Effect of Anthropogenic Disturbances on the Microbial Relationship during Bioremediation of Heavy Metal-Contaminated Sediment
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Preparation
2.2. Strains and Growth Conditions
2.3. Intensified Bioleaching Experiments
2.4. Preparation of Microbial Genomic DNA for Sequencing
2.5. Data Analysis
3. Results
3.1. pH and Oxidation–Reduction Potential (ORP) Variation during Bioleaching
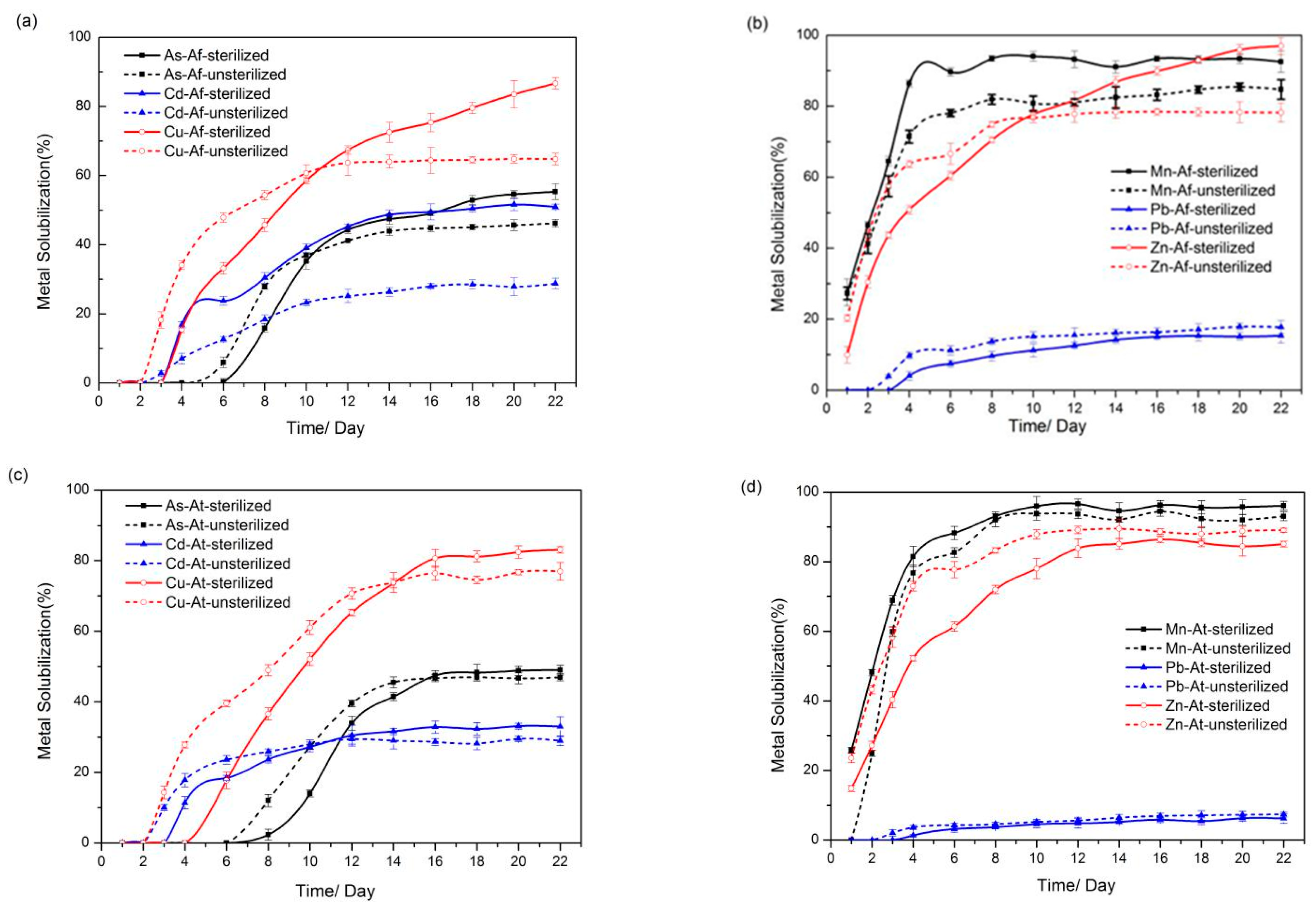
3.2. Solubilization of Heavy Metals
3.3. Microbial Community Structure
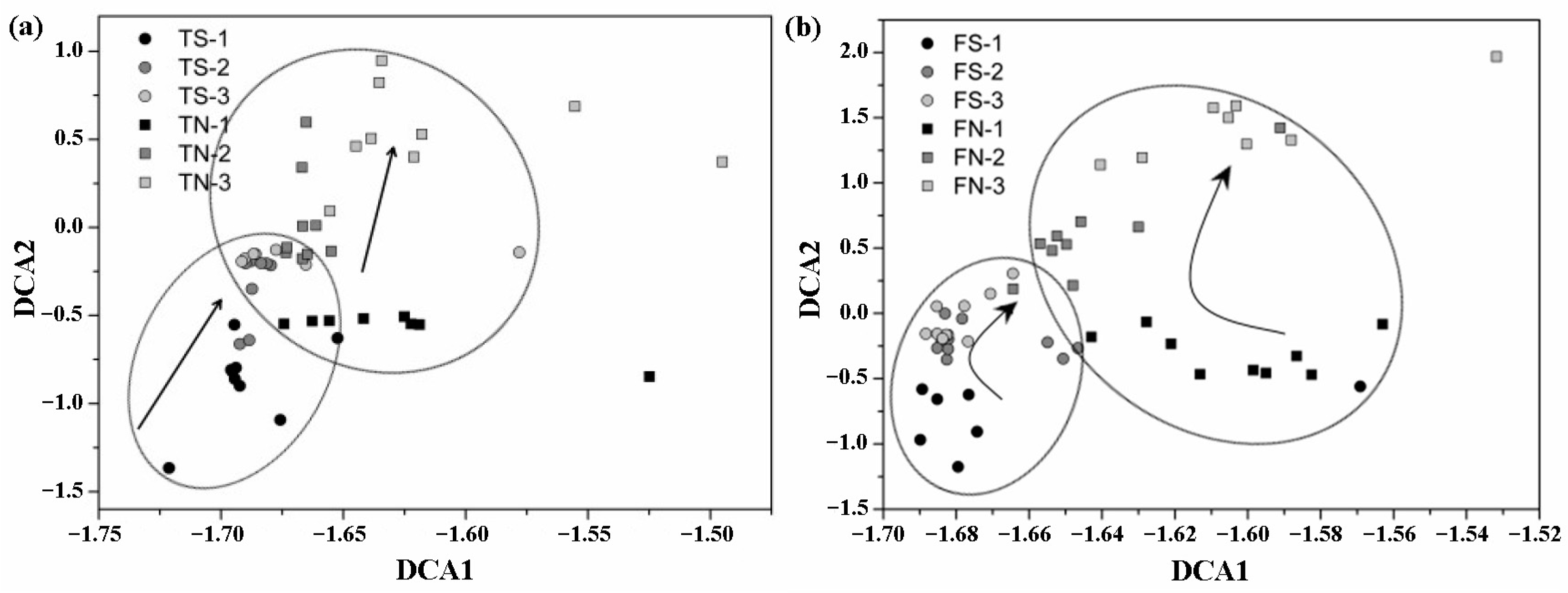
3.4. Analysis of the Altered in Microbial Community with Temporal Turnover
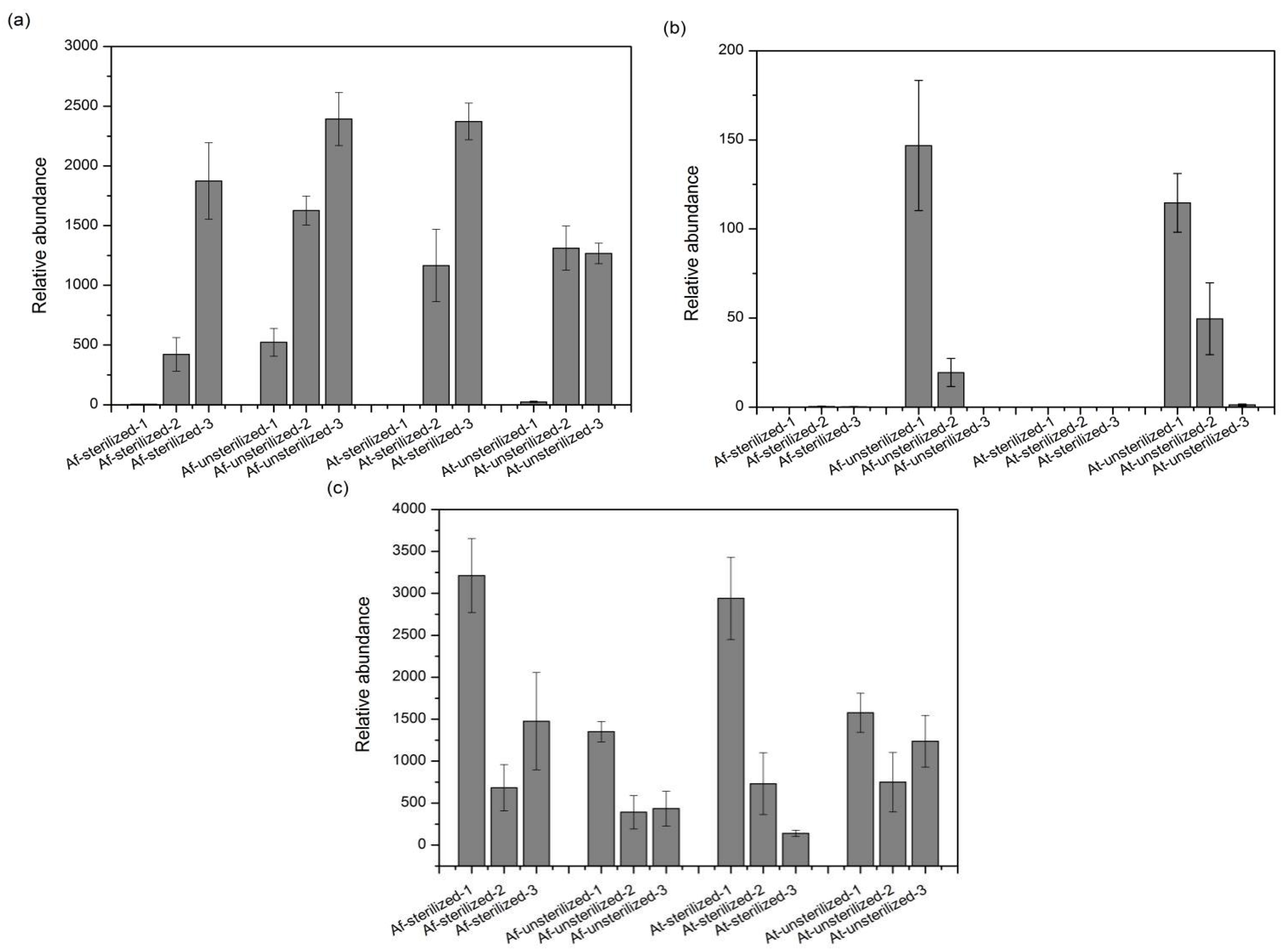
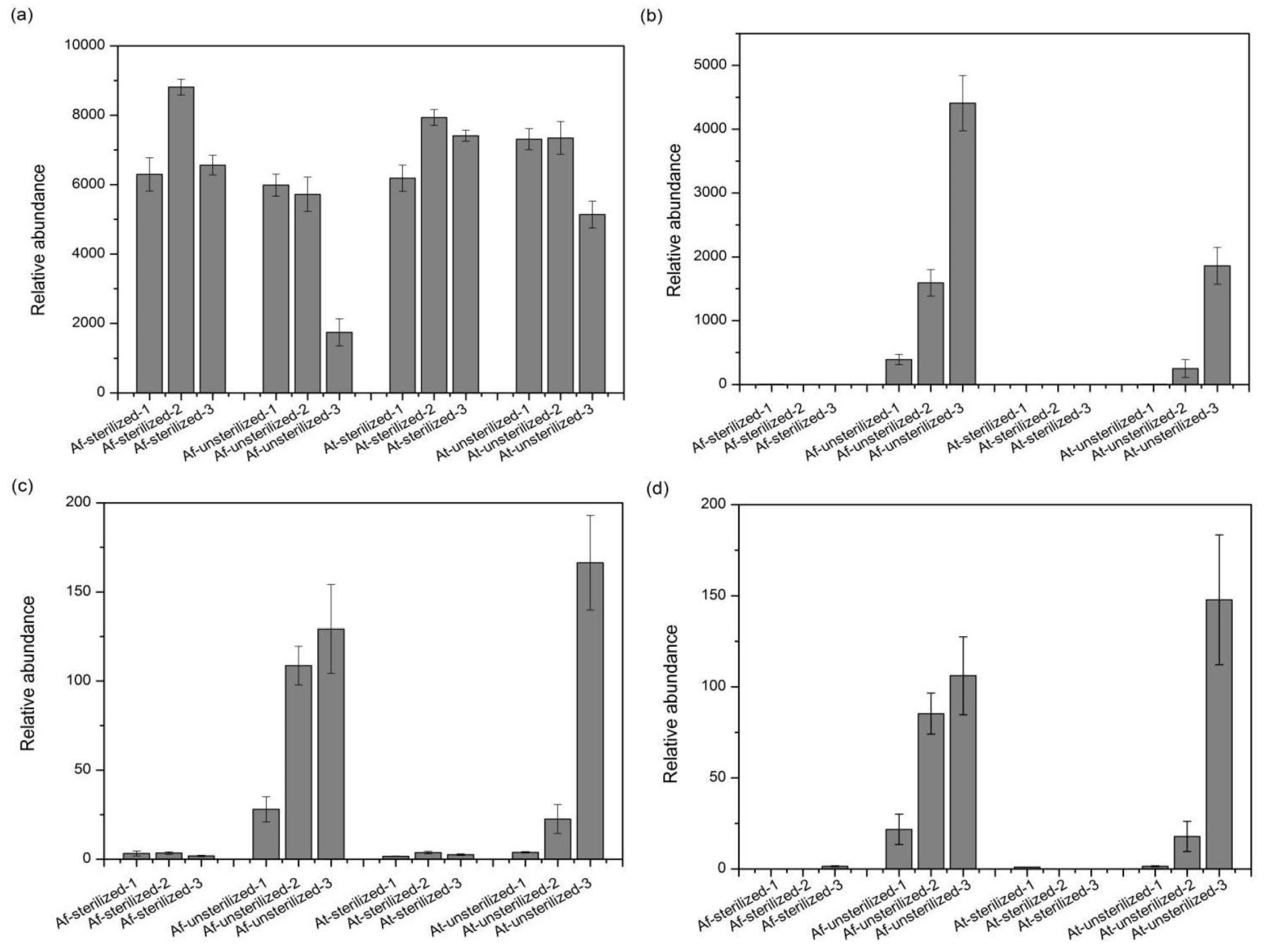
3.5. Species with Significant Variation
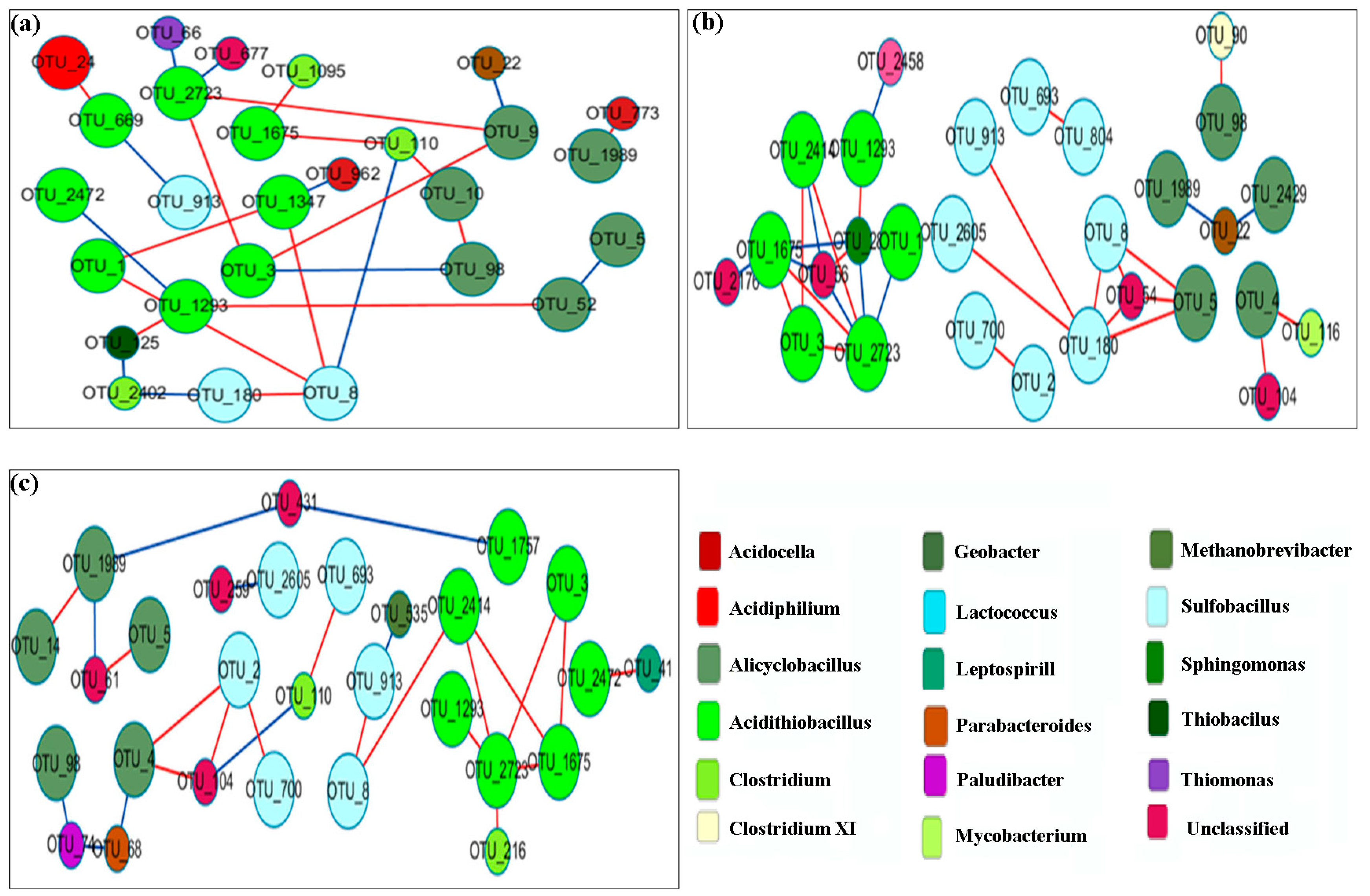
3.6. Network Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rajendran, S.; Priya, T.A.K.; Khoo, K.S.; Hoang, T.K.A.; Ng, H.-S.; Munawaroh, H.S.H.; Karaman, C.; Orooji, Y.; Show, P.L. A critical review on various remediation approaches for heavy metal contaminants removal from contaminated soils. Chemosphere 2022, 287, 132369. [Google Scholar] [CrossRef]
- Faheem, M.; Shabbir, S.; Zhao, J.; Kerr, P.G.; Sultana, N.; Jia, Z. Enhanced Adsorptive Bioremediation of Heavy Metals (Cd2+, Cr6+, Pb2+) by Methane-Oxidizing Epipelon. Microorganisms 2020, 8, 505. [Google Scholar] [CrossRef]
