Abstract
Chlorella vulgaris is one of the most commonly used microalgae in aquaculture feeds. It contains high concentrations of various kinds of nutritional elements that are involved in the physiological regulation of aquaculture animals. However, few studies have been conducted to illustrate their influence on the gut microbiota in fish. In this work, the gut microbiota of Nile tilapia (Oreochromis niloticus) (average weight is 6.64 g) was analyzed by high-throughput sequencing of the 16S rRNA gene after feeding with 0.5% and 2% C. vulgaris additives in diets for 15 and 30 days (average water temperature was 26 °C). We found that the impact of C. vulgaris on the gut microbiota of Nile tilapia was feeding-time dependent. Only by feeding for 30 days (not 15 days) did the addition of 2% C. vulgaris to diets significantly elevate the alpha diversity (Chao1, Faith pd, Shannon, Simpson, and the number of observed species) of the gut microbiota. Similarly, C. vulgaris exerted a significant effect on the beta diversity (Bray–Curtis similarity) of the gut microbiota after feeding for 30 days (not 15 days). During the 15-day feeding trial, LEfSe analysis showed that Paracoccus, Thiobacillus, Dechloromonas, and Desulfococcus were enriched under 2% C. vulgaris treatment. During the 30-day feeding trial, Afipia, Ochrobactrum, Polymorphum, Albidovulum, Pseudacidovorax, and Thiolamprovum were more abundant in 2% C. vulgaris-treated fish. C. vulgaris promoted the interaction of gut microbiota in juvenile Nile tilapia by increasing the abundance of Reyranella. Moreover, during the feeding time of 15 days, the gut microbes interacted more closely than those during the feeding time of 30 days. This work will be valuable for understanding how C. vulgaris in diets impacts the gut microbiota in fish.
1. Introduction
Chlorella vulgaris is one of the most commonly used microalgae in aquaculture feeds [1]. It has been incorporated as an ingredient in the diets of aquaculture animals, such as prawns [2], Nile tilapia [3,4], rainbow trout [5], and African catfish [6]. C. vulgaris contains high concentrations of lutein, beta-carotene, astaxanthin, canthaxanthin, and chlorella growth factor (CGF) [1]. C. vulgaris leads to higher growth, better feed utilization, and increased digestive enzymatic activities in fish when used as a feed additive [7,8,9]. The addition of some amount of C. vulgaris to diets also modulated and increased the antioxidant capacity, innate immunity, and disease resistance of fish [10]. C. vulgaris also contains high-quality protein and lipids, and has been used at higher inclusions (10–20%) in the diet to replace fishmeal [11]. However, few studies have been conducted to illustrate the influence of C. vulgaris on the gut microbiota in fish.
The gut microbiota plays many critical roles in host fish [12,13]. In fish, the early larval and juvenile stages represent a critical period for growth and survival in aquaculture because larvae are vulnerable to diseases, such as infection, during this stage [14]. The gut microbiota in larval and juvenile fish is crucial for protection against pathogens and for nutrition metabolism and physiological functions [15]. Diets have been successfully used to modulate the gut microbiota and further promote the health of vertebrate animals [16,17]. With the rapid development of high-throughput sequencing, the in-depth mechanisms by which C. vulgaris, as a feed additive, impacts the gut microbial communities in aquaculture animals have been illustrated [1]. It was found that the abundance of beneficial bacterial taxa (Firmicutes, Bacteroidetes, and Cetobacterium) in the gut of juvenile largemouth bass (Micropterus salmoides) was significantly increased by feeding with C. vulgaris [18]. Moreover, fishmeal partially substituted with C. vulgaris increased the abundance of Lactobacillus and decreased Escherichia coli abundance in the gut of juvenile narrow clawed crayfish [19]. Notably, although some microalgae, such as Schizochytrium [20], have been studied, we still know little about how C. vulgaris shapes the microbial communities in the gastrointestinal (GI) tract in Nile tilapia (Oreochromis niloticus), especially larval and juvenile fish, until now. Moreover, the digestive systems and immune organs of larval and juvenile fish develop rapidly [14]. We hypothesized that the effect of C. vulgaris meal on the gut microbiota in larval and juvenile fish may be dependent on feeding time. In this work, after feeding with 0.5% and 2% C. vulgaris additives in diets for 15 and 30 days, the gut microbiota in Nile tilapia were analyzed by high-throughput sequencing of the 16S rRNA gene. We aimed to determine how C. vulgaris in diets impacts the gut microbiota in juvenile Nile tilapia at different feeding times.
