Updates in Laboratory Identification of Invasive Fungal Infection in Neonates
Abstract
1. Introduction
2. Blood Culture
3. 1,3-Beta-d-Glucan (BDG)
4. Galactomannan (GM)
5. Multiple Nested PCR
6. Real-Time PCR
7. Droplet Digital PCR (ddPCR)
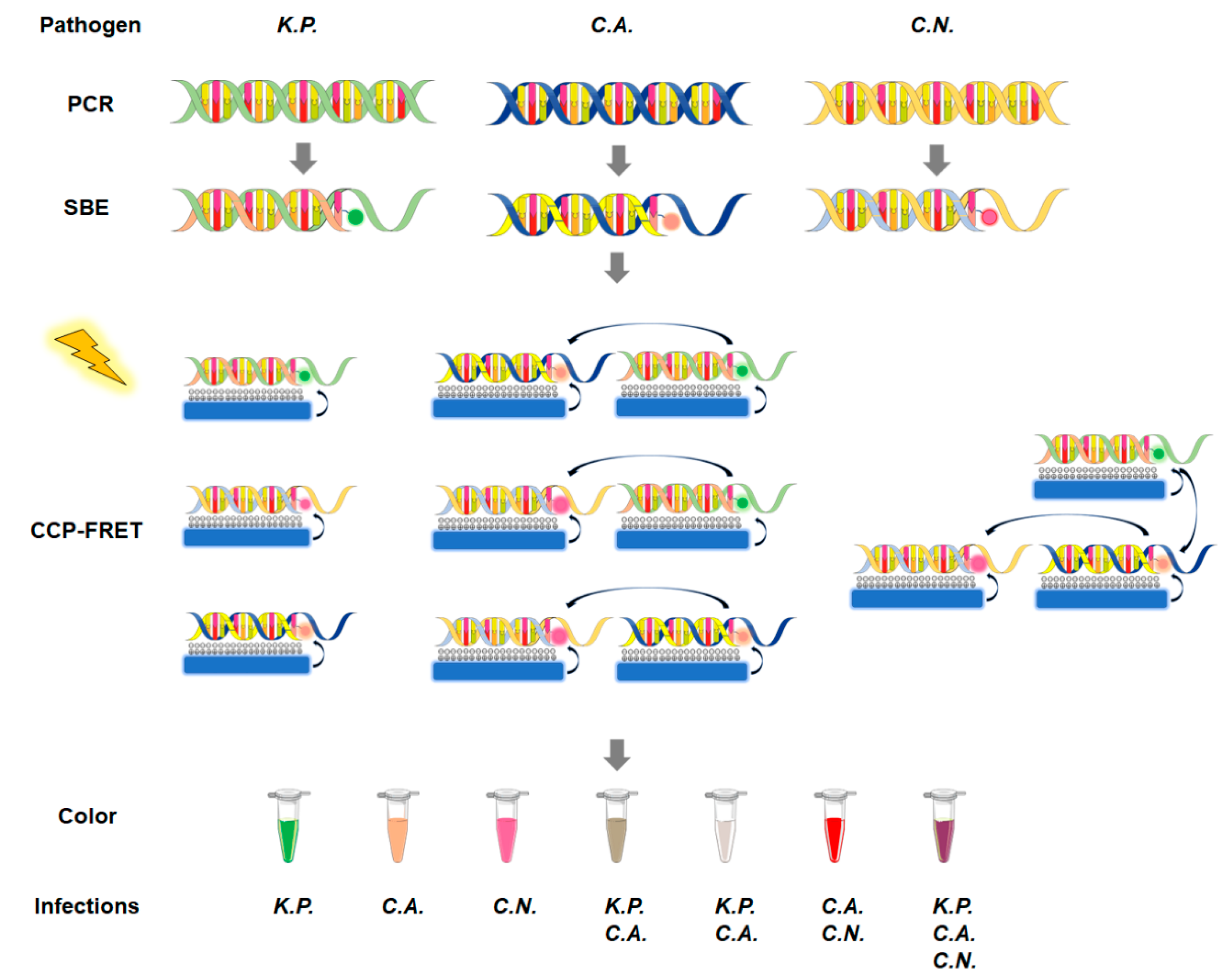
8. CCP-FRET System
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- De Rose, D.U.; Santisi, A.; Ronchetti, M.P.; Martini, L.; Serafini, L.; Betta, P.; Maino, M.; Cavigioli, F.; Cocchi, I.; Pugni, L.; et al. Invasive Candida Infections in Neonates after Major Surgery: Current Evidence and New Directions. Pathogens 2021, 10, 319. [Google Scholar] [CrossRef] [PubMed]
- Ferreras-Antolín, L.; Sharland, M.; Warris, A. Management of Invasive Fungal Disease in Neonates and Children. Pediatr. Infect. Dis. J. 2019, 38, S2–S6. [Google Scholar] [CrossRef] [PubMed]
- Weimer, K.E.D.; Smith, P.B.; Puia-Dumitrescu, M.; Aleem, S. Invasive Fungal Infections in Neonates: A Review. Pediatr. Res. 2021, 91, 404–412. [Google Scholar] [CrossRef] [PubMed]
- Oeser, C.; Vergnano, S.; Naidoo, R.; Anthony, M.; Chang, J.; Chow, P.; Clarke, P.; Embleton, N.; Kennea, N.; Pattnayak, S.; et al. Neonatal Invasive Fungal Infection in England 2004–2010. Clin. Microbiol. Infect. 2014, 20, 936–941. [Google Scholar] [CrossRef] [PubMed]
- Benedict, K.; Roy, M.; Kabbani, S.; Anderson, E.J.; Farley, M.M.; Harb, S.; Harrison, L.H.; Bonner, L.; Wadu, V.L.; Marceaux, K.; et al. Neonatal and Pediatric Candidemia: Results From Population-Based Active Laboratory Surveillance in Four US Locations, 2009–2015. J. Pediatr. Infect. Dis. Soc. 2018, 7, e78–e85. [Google Scholar] [CrossRef] [PubMed]
- Haihua, C.; Jie, C.; Hongyi, T.; Qianqian, M.; Ruijuan, W.; Zhichun, F.; Qiuping, L. Epidemiological Changes in Invasive Fungal Infection in a Neonatal Intensive Care Unit. Chin. J. Perinat. Med. 2017, 20, 577–582. [Google Scholar] [CrossRef]
- Baptista, M.I.; Nona, J.; Ferreira, M.; Sampaio, I.; Abrantes, M.; Tomé, M.T.; Neto, M.T.; Barroso, R.; Serelha, M.; Virella, D. Invasive Fungal Infection in Neonatal Intensive Care Units: A Multicenter Survey. J. Chemother. 2016, 28, 37–43. [Google Scholar] [CrossRef]
- Ishiwada, N.; Kitajima, H.; Morioka, I.; Takeuchi, N.; Endo, M.; Watanabe, A.; Kamei, K. Nationwide Survey of Neonatal Invasive Fungal Infection in Japan. Med. Mycol. 2018, 56, 679–686. [Google Scholar] [CrossRef]
- Demirel, G.; Celik, I.H.; Erdeve, O.; Saygan, S.; Dilmen, U.; Canpolat, F.E. Prophylactic Saccharomyces Boulardii versus Nystatin for the Prevention of Fungal Colonization and Invasive Fungal Infection in Premature Infants. Eur. J. Pediatr. 2013, 172, 1321–1326. [Google Scholar] [CrossRef]
