. Nanopore sequencing of a monkeypox virus strain isolated from a pustular lesion in the Central African Republic. Sci. Rep. 2022, 12, 10768. [Google Scholar] [CrossRef] [PubMed]
- Claro, I.M.; Romano, C.M.; Candido, D.D.S.; Lima, E.L.; Lindoso, J.A.L.; Ramundo, M.S.; Moreira, F.R.R.; Barra, L.A.C.; Borges, L.M.S.; Medeiros, L.A.; et al. Shotgun metagenomic sequencing of the first case of monkeypox virus in Brazil, 2022. Rev. Inst. Med. Trop. Sao Paulo 2022, 64, e48. [Google Scholar] [CrossRef]
- Reynolds, M.G.; Carroll, D.S.; Olson, V.A.; Hughes, C.; Galley, J.; Likos, A.; Montgomery, J.M.; Suu-Ire, R.; Kwasi, M.O.; Jeffrey Root, J.; et al. A silent enzootic of an orthopoxvirus in Ghana, West Africa: Evidence for multi-species involvement in the absence of widespread human disease. Am. J. Trop. Med. Hyg. 2010, 82, 746–754. [Google Scholar] [CrossRef]
- Learned, L.A.; Reynolds, M.G.; Wassa, D.W.; Li, Y.; Olson, V.A.; Karem, K.; Stempora, L.L.; Braden, Z.H.; Kline, R.; Likos, A.; et al. Extended interhuman transmission of monkeypox in a hospital community in the Republic of the Congo, 2003. Am. J. Trop. Med. Hyg. 2005, 73, 428–434. [Google Scholar] [CrossRef]
- Doty, J.B.; Malekani, J.M.; Kalemba, L.N.; Stanley, W.T.; Monroe, B.P.; Nakazawa, Y.U.; Mauldin, M.R.; Bakambana, T.L.; Liyandja Dja Liyandja, T.; Braden, Z.H.; et al. Assessing Monkeypox Virus Prevalence in Small Mammals at the Human-Animal Interface in the Democratic Republic of the Congo. Viruses 2017, 9, 283. [Google Scholar] [CrossRef]
- MacNeil, A.; Reynolds, M.G.; Carroll, D.S.; Karem, K.; Braden, Z.; Lash, R.; Moundeli, A.; Mombouli, J.V.; Jumaan, A.O.; Schmid, D.S.; et al. Monkeypox or varicella? Lessons from a rash outbreak investigation in the Republic of the Congo. Am. J. Trop. Med. Hyg. 2009, 80, 503–507. [Google Scholar] [CrossRef]
- Karem, K.L.; Reynolds, M.; Braden, Z.; Lou, G.; Bernard, N.; Patton, J.; Damon, I.K. characterization of acute-phase humoral immunity to monkeypox: Use of immunoglobulin M enzyme-linked immunosorbent assay for detection of monkeypox infection during the 2003 North American outbreak. Clin. Diagn. Lab. Immunol. 2005, 12, 867–872. [Google Scholar] [CrossRef]
- Breman, J.G.; Bernadou, J.; Nakano, J.H. Poxvirus in West African nonhuman primates: Serological survey results. Bull. World Health Organ. 1977, 55, 605–612. [Google Scholar]
- Gispen, R.; Brand-Saathof, B.B.; Hekker, A.C. Monkeypox-specific antibodies in human and simian sera from the Ivory Coast and Nigeria. Bull. World Health Organ. 1976, 53, 355–360. [Google Scholar] [PubMed]
- Maksyutov, R.A.; Gavrilova, E.V.; Shchelkunov, S.N. Species-specific differentiation of variola, monkeypox, and varicella-zoster viruses by multiplex real-time PCR assay. J. Virol. Methods 2016, 236, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wilkins, K.; McCollum, A.M.; Osadebe, L.; Kabamba, J.; Nguete, B.; Likafi, T.; Balilo, M.P.; Lushima, R.S.; Malekani, J.; et al. Evaluation of the GeneXpert for Human Monkeypox Diagnosis. Am. J. Trop. Med. Hyg. 2017, 96, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Esposito, J.J.; Obijeski, J.F.; Nakano, J.H. Serological relatedness of monkeypox, variola, and vaccinia viruses. J. Med. Virol. 1977, 1, 35–47. [Google Scholar] [CrossRef]
