Abstract
Although high-risk human papillomavirus infection is a well-established risk factor for cervical cancer, other co-factors within the local microenvironment may play an important role in the development of cervical cancer. The current study aimed to characterize the cervicovaginal microbiota in women with premalignant dysplasia or invasive cervical cancer compared with that of healthy women. The study comprised 120 Ethiopian women (60 cervical cancer patients who had not received any treatment, 25 patients with premalignant dysplasia, and 35 healthy women). Cervicovaginal specimens were collected using either an Isohelix DNA buccal swab or an Evalyn brush, and ribosomal RNA sequencing was used to characterize the cervicovaginal microbiota. Shannon and Simpson diversity indices were used to evaluate alpha diversity. Beta diversity was examined using principal coordinate analysis of weighted UniFrac distances. Alpha diversity was significantly higher in patients with cervical cancer than in patients with dysplasia and in healthy women (p < 0.01). Beta diversity was also significantly different in cervical cancer patients compared with the other groups (weighted UniFrac Bray-Curtis, p < 0.01). Microbiota composition differed between the dysplasia and cervical cancer groups. Lactobacillus iners was particularly enriched in patients with cancer, and a high relative abundance of Lactobacillus species was identified in the dysplasia and healthy groups, whereas Porphyromonas, Prevotella, Bacteroides, and Anaerococcus species predominated in the cervical cancer group. In summary, we identified differences in cervicovaginal microbiota diversity, composition, and relative abundance between women with cervical cancer, women with dysplasia, and healthy women. Additional studies need to be carried out in Ethiopia and other regions to control for variation in sample collection.
1. Introduction
Cervical cancer is a major public health problem and is the fourth most common cancer among women globally [1]. In 2020, an estimated 604,237 women globally had cervical cancer, representing 6.5% of all female cancers. About 90% of the new cases and deaths worldwide in 2020 occurred in low- and middle-income countries [2]. In Ethiopia, cervical cancer is the second most common cancer with 6300 annual new cases and about 4884 deaths each year [3].
It is well established that persistent infection with high-risk human papilloma virus (HPV) genotypes causes high-grade cervical intraepithelial neoplasia (CIN2+) and invasive cervical cancer [4]. However, HPV infection alone is insufficient to cause cancer; most genital HPV infections are transient and asymptomatic. Only a small number of women—those chronically carrying oncogenic or high-risk HPV genotypes—eventually develop severe dysplasia (CIN2/3) and, over several decades, cancer [5,6]. Therefore, the most important aspect in cervical cancer prevention is to focus on precancerous lesions and categorize those women according to their risk of persistence or recurrence so as to identify the women at higher risk [7,8].
Mechanisms associated with clearance or persistence of HPV infection are not well understood. Currently, different factors have been found to be associated with persistent HPV infection and its development into cervical dysplasia and/or invasive cancer [8]. These include, architecture of the epithelial surface, mucosal immunity, other sexually transmitted infections, and the cervicovaginal microbiota and microenvironment [9,10]. The prevalence of any HPV or multiple HPV infections is higher among human immunodeficiency virus (HIV) positive women than HIV negative women in sub-Saharan Africa [11]. Lactic acid–producing bacteria and mucosal secretions have been shown to play an important role in trapping pathogens, including HPV, HIV, and others that cause sexually transmitted infections [12,13,14]. The protective role of these lactic acid–producing bacteria, i.e., Lactobacillus species, is disrupted following development of highly diverse vaginal microorganisms associated with proinflammatory responses. The inflammatory response may in turn cause direct damage to the epithelium and facilitate HPV entry, as well as persistent infection [13,15,16].
Although more than 50 microbial species have been identified in the vaginal tract, Lactobacillus species, mainly L. crispatus, L. gasseri, L. iners, and L. jensenii, make up about 70% of the vaginal microbiota [17]. The structure and function of the vaginal microbiota can vary with genetic disposition, ethnicity, diet, hygiene, infections, antibiotic use, sexual activity, physiologic status of the vagina, and especially estrogen levels [17,18]. Studies have indicated that increased CIN grade is associated with increased vaginal microbiota diversity, which may be involved in regulating viral persistence and disease progression [19]. However, specific microbes or microbial communities that can be mechanistically linked to cervical carcinogenesis remain largely unidentified [20]. Recent studies revealed that decreased levels of probiotics, such as Shuttleworthia, Prevotella, Lactobacillus, and Sneathia, along with high levels of pathogenic bacteria, including Dispar, Streptococcus, and Faecalibacterium prausnitzii, could be the direct result of early HPV infection. In addition, some pathogenic bacteria, such as Bifidobacteriaceae, could be involved in cancer progression [21].
With the current rapidly progressing sequencing technologies that characterize microbial communities beyond culture-based or biochemical techniques, cervicovaginal microbiota sequencing and analysis, most specifically 16S ribosomal RNA (rRNA) amplicon sequencing, is becoming the best tool to advance knowledge of the association between microbiota and cervical cancer progression [22]. 16S rRNA gene sequencing produces reliable taxonomic classifications and relative abundances and is a cost-effective method to quantify diversity metrics, as well as provide genus-level identification [23]. However, 16S rRNA sequencing has some disadvantages, including the inability to provide species-level identification and metagenomic functionality. Furthermore, this method includes only bacteria; the virome or other non-bacterial members of the microbiota cannot be identified without targeted screening [24]. These deficiencies of 16S rRNA sequencing can be overcome with whole-genome shotgun sequencing [25].