- Ayari, J.; Barbieri, M.; Agnan, Y.; Sellami, A.; Braham, A.; Dhaha, F.; Charef, A. Trace element contamination in the mine-affected stream sediments of Oued Rarai in north-western Tunisia: A river basin scale assessment. Environ. Geochem. Health 2021, 43, 4027–4042. [Google Scholar] [CrossRef]
- Ayari, J.; Barbieri, M.; Barhoumi, A.; Boschetti, T.; Braham, A.; Dhaha, F.; Charef, A. Trace metal element pollution in media from the abandoned Pb and Zn mine of Lakhouat, Northern Tunisia. J. Geochem. Explor. 2023, 247, 107180. [Google Scholar] [CrossRef]
- Cimermanova, M.; Pristas, P.; Piknova, M. Biodiversity of Actinomycetes from Heavy Metal Contaminated Technosols. Microorganisms 2021, 9, 1635. [Google Scholar] [CrossRef]
- Atuchin, V.V.; Asyakina, L.K.; Serazetdinova, Y.R.; Frolova, A.S.; Velichkovich, N.S.; Prosekov, A.Y. Microorganisms for Bioremediation of Soils Contaminated with Heavy Metals. Microorganisms 2023, 11, 864. [Google Scholar] [CrossRef]
- Wu, H.; Liu, Q.; Ma, J.; Liu, L.; Qu, Y.; Gong, Y.; Yang, S.; Luo, T. Heavy Metal(loids) in typical Chinese tobacco-growing soils: Concentrations, influence factors and potential health risks. Chemosphere 2020, 245, 125591. [Google Scholar] [CrossRef]
- Gan, M.; Zhou, S.; Li, M.; Zhu, J.; Liu, X.; Chai, L. Bioleaching of multiple heavy metals from contaminated sediment by mesophile consortium. Environ. Sci. Pollut. Res. 2015, 22, 5807–5816. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, L.; Kong, L.; Liu, E.; Wang, L.; Zhu, J. Spatial distribution, ecological risk assessment and source identification for heavy metals in surface sediments from Dongping Lake, Shandong, East China. Catena 2015, 125, 200–205. [Google Scholar] [CrossRef]