- Douglas, K.O.; Cayol, C.; Forbes, K.M.; Samuels, T.A.; Vapalahti, O.; Sironen, T.; Gittens-St Hilaire, M. Serological Evidence of Multiple Zoonotic Viral Infections among Wild Rodents in Barbados. Pathogens 2021, 10, 663. [Google Scholar] [CrossRef]
- Gilchuk, I.; Gilchuk, P.; Sapparapu, G.; Lampley, R.; Singh, V.; Kose, N.; Blum, D.L.; Hughes, L.J.; Satheshkumar, P.S.; Townsend, M.B.; et al. Cross-Neutralizing and Protective Human Antibody Specificities to Poxvirus Infections. Cell 2016, 167, 684–694.e689. [Google Scholar] [CrossRef]
- Walls, H.H.; Ziegler, D.W.; Nakano, J.H. Characterization of antibodies to orthopoxviruses in human sera by radioimmunoassay. Bull. World Health Organ. 1981, 59, 253–262. [Google Scholar]
- Macneil, A.; Abel, J.; Reynolds, M.G.; Lash, R.; Fonnie, R.; Kanneh, L.D.; Robert, W.; Lungay, V.K.; Goba, A.; Moses, L.M.; et al. Serologic evidence of human orthopoxvirus infections in Sierra Leone. BMC Res. Notes 2011, 4, 465. [Google Scholar] [CrossRef]
- Guagliardo, S.A.J.; Monroe, B.; Moundjoa, C.; Athanase, A.; Okpu, G.; Burgado, J.; Townsend, M.B.; Satheshkumar, P.S.; Epperson, S.; Doty, J.B.; et al. Asymptomatic Orthopoxvirus Circulation in Humans in the Wake of a Monkeypox Outbreak among Chimpanzees in Cameroon. Am. J. Trop. Med. Hyg. 2020, 102, 206–212. [Google Scholar] [CrossRef]
- Stienlauf, S.; Shoresh, M.; Solomon, A.; Lublin-Tennenbaum, T.; Atsmon, Y.; Meirovich, Y.; Katz, E. Kinetics of formation of neutralizing antibodies against vaccinia virus following re-vaccination. Vaccine 1999, 17, 201–204. [Google Scholar] [CrossRef] [PubMed]
- Jezek, Z.; Nakano, J.H.; Arita, I.; Mutombo, M.; Szczeniowski, M.; Dunn, C. Serological survey for human monkeypox infections in a selected population in Zaire. J. Trop. Med. Hyg. 1987, 90, 31–38. [Google Scholar] [PubMed]
- Leland, D.S.; Ginocchio, C.C. Role of cell culture for virus detection in the age of technology. Clin. Microbiol. Rev. 2007, 20, 49–78. [Google Scholar] [CrossRef] [PubMed]
- Lapa, D.; Carletti, F.; Mazzotta, V.; Matusali, G.; Pinnetti, C.; Meschi, S.; Gagliardini, R.; Colavita, F.; Mondi, A.; Minosse, C.; et al. Monkeypox virus isolation from a semen sample collected in the early phase of infection in a patient with prolonged seminal viral shedding. Lancet Infect. Dis. 2022, 22, 1267–1269. [Google Scholar] [CrossRef]
- Saijo, M.; Ami, Y.; Suzaki, Y.; Nagata, N.; Iwata, N.; Hasegawa, H.; Iizuka, I.; Shiota, T.; Sakai, K.; Ogata, M.; et al. Virulence and pathophysiology of the Congo Basin and West African strains of monkeypox virus in non-human primates. J. Gen. Virol. 2009, 90, 2266–2271. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.W.; Lee, M.; Shin, H.; Choi, C.H.; Choi, M.M.; Kim, J.W.; Yi, H.; Yoo, C.K.; Rhie, G.E. Isolation and identification of monkeypox virus MPXV-ROK-P1-2022 from the first case in the Republic of Korea. Osong Public. Health Res. Perspect. 2022, 13, 308–311. [Google Scholar] [CrossRef]