Recently we followed a community of women who attended HPV-based cervical cancer screening for 2 years and learned that some cleared the virus within 6 months whereas others had persistent infections. In an effort to understand the possible underlying factors, we characterized the cervicovaginal microbiota structure and diversity in women with cervical cancer, women with dysplasia (various CIN grades), and healthy women in central and south-central Ethiopia.
2. Materials and Methods
2.1. Study Area
The study was carried out at Tikur Anbessa Specialized Hospital, Addis Ababa, Ethiopia, and in the rural community of Butajira in south central Ethiopia (Gurage Zone of the Southern Nations, Nationalities, and Peoples’ Region of Ethiopia). The gynecology department at the hospital provided evaluation, examination, surgical treatment, and screening of new and referred cases of cervical cancer.
2.2. Inclusion and Exclusion Criteria
At Tikur Anbessa Specialized Hospital, all eligible women aged 18 years or older who were attending the cervical cancer screening program from October 2019 to February 2020 were screened, and those with histologically confirmed cervical cancer or precancerous lesions were included in the current study. In Butajira, women who underwent cytologic examination for cervical cancer screening and whose results were either normal or cytologically positive for precancerous lesions were included in the current study. Women who were pregnant; had undergone hysterectomy, chemotherapy, or radiotherapy; or had a cognitive or physical impairment that prevented them from giving informed consent or participating were excluded from the study.
This comparative study comprised 120 women: (60 who had cervical cancer and had not received any treatment, 25 who had cytologically or histologically confirmed cervical dysplasia, and 35 who were cytologically or histologically negative for cervical dysplasia (Table 1).

Table 1.
Sociodemographic and clinical characteristics of participants (n = 120).
2.3. Sample Collection and DNA Extraction
Specimens for the current study were collected using one of two devices: 67 specimens were collected using an Isohelix DNA buccal swab, and 53 specimens were collected using an Evalyn brush (Rovers Medical Devices, Oss, The Netherlands). Among the samples collected using Isohelix swabs, 60 were cancer, 6 were dysplasia, and 1 was negative for intraepithelial lesions or malignancy; among those collected using the Evalyn brush, 0 were cancer, 19 were dysplasia, and 34 were negative (Table S1).
The Isohelix cervical swab specimens were obtained by first swabbing the surface of the cervix and then placing the swab into a stabilization buffer, followed by freezing within 30 min at −20 °C. The Evalyn brush is a self-collection device, and women were assisted by trained health workers for sample collection. The brush was placed in a plastic bag and stored in a dry place at room temperature until it could be transported to the HPV laboratory of the Department of Microbiology, Immunology and Parasitology, School of Medicine, Addis Ababa University, for processing.
The Evalyn brushes were processed as previously described [26]. Briefly, the brush was removed from the plastic bag and the tip was pulled off and placed into a 2-mL Eppendorf tube and soaked in 1 mL phosphate-buffered saline overnight to remove the cells from the dry brush. The tube was then centrifuged for 5 min at 2500 rpm and vortexed vigorously for 1 min, and an aliquot of 200 μL of the fluid was used for DNA extraction. DNA extraction was carried out using the Qiagen DNA Extraction Kit (QIAGEN, Hilden, Germany). Both the Isohelix and Evalyn brush samples were shipped to The University of Texas MD Anderson Cancer Center, USA, on dry ice for DNA extraction and 16S rRNA sequencing.
2.4. HPV Genotyping
HPV presence and genotype were determined using the Anyplex II HPV 28 detection kit (Seegene, Seoul, Korea), which uses real-time multiplex PCR with tagging oligonucleotide cleavage and extension technology for simultaneous detection and genotyping of high-risk and low-risk HPV genotypes. Anyplex can detect 28 HPV genotypes including 19 high-risk types (16, 18, 26, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 68, 69, 73, 82) and 9 low-risk types (6, 11, 40, 42, 43, 44, 54, 61, 70).
2.5. 16S rRNA Gene Sequencing and Sequence Data Processing
16S rRNA gene sequencing was performed by the Alkek Center for Metagenomics and Microbiome Research at Baylor College of Medicine (Houston, TX, USA). Sequencing was performed based on methods adapted from the Human Microbiome [27]. DNA was extracted using the MO BIO PowerSoil DNA Isolation Kit (MO BIO Laboratories, Carlsbad, CA USA). The 16S V4 region is the most conserved and variable segment of the genome, which makes it a good target for phylogenetic analyses. This region was amplified by PCR using a 515F-806R primer pair. Sequencing was performed on the Illumina MiSeq platform using the 2 × 250 base pair paired-end protocol, yielding pair-end reads.
16S rRNA sequence reads were processed using the QIIME2 microbiome bioinformatics platform (v2020.11) [28]. FASTQ sequences were imported and demultiplexed as QIIME2 artifacts. Denoising was performed using DADA2 with trim parameters set at 20–245 for forward strands and 8–230 for reverse strands. Trim parameters were set based on quality score plots generated on QIIME2 [29]. Representative sequences were generated using DADA2 for phylogenetic tree construction and taxonomic classification. A pre-trained naïve Bayes classifier was used for phylogenetic reference construction via the q2-feature-classifier plugin. We used a taxonomic classifier trained on the SILVA 138 database 515F/806R region of sequences trimmed to include 250 bases from the 16S V4 region [30].