- Austin, N.; Cleminson, J.; Darlow, B.A.; Mcguire, W. Prophylactic Oral/Topical Non-absorbed Antifungal Agents to Prevent Invasive Fungal Infection in Very Low Birth Weight Infants. Cochrane Database Syst. Rev. 2015, 2015, CD003478. [Google Scholar] [CrossRef]
- Enoch, D.A.; Yang, H.; Aliyu, S.H.; Micallef, C. The Changing Epidemiology of Invasive Fungal Infections. Methods Mol. Biol. 2017, 1508, 17–65. [Google Scholar] [CrossRef]
- Gilligan, P.H. Blood Culture Contamination: A Clinical and Financial Burden. Infect. Control Hosp. Epidemiol. 2013, 34, 22–23. [Google Scholar] [CrossRef] [PubMed]
- Lamoth, F.; Akan, H.; Andes, D.; Cruciani, M.; Marchetti, O.; Ostrosky-Zeichner, L.; Racil, Z.; Clancy, C.J. Assessment of the Role of 1,3-β-d-Glucan Testing for the Diagnosis of Invasive Fungal Infections in Adults. Clin. Infect. Dis. 2021, 72, S102–S108. [Google Scholar] [CrossRef] [PubMed]
- Ramos, J.T.; Villar, S.; Bouza, E.; Bergon-Sendin, E.; Rivilla, A.P.; Collados, C.T.; Andreu, M.; Reyes, C.S.; Campos-Herrero, M.I.; De Heredia, J.L.; et al. Performance of a Quantitative PCR-Based Assay and Beta-d-Glucan Detection for Diagnosis of Invasive Candidiasis in Very-Low-Birth-Weight Preterm Neonatal Patients (CANDINEO Study). J. Clin. Microbiol. 2017, 55, 2752–2764. [Google Scholar] [CrossRef] [PubMed]
- Maaroufi, Y.; Heymans, C.; De Bruyne, J.; Duchateau, V.; Rodriguez-villalobos, H.; Aoun, M. Rapid Detection of Candida Albicans in Clinical Blood Samples by Using a TaqMan-Based PCR Assay. J. Clin. Microbiol. 2003, 41, 3293–3298. [Google Scholar] [CrossRef]
- Mandhaniya, S.; Iqbal, S.; Sharawat, S.K.; Xess, I.; Bakhshi, S. Diagnosis of Invasive Fungal Infections Using Real-Time PCR Assay in Paediatric Acute Leukaemia Induction. Mycoses 2012, 55, 372–379. [Google Scholar] [CrossRef]
- Yang, Q.; He, B.; Chen, C.; Wang, H.; Li, W.; Xue, X.; Qiu, T.; Hao, X.; Lv, F.; Wang, S. A Rapid, Visible, and Highly Sensitive Method for Recognizing and Distinguishing Invasive Fungal Infections via CCP-FRET Technology. ACS Infect. Dis. 2021, 7, 2816–2825. [Google Scholar] [CrossRef]
- Talpaz, M.; Kiladjian, J.J. Fedratinib, a Newly Approved Treatment for Patients with Myeloproliferative Neoplasm-Associated Myelofibrosis. Leukemia 2020, 35, 1–17. [Google Scholar] [CrossRef]
- Arvanitis, M.; Anagnostou, T.; Fuchs, B.B.; Caliendo, A.M.; Mylonakis, E. Molecular and Nonmolecular Diagnostic Methods for Invasive Fungal Infections. Clin. Microbiol. Rev. 2014, 27, 490–526. [Google Scholar] [CrossRef]
- Garey, K.W.; Rege, M.; Pai, M.P.; Mingo, D.E.; Suda, K.J.; Turpin, R.S.; Bearden, D.T. Time to Initiation of Fluconazole Therapy Impacts Mortality in Patients with Candidemia: A Multi-Institutional Study. Clin. Infect. Dis. 2006, 43, 25–31. [Google Scholar] [CrossRef]