- Ke, X.; Gui, S.; Huang, H.; Zhang, H.; Wang, C.; Guo, W. Ecological risk assessment and source identification for heavy metals in surface sediment from the Liaohe River protected area, China. Chemosphere 2017, 175, 473–481. [Google Scholar] [CrossRef]
- Jie, S.; Li, M.; Gan, M.; Zhu, J.; Yin, H.; Liu, X. Microbial functional genes enriched in the Xiangjiang River sediments with heavy metal contamination. BMC Microbiol. 2016, 16, 179. [Google Scholar] [CrossRef]
- Davis, T.A.; Volesky, B.; Mucci, A. A review of the biochemistry of heavy metal biosorption by brown algae. Water Res. 2003, 37, 4311–4330. [Google Scholar] [CrossRef]
- He, L.; Zhong, H.; Liu, G.; Dai, Z.; Brookes, P.C.; Xu, J. Remediation of heavy metal contaminated soils by biochar: Mechanisms, potential risks and applications in China. Environ. Pollut. 2019, 252, 846–855. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Y.; Zhang, Z.; Zhang, H.; Xing, M.; Marhaba, T.; Zhang, W. In situ capping technology for controlling heavy metals release from contaminated sediment. Desalin. Water Treat. 2019, 168, 261–268. [Google Scholar] [CrossRef]
- Chen, Y.; He, P.; Zhang, K.; Wang, X.; Liu, M.; Chen, F.; Gan, M.; Zhu, J. Enhanced Cr(VI) reduction using highly conductive material synthesized by modified chitosan coated with natural iron-manganese minerals. Appl. Surf. Sci. 2023, 611, 155635. [Google Scholar] [CrossRef]