2.6. Statistical Analyses
The Shannon diversity index and Simpson index were used to assess a within-sample (alpha) diversity-based OTU table in QIIME, and results were compared among multiple groups (Kruskal test) or between two groups (pairwise Wilcoxon test). In addition, beta diversity analysis was performed using UniFrac distance metrics and visualized via principal coordinate analysis.
2.7. Ethical Considerations
The study was approved by the Institutional Review Board of the College of Health Sciences at Addis Ababa University and by the National Research Ethics Review Committee of the Ministry of Science and Higher Education-Ethiopia. A material transfer agreement was signed by both institutions to transfer samples to MD Anderson for processing.
3. Results
3.1. Participant Characteristics
The general characteristics of the 120 participants (median age = 40 years) are provided in Table 1. The study included 35 women with normal cytologic or histologic characteristics, 25 with low-grade or high-grade dysplasia, and 60 with cervical cancer (either squamous cell carcinoma or adenocarcinoma). Most participants (n = 84, 70%) were younger than 50 years; 35 (29%) were aged 50 years or older. Twenty-seven women (23%) were HPV-negative and 93 (78%) were HPV-positive (any high-risk HPV genotype). Among those younger than 50 years (n = 84), 28 had cervical cancer, 22 had dysplasia, and 34 had normal cytologic or histologic characteristics. Among those aged 50 years or older, 32 had cervical cancer, 2 had dysplasia, and 1 had normal cytologic or histologic characteristics (Table S2). No participants in our cohort had a low-risk HPV genotype. Cervical dysplasia was classified according to histologic grade of cervical intraepithelial neoplasia (CIN1–3) or according to cytologic grading of squamous intraepithelial lesions (high-grade or low-grade).
3.2. Age-Related Composition and Diversity Changes in Cervicovaginal Microbiota
We observed age-associated alterations in cervical microbiota composition and diversity. Alpha diversity significantly increased with age (Shannon p = 0.00071, Simpson p = 0.0041) (Figure 1A). Those younger than 50 years (n = 84) had significantly increased levels of Lactobacillus (p = 0.00003) and Gardenella (p = 0.01), and those aged 50 years or older (n = 35) had increased levels of Porphyromonas (p = 0.00006), Prevotella (p = 0.0041), Bacteroids (p = 0.0023), and Anaerococcus (p = 0.00097; Figure 1B). However, because the two age groups had an uneven distribution of histologic or cytologic characteristics (Table S2), we compared only cancer patients from each age group (<50 years, n = 28; and ≥50 years, n = 32). There were no differences in alpha (Shannon p = 0.32, Simpson p = 0.39) or beta (p = 0.46) diversities or microbiota composition among the women with cancer (Supplementary Figure S1).
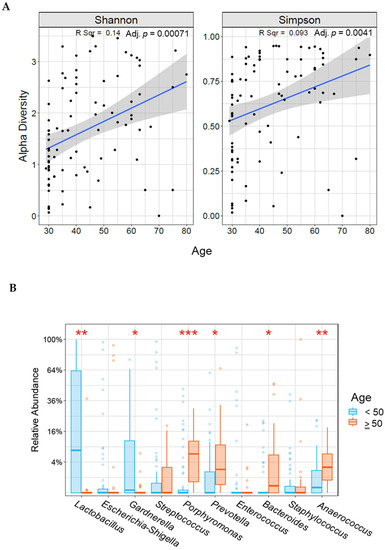
Figure 1.
Correlation between alpha diversity (A) or relative abundance (B) and age in our study population. * indicates statistically significant difference between age groups (* <0.05, ** <0.005, *** <0.0005).
3.3. Composition and Diversity of Cervicovaginal Microbiota among HPV-Positive and HPV-Negative Women
In our study population, we compared the alpha and beta diversity between women who were HPV-negative and those who were HPV-positive. There was no significant difference in alpha diversity between HPV- positive and HPV- negative women (Shannon p = 0.21, Simpson p= 0.18; Supplementary Figure S2A). Principal coordinate analysis ((weighted UniFrac, Bray-Curtis) revealed significant differences in microbial diversity between HPV- positive and HPV- negative women (p = 0.002; Supplementary Figure S2B).
Next, we compared the composition of the most abundant genera and species between HPV- positive and HPV-negative women. Among the top 10 most abundant genera, Porphyromonas (p = 0.0204) and Peptoniphilus (p = 0.0423) were significantly more abundant in the HPV- positive group (Supplementary Figure S3A). However, we observed no significant differences in species abundance between HPV-positive and HPV-negative women (Supplementary Figure S3B).
3.4. Cervicovaginal Microbiota Diversity and Composition among Healthy Women, Women with Cervical Dysplasia, and Women with Cervical Cancer
We compared cervicovaginal microbiota diversity among healthy women, women with dysplasia, and women with cervical cancer. We observed a significant difference in richness and evenness among the three groups (Shannon p= 0.00000054, Simpson p= 0.000005; Figure 2A). In pairwise comparisons, all indices showed that the cervical cancer group had the highest community diversity of cervicovaginal microbiota (p < 0.05, Kruskal-Wallis test). Principal coordinate analysis of the Bray-Curtis distances revealed differences in community structure in the cervical cancer samples compared with dysplasia and negative samples (weighted Bray-Curtis UniFrac, p = 0.001; Figure 2B).