- Lin, P.C.; Chang, C.L.; Chung, Y.H.; Chang, C.C.; Chu, F.Y. Revisiting Factors Associated with Blood Culture Positivity: Critical Factors after the Introduction of Automated Continuous Monitoring Blood Culture Systems. Medicine 2022, 101, E29693. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, C.D.; Samsa, G.P.; Schell, W.A.; Reller, L.B.; Perfect, J.R.; Alexander, B.D. Quantitation of Candida CFU in Initial Positive Blood Cultures. J. Clin. Microbiol. 2011, 49, 2879–2883. [Google Scholar] [CrossRef] [PubMed]
- Clancy, C.J.; Nguyen, M.H. Finding the “Missing 50%” of Invasive Candidiasis: How Nonculture Diagnostics Will Improve Understanding of Disease Spectrum and Transform Patient Care. Clin. Infect. Dis. 2013, 56, 1284–1292. [Google Scholar] [CrossRef] [PubMed]
- Downey, L.C.; Smith, P.B.; Benjamin, D.K.; Cohen-Wolkowiez, M. Recent Advances in the Detection of Neonatal Candidiasis. Curr. Fungal Infect. Rep. 2010, 4, 17–22. [Google Scholar] [CrossRef]
- Lambregts, M.M.C.; Bernards, A.T.; van der Beek, M.T.; Visser, L.G.; de Boer, M.G. Time to Positivity of Blood Cultures Supports Early Re-Evaluation of Empiric Broad-Spectrum Antimicrobial Therapy. PLoS ONE 2019, 14, e0208819. [Google Scholar] [CrossRef]
- Morrell, M.; Fraser, V.J.; Kollef, M.H. Delaying the Empiric Treatment of Candida Bloodstream Infection until Positive Blood Culture Results Are Obtained: A Potential Risk Factor for Hospital Mortality. Antimicrob. Agents Chemother. 2005, 49, 3640–3645. [Google Scholar] [CrossRef]
- Pana, Z.D.; Roilides, E.; Warris, A.; Groll, A.H.; Zaoutis, T. Epidemiology of Invasive Fungal Disease in Children. J. Pediatric Infect. Dis. Soc. 2017, 6, S3–S11. [Google Scholar] [CrossRef]
- Distefano, G.; Curreri, R.; Betta, P.; Romeo, M.; Amato, M. Procalcitonin Serum Levels in Perinatal Bacterial and Fungal Infection of Preterm Infants. Acta Pædiatrica 2004, 93, 216–219. [Google Scholar] [CrossRef]
- Montagna, M.T.; Coretti, C.; Rella, A.; Barbuti, G.; Manca, F.; Montagna, O.; Laforgia, N.; Caggiano, G. The Role of Procalcitonin in Neonatal Intensive Care Unit Patients with Candidemia. Folia Microbiol. 2013, 58, 27–31. [Google Scholar] [CrossRef]
- Yang, Y.C.; Mao, J. Value of Platelet Count in the Early Diagnosis of Nosocomial Invasive Fungal Infections in Premature Infants. Platelets 2017, 29, 65–70. [Google Scholar] [CrossRef]
- Oz, Y.; Kiraz, N. Diagnostic Methods for Fungal Infections in Pediatric Patients: Microbiological, Serological and Molecular Methods. Expert Rev. Anti. Infect. Ther. 2011, 9, 289–298. [Google Scholar] [CrossRef] [PubMed]
- De Pauw, B.; Walsh, T.J.; Donnelly, J.P.; Stevens, D.A.; Edwards, J.E.; Calandra, T.; Pappas, P.G.; Maertens, J.; Lortholary, O.; Kauffman, C.A.; et al. Revised Definitions of Invasive Fungal Disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin. Infect. Dis. 2008, 46, 1813–1821. [Google Scholar] [CrossRef] [PubMed]