- Mutombo, M.; Arita, I.; Jezek, Z. Human monkeypox transmitted by a chimpanzee in a tropical rain-forest area of Zaire. Lancet 1983, 1, 735–737. [Google Scholar] [CrossRef]
- Meyer, H.; Perrichot, M.; Stemmler, M.; Emmerich, P.; Schmitz, H.; Varaine, F.; Shungu, R.; Tshioko, F.; Formenty, P. Outbreaks of disease suspected of being due to human monkeypox virus infection in the Democratic Republic of Congo in 2001. J. Clin. Microbiol. 2002, 40, 2919–2921. [Google Scholar] [CrossRef]
- Mukinda, V.B.; Mwema, G.; Kilundu, M.; Heymann, D.L.; Khan, A.S.; Esposito, J.J. Re-emergence of human monkeypox in Zaire in 1996. Monkeypox Epidemiologic Working Group. Lancet 1997, 349, 1449–1450. [Google Scholar] [CrossRef]
- Sukswai, N.; Khoury, J.D. Immunohistochemistry Innovations for Diagnosis and Tissue-Based Biomarker Detection. Curr. Hematol. Malig. Rep. 2019, 14, 368–375. [Google Scholar] [CrossRef]
- Paran, N.; Yahalom-Ronen, Y.; Shifman, O.; Lazar, S.; Ben-Ami, R.; Yakubovsky, M.; Levy, I.; Wieder-Feinsod, A.; Amit, S.; Katzir, M.; et al. Monkeypox DNA levels correlate with virus infectivity in clinical samples, Israel, 2022. Eurosurveillance 2022, 27, 2200636. [Google Scholar] [CrossRef] [PubMed]
- Pfäfflin, F.; Wendisch, D.; Scherer, R.; Jürgens, L.; Godzick-Njomgang, G.; Tranter, E.; Tober-Lau, P.; Stegemann, M.S.; Corman, V.M.; Kurth, F.; et al. Monkeypox in-patients with severe anal pain. Infection 2022, 51, 483–487. [Google Scholar] [CrossRef] [PubMed]
- Dimitrakoff, J. Monkeypox Virus Infection across 16 Countries—April–June 2022. N. Engl. J. Med. 2022, 387, e69. [Google Scholar] [CrossRef] [PubMed]
- Paparizos, V.; Nicolaidou, E.; Tryfinopoulou, K.; Papa, A.; Rigopoulos, D.; Tsiodras, S.; Stratigos, A. Monkeypox virus infection: First reported case in Greece in a patient with a genital rash. J. Eur. Acad. Dermatol. Venereol. 2022, 37, e350–e351. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, T.L.; Chapman, C.A.; Cameron, K.; Saj, T.; Karesh, W.B.; Wolfe, N.D.; Wong, S.W.; Dubois, M.E.; Slifka, M.K. Serologic evidence for novel poxvirus in endangered red Colobus monkeys, Western Uganda. Emerg. Infect. Dis. 2008, 14, 801–803. [Google Scholar] [CrossRef]
- Orba, Y.; Sasaki, M.; Yamaguchi, H.; Ishii, A.; Thomas, Y.; Ogawa, H.; Hang’ombe, B.M.; Mweene, A.S.; Morikawa, S.; Saijo, M.; et al. Orthopoxvirus infection among wildlife in Zambia. J. Gen. Virol. 2015, 96, 390–394. [Google Scholar] [CrossRef]
- Khodakevich, L.; Jezek, Z.; Kinzanzka, K. Isolation of monkeypox virus from wild squirrel infected in nature. Lancet 1986, 1, 98–99. [Google Scholar] [CrossRef]
- Marennikova, S.S.; Seluhina, E.M.; Mal’ceva, N.N.; Ladnyj, I.D. Poxviruses isolated from clinically ill and asymptomatically infected monkeys and a chimpanzee. Bull. World Health Organ. 1972, 46, 613–620. [Google Scholar]