2. Materials and Methods
2.1. Feed Preparation
C. vulgaris was obtained from the Freshwater Algae Culture Collection at the Institute of Hydrobiology (FACHB) in China. C. vulgaris was cultured with growth media (CaCl2•2H2O 1 g/L, FeSO4•7H2O 0.01 g/L, MgSO4•7H2O 0.2 g/L, NaNO3 2 g/L, K2HPO4 0.2 g/L, yeast extract 1 g/L, glucose 4 g/L) in 10-L sterilized bottles for one week. Afterward, C. vulgaris was centrifuged and air-dried at room temperature for collection. The C. vulgaris and other components of the diets are listed in Table 1. All the dietary components were ground, sifted (60-mesh sieve), and mixed evenly. Afterward, the mixed raw materials were pressed into pellets with a fish feed pelleting machine (SZLH250; HENGFU MACHINERY, China) using a cold pelleting process, and the screen diameter was 2 mm; the pellets were then air-dried (20 °C), and stored at 4–8 °C until use.

Table 1.
Formulation and proximate composition of the experimental diets (%).
2.2. Feeding and Management
A total of 270 healthy Nile tilapia (weight: 6.64 ± 0.88 g) were selected and evenly divided into three groups (control group: fed a basal diet; 0.5% Chl group: fed a basal diet supplemented with 0.5% C. vulgaris; 2% Chl group: fed a basal diet supplemented with 2% C. vulgaris) in nine 300 L fiberglass tanks (three tanks per group). Nile tilapia were maintained in an indoor recirculating freshwater system with temperature control and aeration equipment at Beibu Gulf University (Qinzhou city, China). Water quality was tested daily and properly managed as follows: temperature, 26 ± 1.5 °C; dissolved oxygen, >6 mg L−1; pH, 7.2 ± 0.5; ammonia nitrogen < 0.04 mg L−1. Fish were fed at 2% to 3% body weight by hand twice daily (8:00–9:00 and 17:00–18:00) for 30 days.
2.3. DNA Extraction and 16S rRNA Gene High-Throughput Sequencing of the Gut Microbiota
At the end of the feeding trial, healthy fish with similar weights were collected from each group (six fish from each group). The fish surface was wiped and sterilized with 70% alcohol, and then the fish were placed on a clean dissecting pan. The intestinal contents of the fish were obtained with a sterilized dissecting knife, immediately frozen in liquid nitrogen, and stored in a −80 °C freezer. Then, the total DNA of the gut microbiota in Nile tilapia was extracted from 0.3 g feces (all the feces from fish guts were homogenized before use) with the GenElute™ Stool DNA Isolation Kit (Sigma–Aldrich, St. Louis, MO, USA). After assessing the quality of the DNA, the obtained DNA was used as a template for amplification of the V3–V4 region in the 16S rRNA gene with primers 341F and 909R [21]. Then, high-throughput sequencing was conducted using the Illumina HiSeq2500 platform [22] at Shanghai Majorbio Biopharm Technology Co., Ltd. (Shanghai, China). The raw data were processed with the QIIME 2 pipeline (version 2020.2) following the following procedure: trimming with StreamingTrim [23], merging with MeFiT [24], and chimeric assessment and removal with Greengenes [25]. The clean reads were used to construct amplicon sequence variants (ASVs) with DADA2 in R [26]. The taxonomic annotation of the ASVs was conducted with the SINTAX classifier in usearch (version 11.0.667) [27] against the Silva Database (version 138.1) [28]. A total of 19,439 ASVs were generated from 37 samples, with an average of 84,961 clean reads per sample.