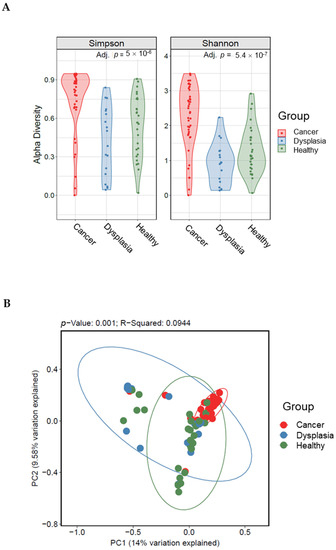
Figure 2.
Alpha (A) and beta (B) diversity indices among women with cervical cancer, women with dysplasia, and healthy women. Beta diversity comparisons were calculated using the principal coordinate analysis discriminate. Abundance profiles in women with cervical cancer (n = 60) were different from those of women with dysplasia (n = 25) and healthy women (n = 35).
Next, we characterized the microbiota taxa abundances in our cohort. A stacked bar plot (Figure 3) showed distinct patterns of taxa in our cohort. Lactobacillus was the dominant genus.
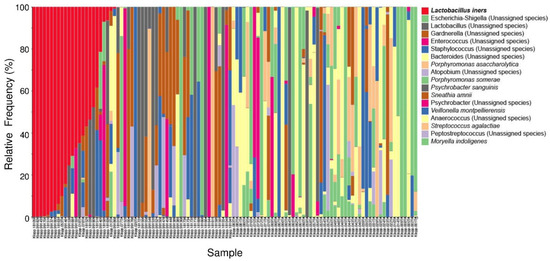
Figure 3.
Stacked bar plot of the top 10 most abundant genus-level bacteria found in our study population. Each bar represents a single participant.
The relative abundance of cervicovaginal microbiota at the genus and species level was also characterized. The top 10 most abundant genera in the samples varied by group. A heat map (Figure 4) reflected the relative abundance of the most prevalent bacterial genera, showing that cervicovaginal microbiota composition changed from less diverse and more Lactobacillus-dominant in dysplasia to more diverse and less Lactobacillus-dominant in cervical cancer. Compared with healthy women and women with dysplasia, many of the women with cervical cancer had more Porphyromonas species (Figure 4 and Figure S4).
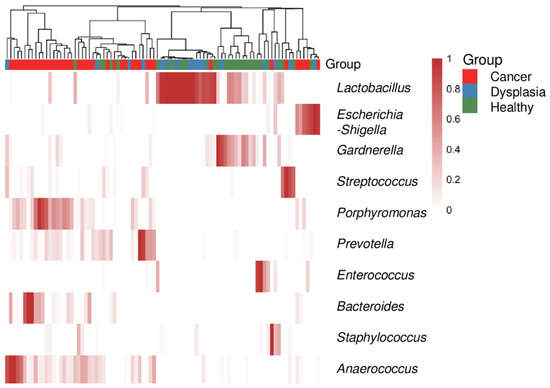
Figure 4.
Heatmap of the top 10 most abundant genus-level bacteria in women with cervical cancer, women with dysplasia, and health women. Each bar represents a single participant.
To identify significant differences in relative abundance at the species and genus level, we further analyzed taxa abundance using a box plot (Figure 5). Lactobacillus predominated in the dysplasia and healthy groups and Porphyromonas, Prevotella, Bacteroides, and Anaerococcus predominated in the cervical cancer group (p < 0.05, Kruskal-Wallis test). At the species level, Lactobacillus iners predominated in the dysplasia group (p < 0.05) and Porphyromonas somerae, Porphyromonas asaccharolytica, and Prevotella timonensis were the most abundant species in the cervical cancer group (p < 0.05; Figure 5).
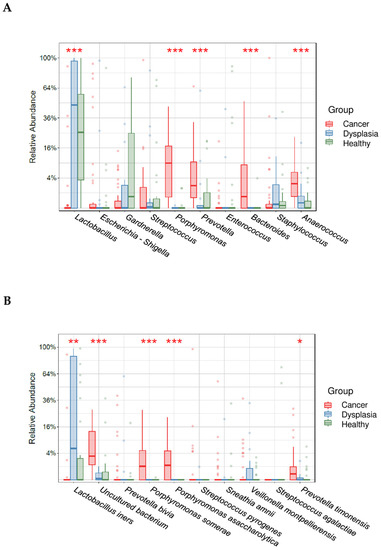
Figure 5.
Relative abundance of the top 10 most abundant genera (A) and species (B) in women with cervical cancer, women with dysplasia, and healthy women. * indicates statistically significant difference between age groups (* <0.05, ** <0.005, *** <0.0005).
4. Discussion
Although persistent infection with high-risk HPV is a well-established risk factor and pre-requisite for cervical cancer, HPV infection is a heterogenous condition with varying outcomes, and the specific impact of other co-factors in cervical carcinogenesis is not yet well identified and characterized. Because the cervicovaginal microbiota differs with geography, ethnicity, and lifestyle, as well as infectious history, it is crucial to characterize the diversity and composition of the microbiota in different populations and the role of the microbiota in the progression of cervical cancer.