- Gu, C.; Cai, M.; He, P.; Zhu, J.; Gan, M. Biogenic carbon encapsulated iron oxides mediated oxalic acid for Cr(VI) reduction in aqueous: Efficient performance, electron transfer and radical mechanisms. Chemosphere 2023, 313, 137557. [Google Scholar] [CrossRef]
- Etesami, H. Bacterial mediated alleviation of heavy metal stress and decreased accumulation of metals in plant tissues: Mechanisms and future prospects. Ecotoxicol. Environ. Saf. 2018, 147, 175–191. [Google Scholar] [CrossRef]
- Darma, A.; Yang, J.; Zandi, P.; Liu, J.; Możdżeń, K.; Xia, X.; Sani, A.; Wang, Y.; Schnug, E. Significance of Shewanella Species for the Phytoavailability and Toxicity of Arsenic—A Review. Biology 2022, 11, 472. [Google Scholar] [CrossRef]
- Zhang, K.; Li, N.; Liao, P.; Jin, Y.; Li, Q.; Gan, M.; Chen, Y.; He, P.; Chen, F.; Peng, M.; et al. Conductive property of secondary minerals triggered Cr(VI) bioreduction by dissimilatory iron reducing bacteria. Environ. Pollut. 2021, 286, 117227. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, T.; Shafi, S.; Bilal, M.; Hussain, T.; Sher, F.; Rizwan, K. Surfactants-based remediation as an effective approach for removal of environmental pollutants—A review. J. Mol. Liq. 2020, 318, 113960. [Google Scholar] [CrossRef]
- Xue, J.; Zhu, E.; Zhu, H.; Liu, D.; Cai, H.; Xiong, C.; Yang, Q.; Shi, Z. Dye adsorption performance of nanocellulose beads with different carboxyl group content. Cellulose 2023, 30, 1623–1636. [Google Scholar] [CrossRef]