2.4. Bioinformatics Analysis
The alpha diversity indices (Chao1, Shannon, Simpson, number of observed species, Faith_pd, and Pielou_e index) of gut microbes were calculated using the phyloseq package in R [29]. The beta diversity of the gut microbiota in fish was calculated using Bray–Curtis dissimilarity [30], and then the results were visualized with nonmetric dimensional scaling (NMDS) using ggplot2 in R [31]. Linear discriminant analysis effect size (LEfSe) analysis of the gut microbiota in Nile tilapia was performed on the Majorbio Cloud Platform with an LDA threshold of 3–4 (FDR < 0.01) [32]. To assess interactions among microbial communities (using operational taxonomic units (OTUs)) within the gut of Nile tilapia in two growth stages (15 days and 30 days), a cooccurrence network analysis was implemented. Networks were calculated using the online tool of the Majorbio Cloud Platform (https://cloud.majorbio.com/page/tools/, accessed on 21 December 2021) [33]. The Pearson correlation coefficient was utilized as an index to evaluate the interaction. Strong (|R| > 0.8) and significant (p < 0.05) interactions were used to build the co-occurrence networks. The networks were rendered using Gephi software (Version 0.9.2).
2.5. Statistical Analysis
One-way analysis of variance (one-way ANOVA) was used to test the differences in alpha diversity variation of gut microbiota among different groups. The statistical analyses were performed in SPSS 24.0 software [34], and p < 0.05 was considered a significance threshold. Analysis of similarities (ANOSIM) was used to test for variations in β-diversity in the gut microbiota between groups in R 4.1.3.
3. Results
3.1. Alpha Diversity of the Gut Microbiota