Recent literature has suggested that the cervicovaginal microbiota plays a mechanistic role in both HPV persistence and cervical cancer progression [31]. In most healthy women of reproductive age, the cervicovaginal microbiota is dominated by Lactobacillus species, and a lack of this dominance is recognized as a cause of adverse reproductive health outcomes [32]. Consistent with these findings, in the current study, we observed a positive relationship between alpha diversity and age (Figure 1A), as well as Lactobacillus dominance in the reproductive years (i.e., women younger than 50 years; Figure 1B). However, among the women with cervical cancer, there was no difference in the diversity or composition of the microbiota by age (Supplementary Figure S1).
The current study was conducted among Ethiopian women with cervical cancer, histologically or cytologically confirmed dysplasia, or no evidence of cancer or dysplasia (healthy women) to characterize the diversity and composition of the cervicovaginal microbiota. To the best of our knowledge, the current study is the first to examine the relationship between the cervicovaginal microbiota and cervical cancer and/or HPV infection among women in Ethiopia. In our study, the cervicovaginal microbiota of most of the healthy women was dominated by Lactobacillus. Alpha and beta diversity was compared between HPV-positive and HPV-negative women regardless of their cervical cancer or dysplasia status, and we observed significant differences in beta diversity between HPV-positive and HPV-negative women, whereas alpha diversity analysis did not differ by HPV status (Supplementary Figure S2).
Because our study participants with cervical cancer had different histologic diagnosis (i.e., squamous cell carcinoma or adenocarcinoma), microbial diversity was observed within the group, suggesting that microbiota diversity and composition may vary by specific cervical cancer diagnosis. Furthermore, genus and species diversity was higher in women with cervical cancer than in healthy women, which suggest that the stable composition of cervicovaginal flora, mainly Lactobacillus supplemented by other bacteria, is destroyed in carcinogenesis, resulting in increased microbial diversity. This finding is supported by other similar studies [17,33].
As indicated from previous studies [12,13,14], depletion of Lactobacillus species from the cervicovaginal microbiota structure leads to proliferation of other pathogens and hence a change of the microbiota composition, including diversity and relative abundance of genera. This explains the higher alpha diversities in the women with cervical cancer in our study compared with those with dysplasia or healthy women (Figure 2A).
Furthermore, among our study groups, relative abundance of some bacterial genera was significantly different. The predominant genus in the healthy group and dysplasia group was Lactobacillus (Figure 3 and Figure 4). In the cervical cancer group, the genus Lactobacillus decreased in relative abundance, and other bacteria, such as Porphyromonas, Prevotella, and Anaerococcus, were the dominant genera (Figure 5). These results are consistent with those of other studies [17,34,35]. For example, Wu et al. demonstrated less Lactobacillus and higher diversity of microbiota were associated with more severe pathological status. Furthermore, Porphyromonas and Prevotella were identified as cervical cancer marker genera.
The role of Porphyromonas and Prevotella in carcinogenesis has been demonstrated in oral cancers [36], with three proposed mechanisms of action: chronic inflammation, anti-apoptotic activity, and carcinogenic metabolites released by these microbes. These bacteria produce inflammatory mediators that facilitate cell proliferation, mutagenesis, oncogene activation, and angiogenesis. Porphyromonas gingivalis, found in oral cavities, has been reported to induce lipopolysaccharides that lead to the production of proinflammatory cytokines such as tumor necrosis factor α (TNF-α) by macrophages and interleukin (IL)-1β and IL-6 by CD4+ T helper cells [37]. Studies have also indicated that Porphyromonas gingivalis can mediate different signaling pathways that influence cell invasion, the cell cycle, anti-apoptosis, and inflammation [36].
Increased relative abundance of Prevotella in human mucosal sites has been shown to be related to various inflammatory disorders. This bacterium was indicated as a major modulator of host inflammatory responses in the female genital tract by increasing the amount of innate cytokines (IL-1α, IL-1β, IL-8, and TNFα) in cervicovaginal fluid, and production of cytokines related to Th17 (IL-23 and IL-17) and Th1 (IL-12p70 and interferon γ) [38].
The increased relative abundance of Porphyromonas, Prevotella, and Anaerococcus in the cervical cancer group in our study indicates that these bacteria may play a substantial role in the development of cervical cancer, supporting the previously proposed mechanisms of chronic inflammation, anti-apoptotic activity, and production of carcinogenic substances. Future studies should assesses the mechanistic relationship of a diverse cervicovaginal microbiota with cervical cancer in women in resource- limited settings, as well as the impacts of different intervention strategies. Currently, there are several treatment outlooks for cervical cancer: including the use of immunotherapy and target therapy, in combination with conventional chemotherapy or in combination with radiotherapy. Therefore, from such studies, cervicovaginal microbiota-derived bacterial markers can be used as a predictive model to predict the progression or regression of precancerous lesions and undertake further research with large sample size and possibility of identifying the right probiotics in women with persistent HPV infection or precancerous lesions of different stages, and invasive cancer that can affect chemotherapy and radiotherapy outcomes [39].