- Agrawal, K.; Ruhil, T.; Gupta, V.K.; Verma, P. Microbial assisted multifaceted amelioration processes of heavy-metal remediation: A clean perspective toward sustainable and greener future. Crit. Rev. Biotechnol. 2023, 1–19. [Google Scholar] [CrossRef]
- Cai, G.; Ebrahimi, M.; Zheng, G.; Kaksonen, A.H.; Morris, C.; O’Hara, I.M.; Zhang, Z. Effect of ferrous iron loading on dewaterability, heavy metal removal and bacterial community of digested sludge by Acidithiobacillus ferrooxidans. J. Environ. Manag. 2021, 295, 113114. [Google Scholar] [CrossRef]
- Moliterni, E.; Rodriguez, L.; Fernández, F.J.; Villaseñor, J. Feasibility of Different Bioremediation Strategies for Treatment of Clayey and Silty Soils Recently Polluted with Diesel Hydrocarbons. Water Air Soil Pollut. 2012, 223, 2473–2482. [Google Scholar] [CrossRef]
- Yang, Y.; Song, Y.; Scheller, H.V.; Ghosh, A.; Ban, Y.; Chen, H.; Tang, M. Community structure of arbuscular mycorrhizal fungi associated with Robinia pseudoacacia in uncontaminated and heavy metal contaminated soils. Soil Biol. Biochem. 2015, 86, 146–158. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590. [Google Scholar] [CrossRef]
- Lavrinienko, A.; Jernfors, T.; Koskimäki, J.J.; Pirttilä, A.M.; Watts, P.C. Does Intraspecific Variation in rDNA Copy Number Affect Analysis of Microbial Communities? Trends Microbiol. 2021, 29, 19–27. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Cole, J.R.; Wang, Q.; Fish, J.A.; Chai, B.; McGarrell, D.M.; Sun, Y.; Brown, C.T.; Porras-Alfaro, A.; Kuske, C.R.; Tiedje, J.M. Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014, 42, D633–D642. [Google Scholar] [CrossRef]