- Guitard, J.; Tabone, M.D.; Senghor, Y.; Cros, C.; Moissenet, D.; Markowicz, K.; Valin, N.; Leverger, G.; Hennequin, C. Detection of β-d-Glucan for the Diagnosis of Invasive Fungal Infection in Children with Hematological Malignancy. J. Infect. 2016, 73, 607–615. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Wu, Y.; Lai, W.; Lu, W.; Mu, X. The Diagnostic Value of (1,3)-β-d-Glucan Alone or Combined with Traditional Inflammatory Markers in Neonatal Invasive Candidiasis. BMC Infect. Dis. 2019, 19. [Google Scholar] [CrossRef]
- Goudjil, S.; Kongolo, G.; Dusol, L.; Imestouren, F.; Cornu, M.; Leke, A.; Chouaki, T. (1-3)-β-d-Glucan Levels in Candidiasis Infections in the Critically Ill Neonate. J. Matern.-Fetal Neonatal Med. 2012, 26, 44–48. [Google Scholar] [CrossRef]
- Cliquennois, P.; Scherdel, P.; Lavergne, R.A.; Flamant, C.; Morio, F.; Cohen, J.F.; Launay, E.; Gras Le Guen, C. Serum (1→3)-β-d-Glucan Could Be Useful to Rule out Invasive Candidiasis in Neonates with an Adapted Cut-Off. Acta Paediatr. 2021, 110, 79–84. [Google Scholar] [CrossRef]
- Ferreras-Antolin, L.; Aziz, N.; Warris, A. Serial (1-3)-Beta-d-Glucan (BDG) Monitoring Shows High Variability among Premature Neonates. Med. Mycol. 2022, 60. [Google Scholar] [CrossRef]
- Goudjil, S.; Chazal, C.; Moreau, F.; Leke, A.; Kongolo, G.; Chouaki, T. Blood Product Transfusions Are Associated with an Increase in Serum (1-3)-Beta-d-Glucan in Infants during the Initial Hospitalization in Neonatal Intensive Care Unit (NICU). J. Matern.-Fetal Neonatal Med. 2016, 30, 933–937. [Google Scholar] [CrossRef]
- Guo, Y.; Li, J. Sandwich-Type Homogeneous Chemiluminescence Immunoassay Based on Nanoparticle toward Detection of Aspergillus Galactomannan Antigen. Talanta 2022, 243, 123392. [Google Scholar] [CrossRef]
- Miceli, M.H.; Maertens, J.; Buvé, K.; Grazziutti, M.; Woods, G.; Rahman, M.; Barlogie, B.; Anaissie, E.J. Immune Reconstitution Inflammatory Syndrome in Cancer Patients with Pulmonary Aspergillosis Recovering from Neutropenia: Proof of Principle, Description, and Clinical and Research Implications. Cancer 2007, 110, 112–120. [Google Scholar] [CrossRef]
- Tong, T.; Shen, J.; Xu, Y. Serum Galactomannan for Diagnosing Invasive Aspergillosis in Pediatric Patients: A Meta-Analysis. Microb. Pathog. 2018, 118, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Siemann, M.; Koch-Dörfler, M.; Gaude, M. False-Positive Results in Premature Infants with the Platelia®Aspergillus Sandwich Enzyme-Linked Immunosorbent Assay. Mycoses 1998, 41, 373–377. [Google Scholar] [CrossRef] [PubMed]