- Hutson, C.L.; Lee, K.N.; Abel, J.; Carroll, D.S.; Montgomery, J.M.; Olson, V.A.; Li, Y.; Davidson, W.; Hughes, C.; Dillon, M.; et al. Monkeypox zoonotic associations: Insights from laboratory evaluation of animals associated with the multi-state US outbreak. Am. J. Trop. Med. Hyg. 2007, 76, 757–768. [Google Scholar] [CrossRef]
- Seang, S.; Burrel, S.; Todesco, E.; Leducq, V.; Monsel, G.; Le Pluart, D.; Cordevant, C.; Pourcher, V.; Palich, R. Evidence of human-to-dog transmission of monkeypox virus. Lancet 2022, 400, 658–659. [Google Scholar] [CrossRef]
- Schroeder, K.; Nitsche, A. Multicolour, multiplex real-time PCR assay for the detection of human-pathogenic poxviruses. Mol. Cell. Probes 2010, 24, 110–113. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Olson, V.A.; Laue, T.; Laker, M.T.; Damon, I.K. Detection of monkeypox virus with real-time PCR assays. J. Clin. Virol. 2006, 36, 194–203. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhao, H.; Wilkins, K.; Hughes, C.; Damon, I.K. Real-time PCR assays for the specific detection of monkeypox virus West African and Congo Basin strain DNA. J. Virol. Methods 2010, 169, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Kalthan, E.; Tenguere, J.; Ndjapou, S.G.; Koyazengbe, T.A.; Mbomba, J.; Marada, R.M.; Rombebe, P.; Yangueme, P.; Babamingui, M.; Sambella, A.; et al. Investigation of an outbreak of monkeypox in an area occupied by armed groups, Central African Republic. Med. Mal. Infect. 2018, 48, 263–268. [Google Scholar] [CrossRef]
- La Rosa, G.; Mancini, P.; Veneri, C.; Ferraro, G.B.; Lucentini, L.; Iaconelli, M.; Suffredini, E. Detection of Monkeypox Virus DNA in Airport Wastewater, Rome, Italy. Emerg. Infect. Dis. 2023, 29, 193–196. [Google Scholar] [CrossRef] [PubMed]
- Shchelkunov, S.N.; Gavrilova, E.V.; Babkin, I.V. Multiplex PCR detection and species differentiation of orthopoxviruses pathogenic to humans. Mol. Cell. Probes 2005, 19, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Neubauer, H.; Reischl, U.; Ropp, S.; Esposito, J.J.; Wolf, H.; Meyer, H. Specific detection of monkeypox virus by polymerase chain reaction. J. Virol. Methods 1998, 74, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Kubista, M.; Andrade, J.M.; Bengtsson, M.; Forootan, A.; Jonák, J.; Lind, K.; Sindelka, R.; Sjöback, R.; Sjögreen, B.; Strömbom, L.; et al. The real-time polymerase chain reaction. Mol. Aspects Med. 2006, 27, 95–125. [Google Scholar] [CrossRef]
- Holland, P.M.; Abramson, R.D.; Watson, R.; Gelfand, D.H. Detection of specific polymerase chain reaction product by utilizing the 5′----3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. USA 1991, 88, 7276–7280. [Google Scholar] [CrossRef]
- Arya, M.; Shergill, I.S.; Williamson, M.; Gommersall, L.; Arya, N.; Patel, H.R. Basic principles of real-time quantitative PCR. Expert. Rev. Mol. Diagn. 2005, 5, 209–219. [Google Scholar] [CrossRef]
- Cardullo, R.A.; Agrawal, S.; Flores, C.; Zamecnik, P.C.; Wolf, D.E. Detection of nucleic acid hybridization by nonradiative fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. USA 1988, 85, 8790–8794. [Google Scholar] [CrossRef] [PubMed]