- Goecks, J.; Nekrutenko, A.; Taylor, J.; The Galaxy Team. Galaxy: A comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010, 11, R86. [Google Scholar] [CrossRef]
- The Galaxy Community. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2022 update. Nucleic Acids Res. 2022, 50, W345–W351. [Google Scholar] [CrossRef]
- Chang, C.-Y.; Chen, S.-Y.; Klipkhayai, P.; Chiemchaisri, C. Bioleaching of heavy metals from harbor sediment using sulfur-oxidizing microflora acclimated from native sediment and exogenous soil. Environ. Sci. Pollut. Res. 2019, 26, 6818–6828. [Google Scholar] [CrossRef]
- Wu, C.; Hu, X.; Wang, H.; Lin, Q.; Shen, C.; Lou, L. Exploring key physicochemical sediment properties influencing bioleaching of heavy metals. J. Hazard. Mater. 2023, 445, 130506. [Google Scholar] [CrossRef]
- Clark, D.A.; Norris, P.R. Acidimicrobium ferrooxidans gen. nov., sp. nov.: Mixed-culture ferrous iron oxidation with Sulfobacillus species. Microbiology 1996, 142, 785–790. [Google Scholar] [CrossRef]
- Huynh, D.; Kaschabek, S.R.; Schlömann, M. Effect of inoculum history, growth substrates and yeast extract addition on inhibition of Sulfobacillus thermosulfidooxidans by NaCl. Res. Microbiol. 2020, 171, 252–259. [Google Scholar] [CrossRef]






| Groups | Sediment Type | Inoculated Bacteria |
|---|---|---|
| FS | sterilized sediment | Acidithiobacillus ferrooxidans |