The current study had some limitations. Two different sample collection devices were used (the Evalyn brush and the Isohelix swab). Although studies have shown that the Evalyn self-sampling device performed equally well compared with samples collected by a clinician according to Illumina MiSeq sequencing of the 16S rRNA gene [40,41], the different sampling strategies may not yield comparable cervicovaginal microbiota composition and diversity. In addition, we did not consider the detailed sexual, behavioral, and clinical characteristics of the study participants. Furthermore, we used cytology, not histology, for the classification of dysplasia.
5. Conclusions
In conclusion, our study showed differences in cervicovaginal microbiota diversity, composition, and relative abundance between women with cervical cancer, women with dysplasia, and healthy women. The diversity and composition of the cervicovaginal microbiota increased from dysplasia to cancer, and increased levels of L. iners were found in women with dysplasia compared with healthy women. Differences in methods of sample collection may account for observed differences in composition and diversity. Other studies are needed to further validate the role of the cervicovaginal microbiota in development of cervical cancer.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms11040833/s1, Figure S1: Alpha (A) and beta (B) diversity and microbial composition (C) among patients with cervical cancer who were younger than 50 years or aged 50 years or older; Figure S2: Alpha (A) and beta (B) diversity among HPV-positive and HPV-negative women in our study population; Figure S3: Microbial composition at the genus and species level among HPV-positive and HPV-negative women in our study population; Figure S4: Stacked bar plot of the top 10 most abundant genus-level bacteria in women with cervical cancer, women with dysplasia, and healthy women. Each bar represents a single participant; Table S1: Histological/Cytological status of study participants based on age category; Table S2: Histological/Cytological status of study participants based on type of sample collection device.
Author Contributions
A.H.K. and T.A. are joint senior authors. All authors were involved with subject identification and data collection, interpretation of the statistical analysis, and review and approval of the final manuscript. The study concept was developed by B.T., K.Y.-C., M.B.E.A., A.H.K. and T.A.; B.T., K.Y.-C. and M.B.E.A. drafted the manuscript. E.F., Z.C., M.G., A.M. and A.A. collected and supervised data collection process. L.E.C., T.C.N., E.J.L. and M.M., contributed to data collection and interpretation; J.A., E.J.K. and A.M.K. contributed to supervision of sample analyses, revision of the study data analyses and revisions of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Sister Institution Network Fund (SINF) 2019-20 (AHK) and was partially supported by the generous philanthropic contributions to The University of Texas MD Anderson Cancer Center Moon Shots Program (AHK). Funding was also obtained from Else Kroener-Fresenius-Stiftung grant 2017-HA03. Furthermore, this study was supported by Addis Ababa University, grant/award number: VPRTT/PY-041/2018.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of the College of Health Sciences, Addis Ababa University (079/20/DMIP) and by the National Research Ethics Review Committee (NRERC) (SHE/RAAA/9.1/339/19/11).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Data are available upon reasonable request. The data supporting this study’s findings are available from the corresponding author, [BT], upon reasonable request.
Acknowledgments
We thank Erica Goodoff, Senior Scientific Editor in the Research Medical Library at The University of Texas MD Anderson Cancer Center, for editing this article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lee, J.E.; Lee, S.; Lee, H.; Song, Y.M.; Lee, K.; Han, M.J.; Sung, J.; Ko, G.P. Association of the Vaginal Microbiota with Human Papillomavirus Infection in a Korean Twin Cohort. PLoS ONE 2013, 8, e63514. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Bruni, L.; Saura-Lázaro, A.; Montoliu, A.; Brotons, M.; Alemany, L.; Diallo, M.S.; Afsar, O.Z.; LaMontagne, D.S.; Mosina, L.; Contreras, M.; et al. HPV vaccination introduction worldwide and WHO and UNICEF estimates of national HPV immunization coverage 2010–2019. Prev. Med. 2021, 144, 106399. [Google Scholar] [CrossRef]