- Kutyavin, I.V.; Afonina, I.A.; Mills, A.; Gorn, V.V.; Lukhtanov, E.A.; Belousov, E.S.; Singer, M.J.; Walburger, D.K.; Lokhov, S.G.; Gall, A.A.; et al. 3′-minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures. Nucleic Acids Res. 2000, 28, 655–661. [Google Scholar] [CrossRef]
- Martins-Filho, P.R.; Tanajura, D.M.; Alves Dos Santos, C. Polymerase chain reaction positivity and cycle threshold values in biological samples from patients with monkeypox: A meta-analysis. Travel. Med. Infect. Dis. 2022, 50, 102448. [Google Scholar] [CrossRef]
- Ma, A.; Langer, J.; Hanson, K.E.; Bradley, B.T. Characterization of the Cytopathic Effects of Monkeypox Virus Isolated from Clinical Specimens and Differentiation from Common Viral Exanthems. J. Clin. Microbiol. 2022, 60, e0133622. [Google Scholar] [CrossRef] [PubMed]
- Shchelkunov, S.N.; Totmenin, A.V.; Safronov, P.F.; Mikheev, M.V.; Gutorov, V.V.; Ryazankina, O.I.; Petrov, N.A.; Babkin, I.V.; Uvarova, E.A.; Sandakhchiev, L.S.; et al. Analysis of the monkeypox virus genome. Virology 2002, 297, 172–194. [Google Scholar] [CrossRef] [PubMed]
- Dumont, C.; Irenge, L.M.; Magazani, E.K.; Garin, D.; Muyembe, J.J.; Bentahir, M.; Gala, J.L. Simple technique for in field samples collection in the cases of skin rash illness and subsequent PCR detection of orthopoxviruses and varicella zoster virus. PLoS ONE 2014, 9, e96930. [Google Scholar] [CrossRef] [PubMed]
- Nolen, L.D.; Osadebe, L.; Katomba, J.; Likofata, J.; Mukadi, D.; Monroe, B.; Doty, J.; Hughes, C.M.; Kabamba, J.; Malekani, J.; et al. Extended Human-to-Human Transmission during a Monkeypox Outbreak in the Democratic Republic of the Congo. Emerg. Infect. Dis. 2016, 22, 1014–1021. [Google Scholar] [CrossRef] [PubMed]
- Aguilera-Alonso, D.; Alonso-Cadenas, J.A.; Roguera-Sopena, M.; Lorusso, N.; Miguel, L.G.S.; Calvo, C. Monkeypox virus infections in children in Spain during the first months of the 2022 outbreak. Lancet Child. Adolesc. Health 2022, 6, e22–e23. [Google Scholar] [CrossRef]
- Jezek, Z.; Arita, I.; Mutombo, M.; Dunn, C.; Nakano, J.H.; Szczeniowski, M. Four generations of probable person-to-person transmission of human monkeypox. Am. J. Epidemiol. 1986, 123, 1004–1012. [Google Scholar] [CrossRef]
- de Nicolas-Ruanes, B.; Vivancos, M.J.; Azcarraga-Llobet, C.; Moreno, A.M.; Rodriguez-Dominguez, M.; Berna-Rico, E.D.; Garcia-Mouronte, E.; Carron-Herrero, A.; McGee, A.; Galan, J.C.; et al. Monkeypox virus case with maculopapular exanthem and proctitis during the Spanish outbreak in 2022. J. Eur. Acad. Dermatol. Venereol. 2022, 36, e658–e660. [Google Scholar] [CrossRef]
- Nolasco, S.; Vitale, F.; Geremia, A.; Tramuto, F.; Maida, C.M.; Sciuto, A.; Coco, C.; Manuele, R.; Frasca, E.; Frasca, M.; et al. First case of monkeypox virus, SARS-CoV-2 and HIV co-infection. J. Infect. 2023, 86, e21–e23. [Google Scholar] [CrossRef] [PubMed]
- Tarín-Vicente, E.J.; Alemany, A.; Agud-Dios, M.; Ubals, M.; Suñer, C.; Antón, A.; Arando, M.; Arroyo-Andrés, J.; Calderón-Lozano, L.; Casañ, C.; et al. Clinical presentation and virological assessment of confirmed human monkeypox virus cases in Spain: A prospective observational cohort study. Lancet 2022, 400, 661–669. [Google Scholar] [CrossRef] [PubMed]