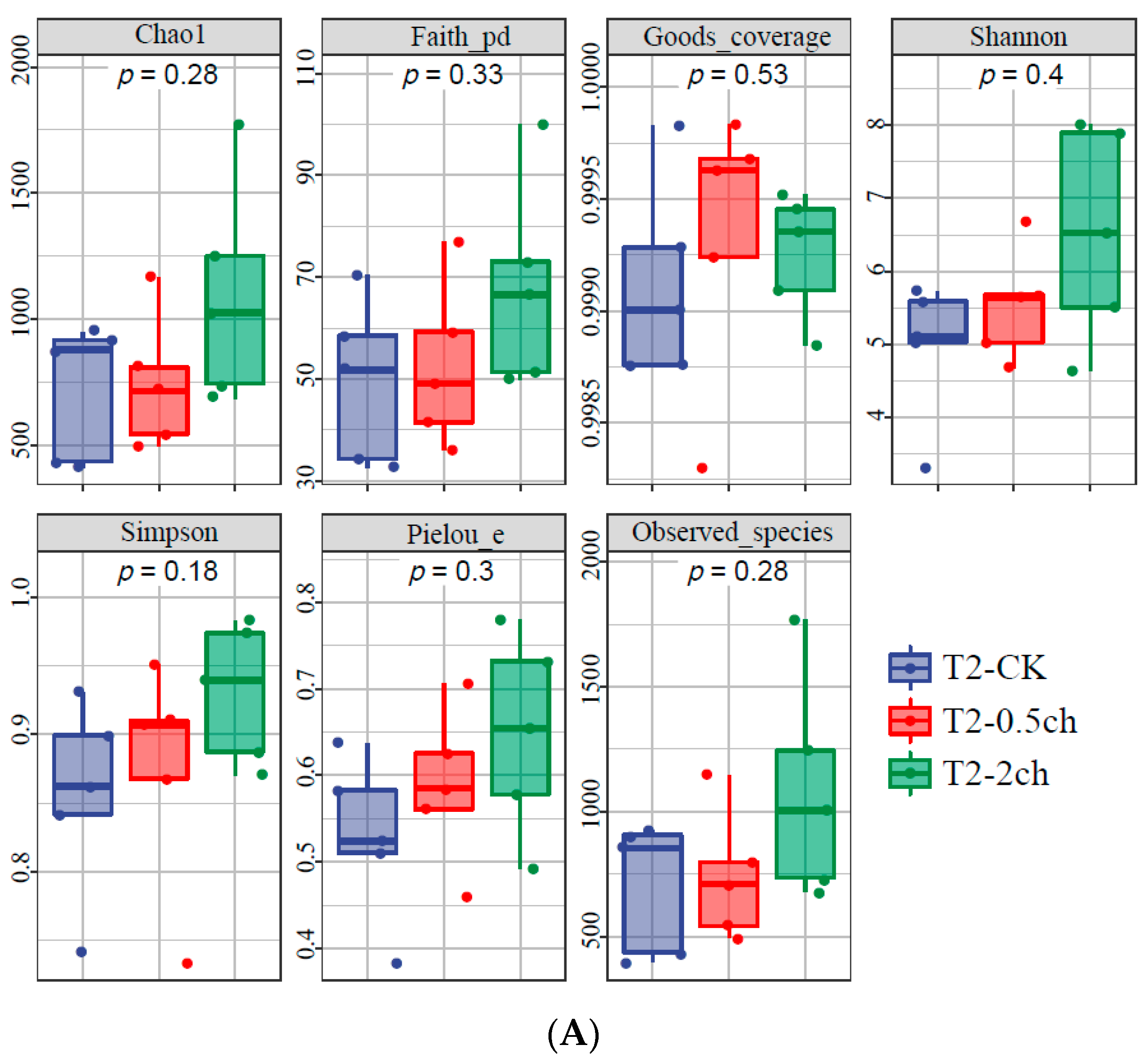
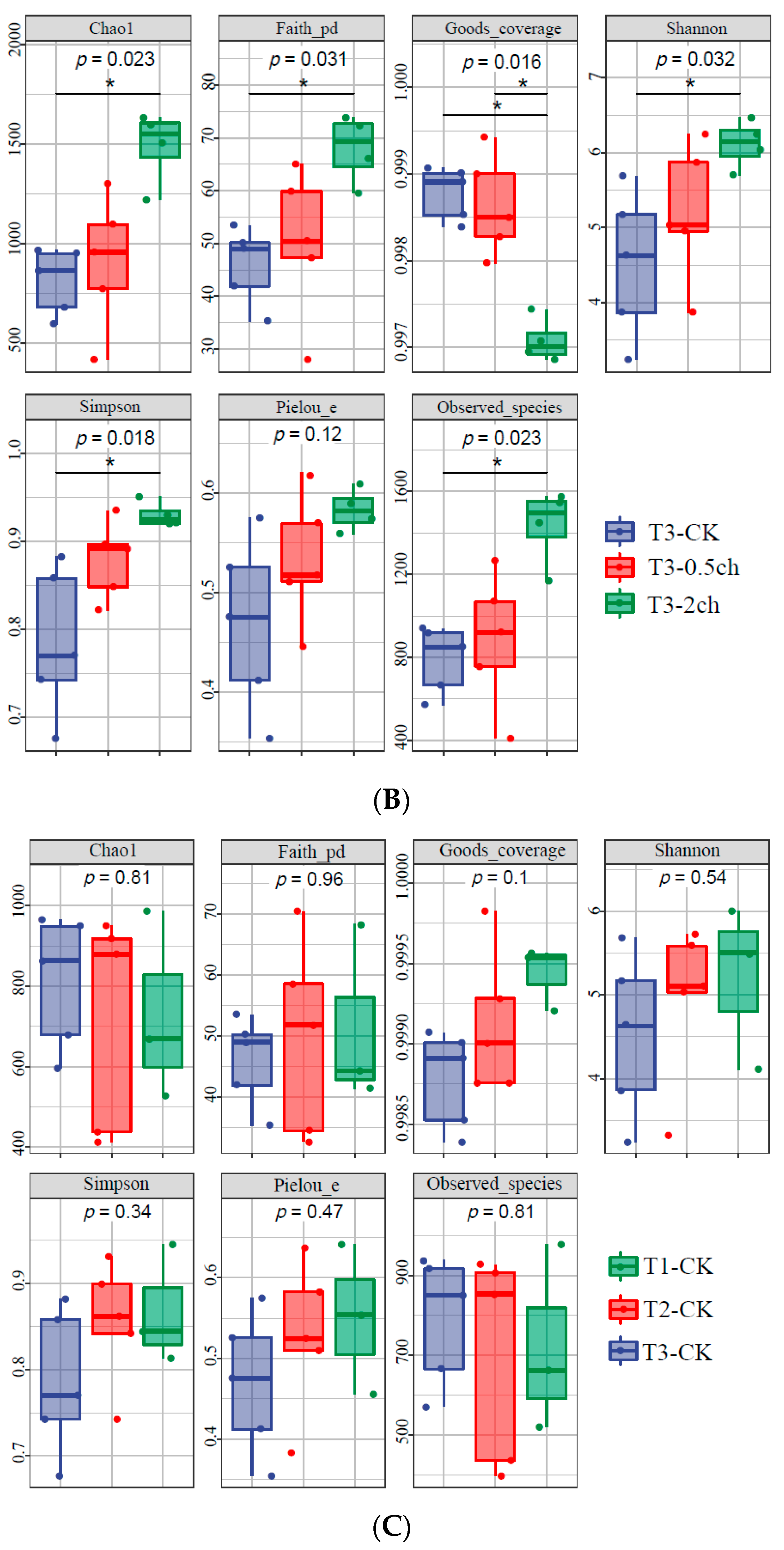
The alpha diversity of the microbial community in the gut of Nile tilapia was detected and analyzed. The alpha diversity of the gut microbiota in Nile tilapia fed C. vulgaris for 15 days (Figure 1A) and 30 days (Figure 1B) was compared. During this 30-day feeding trial, 2% C. vulgaris significantly increased the Chao1, Faith pd, Shannon, Simpson, and the number of observed species (Figure 1B) (ANOVA, p < 0.05). The alpha diversity of the gut microbiota at different feeding times (0 days, 15 days, and 30 days) was compared (Figure 1C). The Shannon and Simpson indices in the gut microbiota of fish fed for 0 and 15 days were higher than those in the 30-day group.
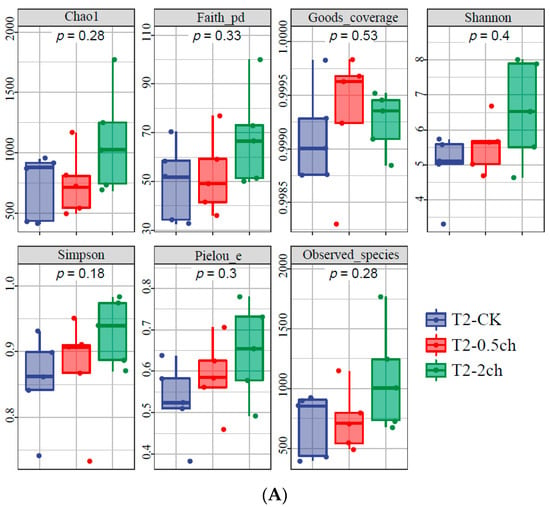
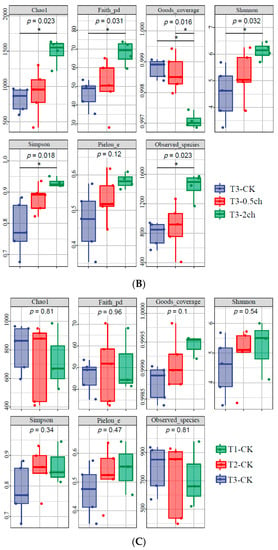
Figure 1.