- Fisher, B.T.; Westling, T.; Boge, C.L.K.; Zaoutis, T.E.; Dvorak, C.C.; Nieder, M.; Zerr, D.M.; Wingard, J.R.; Villaluna, D.; Esbenshade, A.J.; et al. Prospective Evaluation of Galactomannan and (1→3) β-d-Glucan Assays as Diagnostic Tools for Invasive Fungal Disease in Children, Adolescents, and Young Adults With Acute Myeloid Leukemia Receiving Fungal Prophylaxis. J. Pediatr. Infect. Dis. Soc. 2021, 10, 864–871. [Google Scholar] [CrossRef] [PubMed]
- Roilides, E. Early Diagnosis of Invasive Aspergillosis in Infants and Children. Med. Mycol. 2006, 44, 199–205. [Google Scholar] [CrossRef]
- Taira, C.L.; Okay, T.S.; Delgado, A.F.; Ceccon, M.E.J.R.; de Almeida, M.T.G.; Del Negro, G.M.B. A Multiplex Nested PCR for the Detection and Identification of Candida Species in Blood Samples of Critically Ill Paediatric Patients. BMC Infect. Dis. 2014, 14. [Google Scholar] [CrossRef]
- Foongladda, S.; Mongkol, N.; Petlum, P.; Chayakulkeeree, M. Multi-Probe Real-Time PCR Identification of Four Common Candida Species in Blood Culture Broth. Mycopathologia 2014, 177, 251–261. [Google Scholar] [CrossRef]
- Bassetti, M.; Garnacho-Montero, J.; Calandra, T.; Kullberg, B.; Dimopoulos, G.; Azoulay, E.; Chakrabarti, A.; Kett, D.; Leon, C.; Ostrosky-Zeichner, L.; et al. Intensive Care Medicine Research Agenda on Invasive Fungal Infection in Critically Ill Patients. Intensive Care Med. 2017, 43, 1225–1238. [Google Scholar] [CrossRef]
- Huppler, A.R.; Fisher, B.T.; Lehrnbecher, T.; Walsh, T.J.; Steinbach, W.J. Role of Molecular Biomarkers in the Diagnosis of Invasive Fungal Diseases in Children. J. Pediatr. Infect. Dis. Soc. 2017, 6, S32–S44. [Google Scholar] [CrossRef]
- Nguyen, M.H.; Wissel, M.C.; Shields, R.K.; Salomoni, M.A.; Hao, B.; Press, E.G.; Shields, R.M.; Cheng, S.; Mitsani, D.; Vadnerkar, A.; et al. Performance of Candida Real-Time Polymerase Chain Reaction, β-d-Glucan Assay, and Blood Cultures in the Diagnosis of Invasive Candidiasis. Clin. Infect. Dis. 2012, 54, 1240–1248. [Google Scholar] [CrossRef]
- Prakash, P.Y.; Irinyi, L.; Halliday, C.; Chen, S.; Robert, V.; Meyer, W. Online Databases for Taxonomy and Identification of Pathogenic Fungi and Proposal for a Cloud-Based Dynamic Data Network Platform. J. Clin. Microbiol. 2017, 55, 1011. [Google Scholar] [CrossRef]
- Vu, D.; Groenewald, M.; de Vries, M.; Gehrmann, T.; Stielow, B.; Eberhardt, U.; Al-Hatmi, A.; Groenewald, J.Z.; Cardinali, G.; Houbraken, J.; et al. Large-Scale Generation and Analysis of Filamentous Fungal DNA Barcodes Boosts Coverage for Kingdom Fungi and Reveals Thresholds for Fungal Species and Higher Taxon Delimitation. Stud. Mycol. 2019, 92, 135. [Google Scholar] [CrossRef] [PubMed]