- Pipitò, L.; Cascio, A. Monkeypox virus infection and creatine phosphokinase increase: A case from Italy. Travel. Med. Infect. Dis. 2022, 50, 102412. [Google Scholar] [CrossRef]
- Peiró-Mestres, A.; Fuertes, I.; Camprubí-Ferrer, D.; Marcos, M.; Vilella, A.; Navarro, M.; Rodriguez-Elena, L.; Riera, J.; Català, A.; Martínez, M.J.; et al. Frequent detection of monkeypox virus DNA in saliva, semen, and other clinical samples from 12 patients, Barcelona, Spain, May to June 2022. Eurosurveillance 2022, 27, 2200503. [Google Scholar] [CrossRef]
- Vallée, A.; Farfour, E.; Zucman, D. Monkeypox virus: A novel sexually transmitted disease? A case report from France. Travel. Med. Infect. Dis. 2022, 49, 102394. [Google Scholar] [CrossRef] [PubMed]
- Boesecke, C.; Monin, M.B.; van Bremen, K.; Schlabe, S.; Hoffmann, C. Severe monkeypox-virus infection in undiagnosed advanced HIV infection. Infection 2022, 50, 1633–1634. [Google Scholar] [CrossRef]
- Vaughan, A.; Aarons, E.; Astbury, J.; Brooks, T.; Chand, M.; Flegg, P.; Hardman, A.; Harper, N.; Jarvis, R.; Mawdsley, S.; et al. Human-to-Human Transmission of Monkeypox Virus, United Kingdom, October 2018. Emerg. Infect. Dis. 2020, 26, 782–785. [Google Scholar] [CrossRef]
- Okareh, O.T.; Morakinyo, O. Monkeypox in Nigeria: A case report of re-emerged disease outbreak. J. Microbiol. Exp. 2018, 6, 89–91. [Google Scholar] [CrossRef]
- Miura, F.; van Ewijk, C.E.; Backer, J.A.; Xiridou, M.; Franz, E.; Op de Coul, E.; Brandwagt, D.; van Cleef, B.; van Rijckevorsel, G.; Swaan, C.; et al. Estimated incubation period for monkeypox cases confirmed in the Netherlands, May 2022. Eurosurveillance 2022, 27, 2200448. [Google Scholar] [CrossRef]
- Reynolds, M.G.; Yorita, K.L.; Kuehnert, M.J.; Davidson, W.B.; Huhn, G.D.; Holman, R.C.; Damon, I.K. Clinical manifestations of human monkeypox influenced by route of infection. J. Infect. Dis. 2006, 194, 773–780. [Google Scholar] [CrossRef]
- Turco, M.; Mancuso, F.R.; Pisano, L. A monkeypox virus infection mimicking primary syphilis. Br. J. Dermatol. 2022, 187, e194–e195. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, R.P.; Gordon, M.L. A Systematic Review Analyzing the Prevalence and Circulation of Influenza Viruses in Swine Population Worldwide. Pathogens 2020, 9, 355. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, R.P.; Gordon, M.L. Deciphering transmission dynamics and spillover of avian influenza viruses from avian species to swine populations globally. Virus Genes 2021, 57, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, R.P.; Gordon, M.L. A systematic review of influenza A virus prevalence and transmission dynamics in backyard swine populations globally. Porc. Health Manag. 2022, 8, 10. [Google Scholar] [CrossRef]
- Chauhan, R.P.; Gordon, M.L. Review of genome sequencing technologies in molecular characterization of influenza A viruses in swine. J. Vet. Diagn. Investig. 2022, 34, 177–189. [Google Scholar] [CrossRef]