The alpha diversity of the microbial community in the gut of Nile tilapia (Oreochromis niloticus). (A). The alpha diversity of the microbiota in Nile tilapia was fed 0.5% and 2% C. vulgaris for 15 days. T2-CK, control group; T2-0.5ch, 0.5% C. vulgaris-treated group; T2-2ch, 2% C. vulgaris-treated group. (B). The alpha diversity of the microbiota in Nile tilapia fed 0.5% and 2% C. vulgaris for 30 days. T3-CK, control group; T3-0.5ch, 0.5% C. vulgaris-treated group; T3-2ch, 2% C. vulgaris-treated group. (C). The alpha diversity of the microbiota in Nile tilapia during different feeding times. T1-CK, before the feeding trial; T2-CK, fed for 15 days without C. vulgaris; T2-CK, fed for 30 days without C. vulgaris. Symbol * means p < 0.01.
3.2. NMDS Analysis of the Gut Microbiota
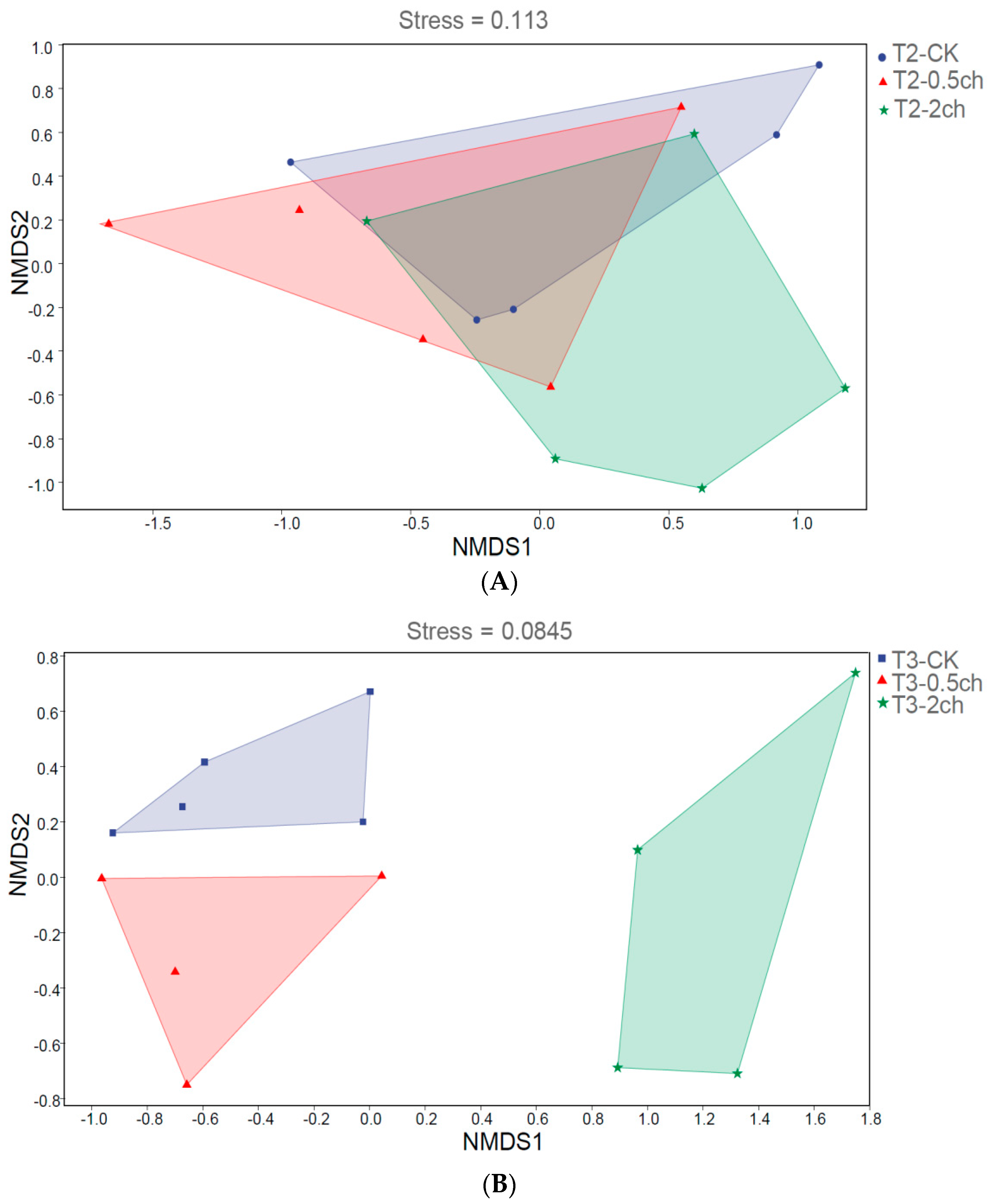
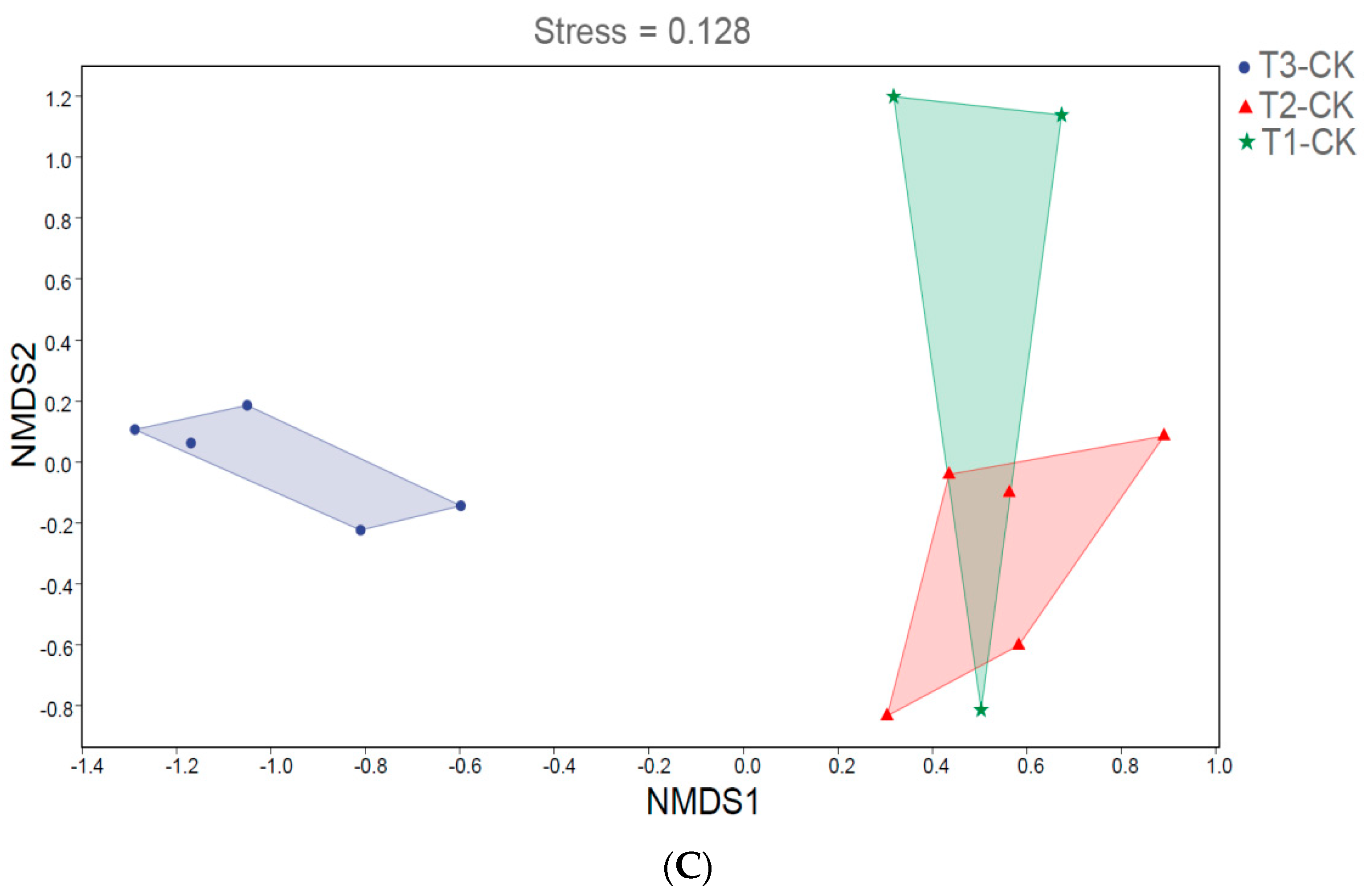