- Wellinghausen, N.; Siegel, D.; Winter, J.; Gebert, S. Rapid Diagnosis of Candidaemia by Real-Time PCR Detection of Candida DNA in Blood Samples. J. Med. Microbiol. 2009, 58, 1106–1111. [Google Scholar] [CrossRef] [PubMed]
- Mussap, M.; Molinari, M.P.; Senno, E.; Gritti, P.; Soro, B.; Mannelli, S.; Fabris, C. New Diagnostic Tools for Neonatal Sepsis: The Role of a Real-Time Polymerase Chain Reaction for the Early Detection and Identification of Bacterial and Fungal Species in Blood Samples. J. Chemother. 2016, 19 (Suppl. S2), 31–34. [Google Scholar] [CrossRef]
- Oeser, C.; Pond, M.; Butcher, P.; Russell, A.B.; Henneke, P.; Laing, K.; Planche, T.; Heath, P.T.; Harris, K. PCR for the Detection of Pathogens in Neonatal Early Onset Sepsis. PLoS ONE 2020, 15, e226817. [Google Scholar] [CrossRef]
- Valero, C.; De La Cruz-Villar, L.; Zaragoza, Ó.; Buitrago, M.J. New Panfungal Real-Time PCR Assay for Diagnosis of Invasive Fungal Infections. J. Clin. Microbiol. 2016, 54, 2910–2918. [Google Scholar] [CrossRef] [PubMed]
- Kojabad, A.A.; Farzanehpour, M.; Galeh, H.E.G.; Dorostkar, R.; Jafarpour, A.; Bolandian, M.; Nodooshan, M.M. Droplet Digital PCR of Viral DNA/RNA, Current Progress, Challenges, and Future Perspectives. J. Med. Virol. 2021, 93, 4182–4197. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, L.B.; Coleman, V.A.; Hindson, C.M.; Herrmann, J.; Hindson, B.J.; Bhat, S.; Emslie, K.R. Evaluation of a Droplet Polymerase Chain Reaction Format for DNA Copy Number Quantification. Anal. Chem. 2012, 84, 1003. [Google Scholar] [CrossRef]
- Li, H.T.; Lin, B.C.; Huang, Z.F.; Yang, C.Z.; Huang, W.M. [Clinical Value of Droplet Digital PCR in Rapid Diagnosis of Invasive Fungal Infection in Neonates]. Zhongguo Dang Dai Er Ke Za Zhi 2019, 21, 45–51. [Google Scholar] [CrossRef]
- Zhu, C.; Yang, Q.; Liu, L.; Wang, S. Visual Optical Discrimination and Detection of Microbial Pathogens Based on Diverse Interactions of Conjugated Polyelectrolytes with Cells. J. Mater. Chem. 2011, 21, 7905–7912. [Google Scholar] [CrossRef]
- Jeong, J.-E.; Jung, I.H.; Kim, B.; Le, V.S.; Woo, H.Y.; Jeong, J.-E.; Kim, B.; Le, V.S.; Jung, I.H.; Woo, Y. Modulation of Charge Density of Cationic Conjugated Polyelectrolytes for Improving the FRET-Induced Sensory Signal with Enhanced On/Off Ratio. Macromol. Chem. Phys. 2016, 217, 459–466. [Google Scholar] [CrossRef]
- Jiang, G.; Susha, A.S.; Lutich, A.A.; Stefani, F.D.; Feldmann, J.; Rogach, A.L. Cascaded FRET in Conjugated Polymer/Quantum Dot/Dye-Labeled DNA Complexes for DNA Hybridization Detection. ACS Nano 2009, 3, 4127–4131. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.; Li, Z.; He, F.; Wang, S. A Sensitive and Homogeneous SNP Detection Using Cationic Conjugated Polymers. J. Am. Chem. Soc. 2007, 129, 4154–4155. [Google Scholar] [CrossRef] [PubMed]