- Walboomers, J.M.; Jacobs, M.V.; Manos, M.M.; Bosch, F.X.; Kummer, J.A.; Shah, K.V.; Snijders, P.J.; Peto, J.; Meijer, C.J.; Muñoz, N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 1999, 189, 12–19. [Google Scholar] [CrossRef]
- Kyrgiou, M.; Mitra, A.; Moscicki, A.B. Does the vaginal microbiota play a role in the development of cervical cancer? Transl. Res. 2017, 179, 168–182. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, A.C.; Burk, R.; Herrero, R.; Hildesheim, A.; Bratti, C.; Sherman, M.E.; Solomon, D.; Guillen, D.; Alfaro, M.; Viscidi, R.; et al. The natural history of human papillomavirus infection and cervical intraepithelial neoplasia among young women in the Guanacaste cohort shortly after initiation of sexual life. Sex. Transm. Dis. 2007, 34, 494–502. [Google Scholar] [CrossRef] [PubMed]
- Giannini, A.; Bogani, G.; Vizza, E.; Chiantera, V.; Laganà, A.S.; Muzii, L.; Salerno, M.G.; Caserta, D.; D’Oria, O. Advances on Prevention and Screening of Gynecologic Tumors: Are We Stepping Forward? Healthcare 2022, 10, 1605. [Google Scholar] [CrossRef]
- Veldhuijzen, N.J.; Snijders, P.J.F.; Reiss, P.; Meijer, C.J.L.M.; van de Wijgert, J.H.H.M. Factors affecting transmission of mucosal human papillomavirus. Lancet Infect. Dis. 2010, 10, 862–874. [Google Scholar] [CrossRef]
- Chang, A.H.; Parsonnet, J. Role of bacteria in oncogenesis. Clin. Microbiol. Rev. 2010, 23, 837–857. [Google Scholar] [CrossRef]
- Pyeon, D.; Pearce, S.M.; Lank, S.M.; Ahlquist, P.; Lambert, P.F. Establishment of human papillomavirus infection requires cell cycle progression. PLoS Pathog. 2009, 5, e1000318. [Google Scholar] [CrossRef]
- Okoye, J.O.; Ofodile, C.A.; Adeleke, O.K.; Obioma, O. Prevalence of high-risk HPV genotypes in sub-Saharan Africa according to HIV status: A 20-year systematic review. Epidemiol. Health 2021, 43, e2021039. [Google Scholar] [CrossRef] [PubMed]
- Aldunate, M.; Srbinovski, D.; Hearps, A.C.; Latham, C.F.; Ramsland, P.A.; Gugasyan, R.; Cone, R.A.; Tachedjian, G. Antimicrobial and immune modulatory effects of lactic acid and short chain fatty acids produced by vaginal microbiota associated with eubiosis and bacterial vaginosis. Front. Physiol. 2015, 6, 164. [Google Scholar] [CrossRef] [PubMed]
- Borgdorff, H.; Tsivtsivadze, E.; Verhelst, R.; Marzorati, M.; Jurriaans, S.; Ndayisaba, G.F.; Schuren, F.H.; Van De Wijgert, J.H.H.M. Lactobacillus-dominated cervicovaginal microbiota associated with reduced HIV/STI prevalence and genital HIV viral load in african women. ISME J. 2014, 8, 1781–1793. [Google Scholar] [CrossRef]
- Dareng, E.O.; Ma, B.; Adebamowo, S.N.; Famooto, A.; Ravel, J.; Pharoah, P.P.; Adebamowo, C.A. Vaginal microbiota diversity and paucity of Lactobacillus species are associated with persistent hrHPV infection in HIV negative but not in HIV positive women. Sci. Rep. 2020, 10, 19095. [Google Scholar] [CrossRef] [PubMed]
- King, C.C.; Jamieson, D.J.; Wiener, J.; Cu-Uvin, S.; Klein, R.S.; Rompalo, A.M.; Shah, K.V.; Sobel, J.D. Bacterial vaginosis and the natural history of human papillomavirus. Infect. Dis. Obstet. Gynecol. 2011, 2011, 319460. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.L.; You, K.; Qiao, J.; Zhao, Y.M.; Geng, L. Bacterial vaginosis is conducive to the persistence of HPV infection. Int. J. STD AIDS 2012, 23, 581–584. [Google Scholar] [CrossRef]
- Wu, S.; Ding, X.; Kong, Y.; Acharya, S.; Wu, H.; Huang, C.; Liang, Y.; Nong, X.; Chen, H. The feature of cervical microbiota associated with the progression of cervical cancer among reproductive females. Gynecol. Oncol. 2021, 163, 348–357. [Google Scholar] [CrossRef]
- Kaur, H.; Merchant, M.; Haque, M.M.; Mande, S.S. Crosstalk Between Female Gonadal Hormones and Vaginal Microbiota Across Various Phases of Women’s Gynecological Lifecycle. Front. Microbiol. 2020, 11, 551. [Google Scholar] [CrossRef]
- International Agency for Research on Cancer (IARC). Cervix Uteri Factsheet. Globocan. 2020, Volume 419, pp. 1–10. Available online: https://gco.iarc.fr/today/online-analysis-map?v=2020&mode=population&mode_population=continents&population=900&populations=900&key=asr&sex=0&cancer=39&type=0&statistic=5&prevalence=0&population_group=0&ages_group%5B%5D=0&ages_group%5B%5D=17&nb_items=10&gr (accessed on 20 December 2022).