| FN | unsterilized sediment | Acidithiobacillus ferrooxidans |
| TS | sterilized sediment | Acidithiobacillus thiooxidans |
| TN | unsterilized sediment | Acidithiobacillus thiooxidans |
| Item | Diversity | Evenness | Chao | Number of OTUs |
|---|---|---|---|---|
| Value | 5.73008 | 0.80073 | 2071.197 | 2506 |
| Index | A. ferrooxidans + Sterilized Sediment | A. ferrooxidans + Unsterilized Sediment | ||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 1 | 2 | 3 | |
| Diversity | 1.00745 | 0.44935 | 0.88317 | 1.54914 | 1.32424 | 1.50363 |
| Evenness | 0.31305 | 0.28736 | 0.28511 | 0.36462 | 0.32895 | 0.36427 |
| Chao | 40.00521 | 39.10833 | 57.80833 | 120.71262 | 110.67991 | 137.32339 |
| Number of OTUs | 81 | 34 | 80 | 176 | 116 | 137 |
| Index | A. thiooxidans + sterilized sediment | A. thiooxidans + unsterilized sediment | ||||
| 1 | 2 | 3 | 1 | 2 | 3 | |
| Diversity | 0.95649 | 0.62382 | 0.67928 | 1.19243 | 0.99909 | 1.40654 |
| Evenness | 0.34091 | 0.21024 | 0.21861 | 0.27989 | 0.24985 | 0.33742 |
| Chao | 33.50001 | 26.65953 | 88.11191 | 215.86589 | 88.60981 | 219.89771 |
| Number of OTUs | 53 | 54 | 116 | 230 | 125 | 247 |
| Index | Number of Original OTUs | Similarity Threshold | Total Nodes | Total Links | R Square of Power-Law | Average Clustering Coefficient (avgCC) | Average Path Distance (GD) | Module | Modularity |
|---|---|---|---|---|---|---|---|---|---|
| Sediment | 2506 | 0.88 | 345 | 523 | 0.921 | 0.157 | 3.406 | 43 | 0.736 |
| FN-1 | 176 | 0.81 | 27 | 22 | 0.74 | 0.16 | 0.316 | 7 | 0.764 |