- Feng, F.; Liu, L.; Wang, S. Fluorescent Conjugated Polymer-Based FRET Technique for Detection of DNA Methylation of Cancer Cells. Nat. Protoc. 2010, 5, 1255–1264. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Zhang, J.; Lv, F.; Cheng, Y.; Wang, B.; Feng, L.; Liu, L.; Wang, S. Multiplex Detection of DNA Mutations by the Fluorescence Fingerprint Spectrum Technique. Angew. Chem. Int. Ed. 2013, 52, 13020–13023. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Yang, Q.; Lv, F.; Liu, L.; Wang, S. Visual Detection of DNA Mutation Using Multicolor Fluorescent Coding. ACS Appl. Mater. Interfaces 2012, 4, 2885–2890. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Zhang, H.; Huang, Y.; Li, M.; Zhao, H.; Yang, Z.; Zhao, H.; Liu, Q.; Fu, Z.; Di, Y.; et al. Sensitive Detection of Single-Nucleotide Polymorphisms by Conjugated Polymers for Personalized Treatment of Hypertension. Sci. Transl. Med. 2023, 15, eabq5753. [Google Scholar] [CrossRef]
| Methodology | Advantages | Disadvantages | References |
|---|---|---|---|
| Blood culture | Detection of fungal pathogen | Long turn-around time Low sensitivity | [18,19,20] |
| BDG | Detection of relevant infection Time-saving | Nonspecific Poor sensitivity High false-positivity | [31,34] |
| Galactomannan | Detection of Aspergillus infection Noninvasive Time-saving | Low sensitivity High false-positivity | [41,42] |
| Multiple nested PCR | Detection of multiple fungi simultaneously High sensitivity Time-saving | High false-positivity Lack of a standardized methodology | [47,48] |
| Real-time PCR | High specificity High sensitivity Time-saving | Contamination Lack of a standardized methodology | [14,53,54] |
| ddPCR | High sensitivity Time-saving | Lack of a standardized methodology | [57] |
| CCP-FRET system | High sensitivity Multiple fungi detection High specificity Time-saving | Careful selection of primers and optimization | [17] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, B.; Yang, Q. Updates in Laboratory Identification of Invasive Fungal Infection in Neonates. Microorganisms 2023, 11, 1001. https://doi.org/10.3390/microorganisms11041001
He B, Yang Q. Updates in Laboratory Identification of Invasive Fungal Infection in Neonates. Microorganisms. 2023; 11(4):1001. https://doi.org/10.3390/microorganisms11041001
Chicago/Turabian StyleHe, Binghong, and Qiong Yang. 2023. "Updates in Laboratory Identification of Invasive Fungal Infection in Neonates" Microorganisms 11, no. 4: 1001. https://doi.org/10.3390/microorganisms11041001
APA StyleHe, B., & Yang, Q. (2023). Updates in Laboratory Identification of Invasive Fungal Infection in Neonates. Microorganisms, 11(4), 1001. https://doi.org/10.3390/microorganisms11041001