- Mitra, A.; MacIntyre, D.A.; Lee, Y.S.; Smith, A.; Marchesi, J.R.; Lehne, B.; Bhatia, R.; Lyons, D.; Paraskevaidis, E.; Li, J.V.; et al. Cervical intraepithelial neoplasia disease progression is associated with increased vaginal microbiome diversity. Sci. Rep. 2015, 5, 16865. [Google Scholar] [CrossRef]
- Piyathilake, C.J.; Ollberding, N.J.; Kumar, R.; Macaluso, M.; Alvarez, R.D.; Morrow, C.D. Cervical microbiota associated with higher grade cervical intraepithelial neoplasia in women infected with high-risk human papillomaviruses. Cancer Prev. Res. 2016, 9, 357–366. [Google Scholar] [CrossRef]
- Liu, J.; Luo, M.; Zhang, Y.; Cao, G.; Wang, S. Association of high-risk human papillomavirus infection duration and cervical lesions with vaginal microbiota composition. Ann. Transl. Med. 2020, 8, 1161. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Kouzy, R.; Jaoude, J.A.; Noticewala, S.S.; Delgado Medrano, A.Y.; Klopp, A.H.; Taniguchi, C.M.; Colbert, L.E. Microbiome factors in HPV-driven carcinogenesis and cancers. PLoS Pathog. 2020, 16, e1008524. [Google Scholar] [CrossRef] [PubMed]
- D’Amore, R.; Ijaz, U.Z.; Schirmer, M.; Kenny, J.G.; Gregory, R.; Darby, A.C.; Shakya, M.; Podar, M.; Quince, C.; Hall, N. A comprehensive benchmarking study of protocols and sequencing platforms for 16S rRNA community profiling. BMC Genom. 2016, 17, 55. [Google Scholar] [CrossRef]
- Biegert, G.; El Alam, M.B.; Karpinets, T.; Wu, X.; Sims, T.T.; Yoshida-Court, K.; Lynn, E.J.; Yue, J.; Medrano, A.D.; Petrosino, J.; et al. Diversity and composition of gut microbiome of cervical cancer patients: Do results of 16S rRNA sequencing and whole genome sequencing approaches align? J. Microbiol. Methods 2021, 185, 106213. [Google Scholar] [CrossRef]
- Ranjan, R.; Rani, A.; Metwally, A.; McGee, H.S.; Perkins, D.L. Analysis of the microbiome: Advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem. Biophys. Res. Commun. 2016, 469, 967–977. [Google Scholar] [CrossRef] [PubMed]
- Teka, B.; Gizaw, M.; Ruddies, F.; Addissie, A.; Chanyalew, Z.; Skof, A.S.; Thies, S.; Mihret, A.; Kantelhardt, E.J.; Kaufmann, A.M.; et al. Population-based human papillomavirus infection and genotype distribution among women in rural areas of South Central Ethiopia. Int. J. Cancer 2021, 148, 723–730. [Google Scholar] [CrossRef] [PubMed]
- Methé, B.A.; Nelson, K.E.; Pop, M.; Creasy, H.H.; Giglio, M.G.; Huttenhower, C.; Gevers, D.; Petrosino, J.F.; Abubucker, S.; Badger, J.H.; et al. A framework for human microbiome research. Nature 2012, 486, 215–221. [Google Scholar] [CrossRef]
- Bolyen, E.; Ram Rideout, J.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 848–857. [Google Scholar] [CrossRef]
- Nearing, J.T.; Douglas, G.M.; Comeau, A.M.; Langille, M.G.I. Denoising the Denoisers: An independent evaluation of microbiome sequence error- correction approaches. Peer J. 2018, 2018, e5364. [Google Scholar] [CrossRef]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glöckner, F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef]
- Łaniewski, P.; Barnes, D.; Goulder, A.; Cui, H.; Roe, D.J.; Chase, D.M.; Herbst-Kralovetz, M.M. Linking cervicovaginal immune signatures, HPV and microbiota composition in cervical carcinogenesis in non-Hispanic and Hispanic women. Sci. Rep. 2018, 8, 7593. [Google Scholar] [CrossRef]
- Ravel, J.; Gajer, P.; Abdo, Z.; Schneider, G.M.; Koenig, S.S.K.; McCulle, S.L.; Karlebach, S.; Gorle, R.; Russell, J.; Tacket, C.O.; et al. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. USA 2011, 108, 4680–4687. [Google Scholar] [CrossRef]
- Qingqing, B.; Jie, Z.; Songben, Q.; Juan, C.; Lei, Z.; Mu, X. Cervicovaginal microbiota dysbiosis correlates with HPV persistent infection. Microb. Pathog. 2021, 152, 104617. [Google Scholar] [CrossRef]
- Sims, T.T.; Colbert, L.E.; Zheng, J.; Delgado Medrano, A.Y.; Hoffman, K.L.; Ramondetta, L.; Jazaeri, A.; Jhingran, A.; Schmeler, K.M.; Daniel, C.R.; et al. Gut microbial diversity and genus-level differences identified in cervical cancer patients versus healthy controls. Gynecol. Oncol. 2019, 155, 237–244. [Google Scholar] [CrossRef]
- Chen, Y.; Qiu, X.; Wang, W.; Li, D.; Wu, A.; Hong, Z.; Di, W.; Qiu, L. Human papillomavirus infection and cervical intraepithelial neoplasia progression are associated with increased vaginal microbiome diversity in a Chinese cohort. BMC Infect. Dis. 2020, 20, 629. [Google Scholar] [CrossRef]
- Karpiński, T.M. Role of oral microbiota in cancer development. Microorganisms 2019, 7, 20. [Google Scholar] [CrossRef]
- Amano, A.; Nakagawa, I.; Okahashi, N.; Hamada, N. Variations of Porphyromonas gingivalis fimbriae in relation to microbial pathogenesis. J. Periodontal Res. 2004, 39, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Anahtar, M.N.; Byrne, E.H.; Doherty, K.E.; Bowman, B.A.; Yamamoto, S.; Soumillon, M.; Padavattan, N.; Ismail, N.; Moodley, A.; Sabatini, M.E.; et al. Inflammatory responses in the female genital tract. Immunity 2016, 42, 965–976. [Google Scholar] [CrossRef]
- D’oria, O.; Corrado, G.; Laganà, A.S.; Chiantera, V.; Vizza, E.; Giannini, A. New Advances in Cervical Cancer: From Bench to Bedside. Int. J. Environ. Res. Public Health 2022, 19, 7094. [Google Scholar] [CrossRef] [PubMed]
- Virtanen, S.; Kalliala, I.; Nieminen, P.; Salonen, A. Comparative analysis of vaginal microbiota sampling using 16S rRNA gene analysis. PLoS ONE 2017, 12, e0181477. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).