| FN-2 | 116 | 0.81 | 23 | 16 | 0.99 | 0.13 | 0.134 | 8 | 0.851 |
| FN-3 | 136 | 0.81 | 45 | 43 | 0.97 | 0.219 | 0.226 | 10 | 0.829 |
| TN-1 | 230 | 0.78 | 41 | 49 | 0.85 | 0.128 | 3.447 | 6 | 0.62 |
| TN-2 | 125 | 0.78 | 33 | 33 | 0.85 | 0.184 | 0.265 | 9 | 0.707 |
| TN-3 | 247 | 0.78 | 39 | 36 | 0.9 | 0.082 | 0.638 | 8 | 0.771 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Q.; Jie, S.; Lei, P.; Gan, M.; He, P.; Zhu, J.; Zhou, Q. Effect of Anthropogenic Disturbances on the Microbial Relationship during Bioremediation of Heavy Metal-Contaminated Sediment. Microorganisms 2023, 11, 1185. https://doi.org/10.3390/microorganisms11051185
Yang Q, Jie S, Lei P, Gan M, He P, Zhu J, Zhou Q. Effect of Anthropogenic Disturbances on the Microbial Relationship during Bioremediation of Heavy Metal-Contaminated Sediment. Microorganisms. 2023; 11(5):1185. https://doi.org/10.3390/microorganisms11051185
Chicago/Turabian StyleYang, Quanliu, Shiqi Jie, Pan Lei, Min Gan, Peng He, Jianyu Zhu, and Qingming Zhou. 2023. "Effect of Anthropogenic Disturbances on the Microbial Relationship during Bioremediation of Heavy Metal-Contaminated Sediment" Microorganisms 11, no. 5: 1185. https://doi.org/10.3390/microorganisms11051185
APA StyleYang, Q., Jie, S., Lei, P., Gan, M., He, P., Zhu, J., & Zhou, Q. (2023). Effect of Anthropogenic Disturbances on the Microbial Relationship during Bioremediation of Heavy Metal-Contaminated Sediment. Microorganisms, 11(5), 1185. https://doi.org/10.3390/microorganisms11051185