- Chauhan, R.P.; Gordon, M.L. An overview of influenza A virus genes, protein functions, and replication cycle highlighting important updates. Virus Genes 2022, 58, 255–269. [Google Scholar] [CrossRef]
- Chauhan, R.P.; San, J.E.; Gordon, M.L. Metagenomic Analysis of RNA Fraction Reveals the Diversity of Swine Oral Virome on South African Backyard Swine Farms in the uMgungundlovu District of KwaZulu-Natal Province. Pathogens 2022, 11, 927. [Google Scholar] [CrossRef]
- Chauhan, R.P.; Gordon, M.L. Characterization of a Near Full-Length Hepatitis E Virus Genome of Subtype 3c Generated from Naturally Infected South African Backyard Pigs. Pathogens 2022, 11, 1030. [Google Scholar] [CrossRef]
- Blome, S.; Franzke, K.; Beer, M. African swine fever—A review of current knowledge. Virus Res. 2020, 287, 198099. [Google Scholar] [CrossRef]
- Rudova, N.; Buttler, J.; Kovalenko, G.; Sushko, M.; Bolotin, V.; Muzykina, L.; Zinenko, O.; Stegniy, B.; Dunaiev, Y.; Sytiuk, M.; et al. Genetic Diversity of Porcine Circovirus 2 in Wild Boar and Domestic Pigs in Ukraine. Viruses 2022, 14, 924. [Google Scholar] [CrossRef]
- Lederman, E.R.; Reynolds, M.G.; Karem, K.; Braden, Z.; Learned-Orozco, L.A.; Wassa-Wassa, D.; Moundeli, O.; Hughes, C.; Harvey, J.; Regnery, R.; et al. Prevalence of antibodies against orthopoxviruses among residents of Likouala region, Republic of Congo: Evidence for monkeypox virus exposure. Am. J. Trop. Med. Hyg. 2007, 77, 1150–1156. [Google Scholar] [CrossRef] [PubMed]
- Zaucha, G.M.; Jahrling, P.B.; Geisbert, T.W.; Swearengen, J.R.; Hensley, L. The pathology of experimental aerosolized monkeypox virus infection in cynomolgus monkeys (Macaca fascicularis). Lab. Investig. 2001, 81, 1581–1600. [Google Scholar] [CrossRef] [PubMed]
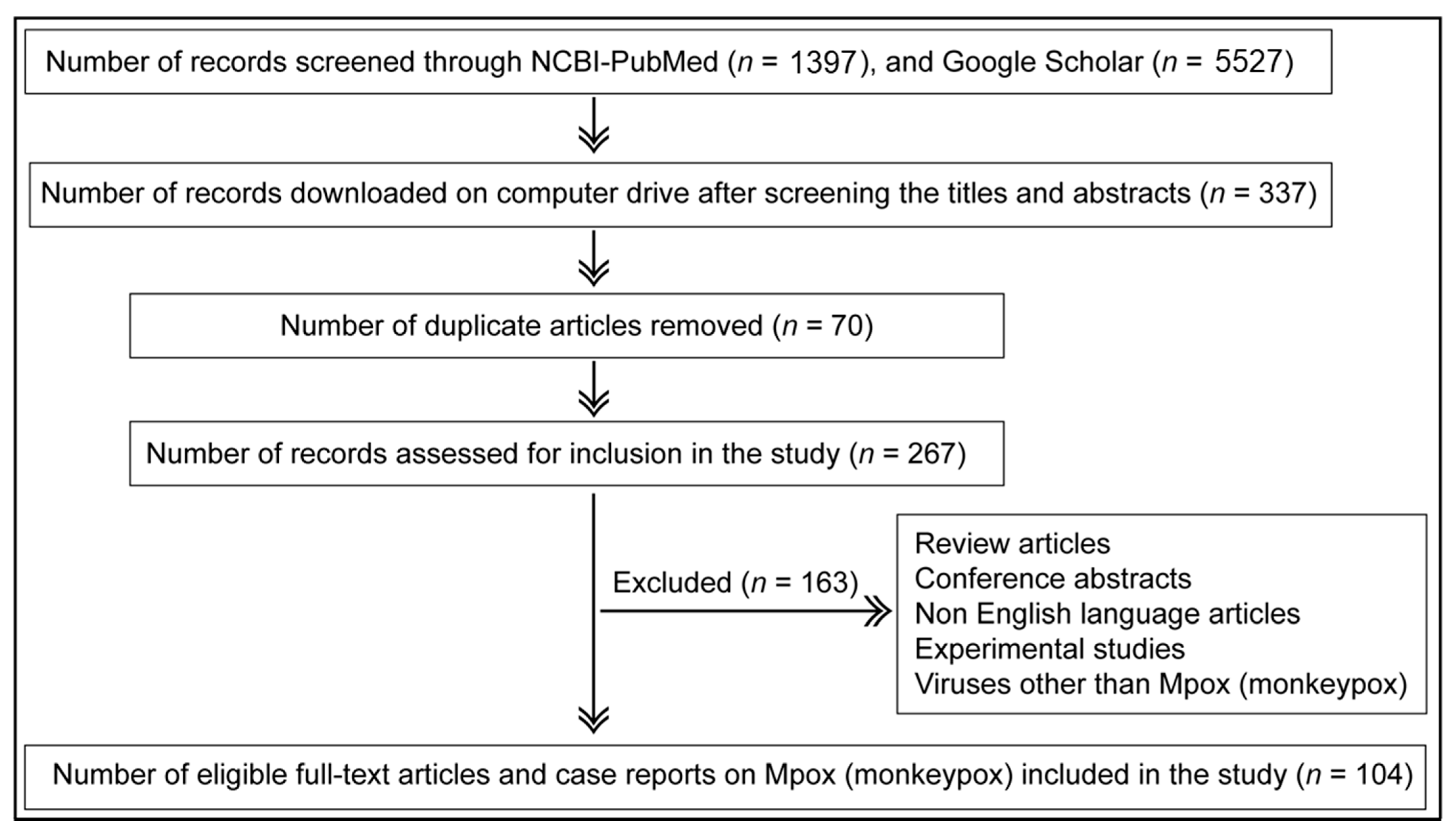
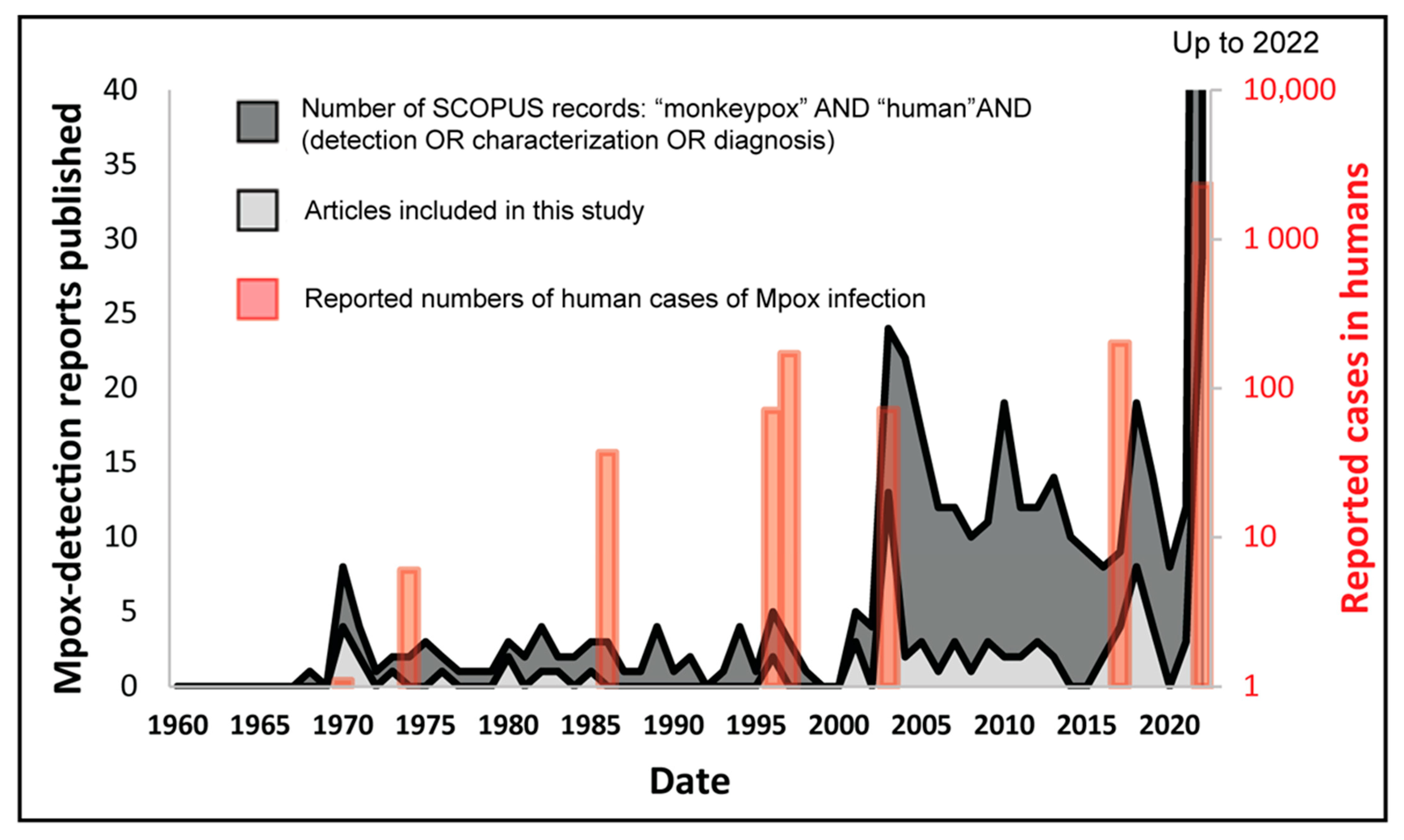
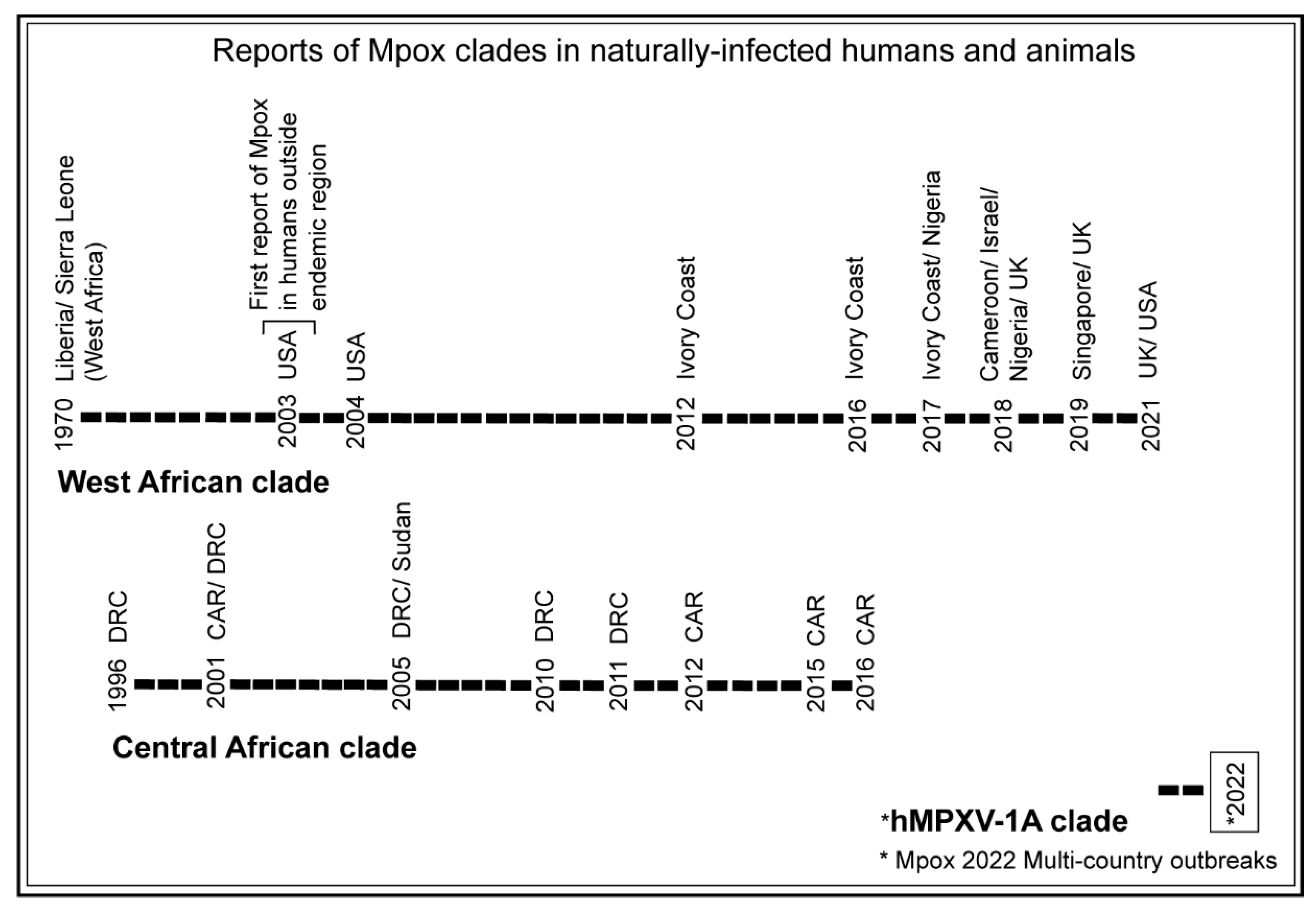
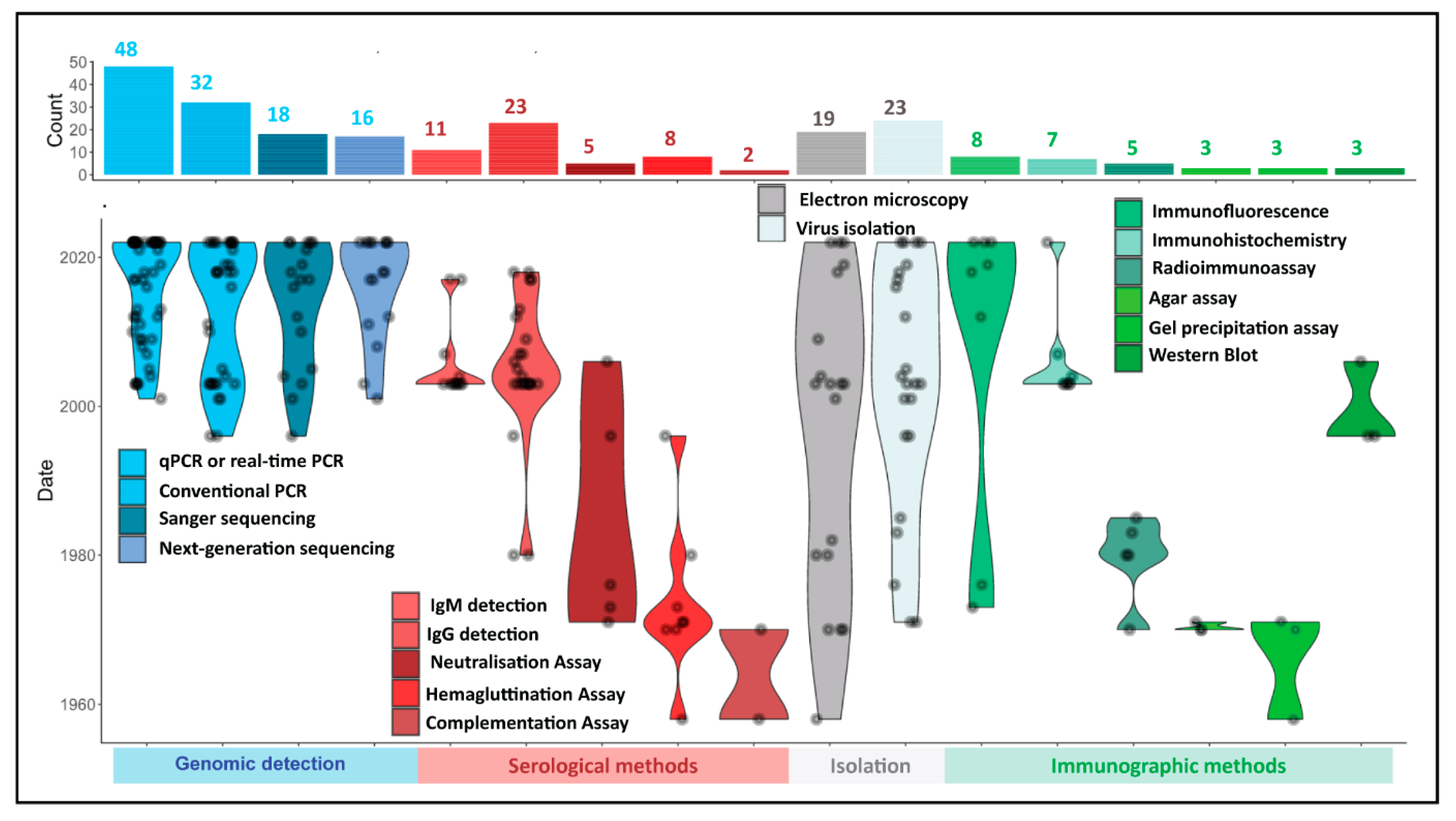
| Search Terms Used | # Titles Screened in NCBI-PubMed | # Titles Screened in Google Scholar |
|---|---|---|
| Monkeypox virus outbreak | 328 | 430 |
| Detection of monkeypox virus disease | 83 | 390 |
| Monkeypox virus in Africa | 240 | 330 |
| Epidemiology of monkeypox virus disease | 223 | 270 |
| Molecular detection of monkeypox virus | 33 | 310 |
| Serological detection of monkeypox virus | 10 | 270 |
| Microarray for monkeypox virus | 15 | 210 |
| Real-time PCR for monkeypox virus | 43 | 360 |
| Antigenic detection of monkeypox virus | 29 | 250 |
| Genomic sequencing of monkeypox virus | 83 | 220 |
| Rapid detection of monkeypox virus | 28 | 260 |
| Point of care detection of monkeypox virus | 3 | 150 |
| ELISA for monkeypox virus detection | 16 | 190 |
| Monkeypox virus outbreak 2022 | 263 | 490 |
| Target (Specificity) | ORF or Genomic Region/Primer and Probe ID | Oligonucleotide Sequence 5′–3′ | References (Earliest Citation) |
|---|---|---|---|
| General OPXV-detecting real-time PCR | |||
| P4A: Major core protein 4a precursor (OPXV family) | P4A/Forward | TAATACTTCGATTGCTCATCCAGG | [155] |
| P4A/Reverse | ACTTCTCACAAATGGATTTGAAAATC | ||
| P4A/Probe | FAM-TCCTTTACGTGATAAATCAT-NFQ MGB | ||
| E9L: DNA polymerase gene (OPXV family; non-variola) | E9L/Forward | TCAACTGAAAAGGCCATCTATGA | [13,64,114,153,156] |
| E9L/Reverse | GAGTATAGAGCACTATTTCTAAATCCCA | ||
| E9L/NVAR-Probe | TET-CCATGCAATATACGTACAAGATA- GTAGCCAAC-QSY7 | ||
| Mpox-specific real-time PCR | |||
| G2R: TNF receptor gene (Mpox-generic) | G2R/G-Forward | GGAAAATGTAAAGACAACGAATACAG | [3,8,114,157,158] |
| G2R/G-Reverse | GCTATCACATAATCTGGAAGCGTA | ||
| G2R/G-Probe | FAM-AAGCCGTAATCTATGTTGTCTATC-GTGTCC-BHQ1 | ||
| G2R: TNF receptor gene (West African-specific Mpox clade) | G2R/WA-Forward | CACACCGTCTCTTCCACAGA | [3,8,114,157,158] |
| G2R/WA-Reverse | GATACAGGTTAATTTCCACATCG | ||
| G2R/WA-Probe | FAM-AACCCGTCGTAACCAG-CAATACATTT-BHQ1 | ||
| C3L: Complement binding protein Central African (formerly Congo Basin)-specific Mpox clade | C3L/CB-Forward | TGTCTACCTGGATACAGAAAGCAA | [3,8,114,157,158] |
| C3L/CB-Reverse | GGCATCTCCGTTTAATACATTGAT | ||
| C3L/CB-Probe | FAM-CCCATATATGCTAAATGTACCGGT-ACCGGA-BHQ1 | ||
| B6R: Envelope protein gene (Mpox) | B6R/Forward | ATTGGTCATTATTTTTGTCACAGGAACA | [13,64,114,153,156] |
| B6R/Reverse | AATGGCGTTGACAATTATGGGTG | ||
| B6R/Probe | MGB-AGAGATTAGAAATA-FAM | ||
| F3L: Interferon resistance gene (Mpox) | F3L/F290-Forward | CTCATTGATTTTTCGCGGGATA | [40,89,109,159] |
| F3L/R396-Reverse | GACGATACTCCTCCTCGTTGGT | ||
| F3L/p333S-MGB-Probe | FAM-CATCAGAATCTGTAGGCCGT-MGB NFQ | ||
| N3R: Conserved (unclassified function) (Mpox) | N3R/F319-Forward | AACAACCGTCCTACAATTAAACAACA | [40,89,109,159] |
| N3R/R457-Reverse | CGCTATCGAACCATTTTTGTAGTCT | ||
| N3R/p352S-MGB-Probe | FAM-TATAACGGCGAAGAATATACT-MGB NFQ | ||
| Conventional Mpox PCR | |||
| E5R: Nucleic acid-independent nucleoside triphosphatase (Mpox) | E5R/Forward | ATGTTGATATTAATAATCGTATTGTGGTT | [4,160] |
| E5R/Reverse | AAAGTCAATACACTCTTAAAGATTCTCAA | ||
| ATI: A-type inclusion body protein (Mpox) | ATI/Gabon 1- Forward | GAGAGAATCTCTTGATAT | [142,161] |
| ATI/Gabon 2- Reverse | ATTCTAGATTGTAATC | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chauhan, R.P.; Fogel, R.; Limson, J. Overview of Diagnostic Methods, Disease Prevalence and Transmission of Mpox (Formerly Monkeypox) in Humans and Animal Reservoirs. Microorganisms 2023, 11, 1186. https://doi.org/10.3390/microorganisms11051186
Chauhan RP, Fogel R, Limson J. Overview of Diagnostic Methods, Disease Prevalence and Transmission of Mpox (Formerly Monkeypox) in Humans and Animal Reservoirs. Microorganisms. 2023; 11(5):1186. https://doi.org/10.3390/microorganisms11051186
Chicago/Turabian StyleChauhan, Ravendra P., Ronen Fogel, and Janice Limson. 2023. "Overview of Diagnostic Methods, Disease Prevalence and Transmission of Mpox (Formerly Monkeypox) in Humans and Animal Reservoirs" Microorganisms 11, no. 5: 1186. https://doi.org/10.3390/microorganisms11051186
APA StyleChauhan, R. P., Fogel, R., & Limson, J. (2023). Overview of Diagnostic Methods, Disease Prevalence and Transmission of Mpox (Formerly Monkeypox) in Humans and Animal Reservoirs. Microorganisms, 11(5), 1186. https://doi.org/10.3390/microorganisms11051186