We analyzed the influence of 0.5% and 2% C. vulgaris and growth time on the beta diversity of the gut microbiota in Nile tilapia by NMDS analysis (Figure 2). During the 15-day feeding trial, we did not observe a distinct discrepancy in the microbial community among the 0.5% C. vulgaris-treated, 2% C. vulgaris-treated, and non-Chlorella-treated groups (Figure 2A). However, the gut microbiota in Nile tilapia at 0 days and 15 days was different from that at 30 days (Figure 2C). As the figure shows, the microbial communities during the 0-day and 15-day feeding trials were closely clustered, while the microbiota during the 30-day feeding trials were distributed in another cluster. During the 30-day feeding trial, the gut microbiota in the 2% C. vulgaris-treated, 0.5% C. vulgaris-treated, and non-Chlorella-treated groups were significantly separated from each other (ANOSIM, R = 0.452, p = 0.001) (Figure 2B).
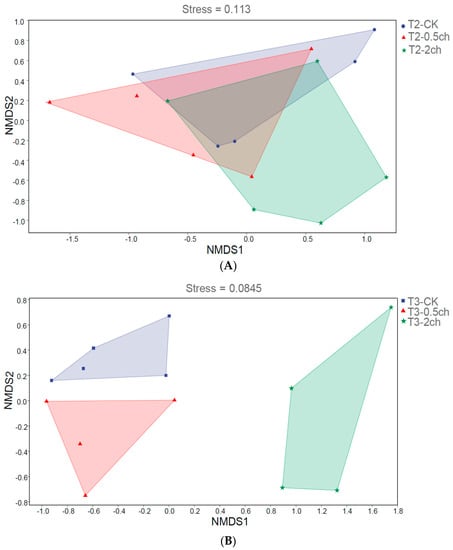
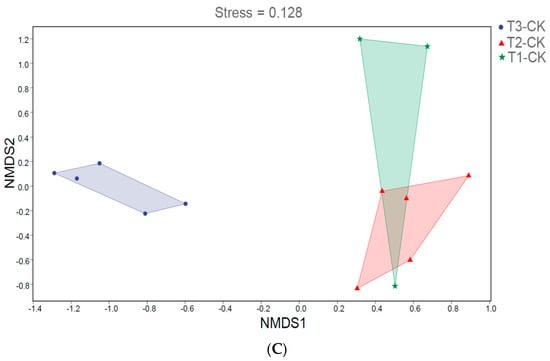
Figure 2.
Nonmetric multidimensional scaling (NMDS) analysis of gut microbiota in Nile tilapia. (A). NMDS analysis of the microbiota in Nile tilapia fed 0.5% and 2% C. vulgaris for 15 days. T2-CK, control group; T2-0.5ch, 0.5% C. vulgaris-treated group; T2-2ch, 2% C. vulgaris-treated group. (B). NMDS analysis of the microbiota in Nile tilapia fed 0.5% and 2% C. vulgaris for 30 days. T3-CK, control group; T3-0.5ch, 0.5% C. vulgaris-treated group; T3-2ch, 2% C. vulgaris-treated group. (C). NMDS analysis of the microbiota in Nile tilapia during different feeding times. T1-CK, before feeding; T2-CK, fed for 15 days without C. vulgaris; T2-CK, fed for 30 days without C. vulgaris.
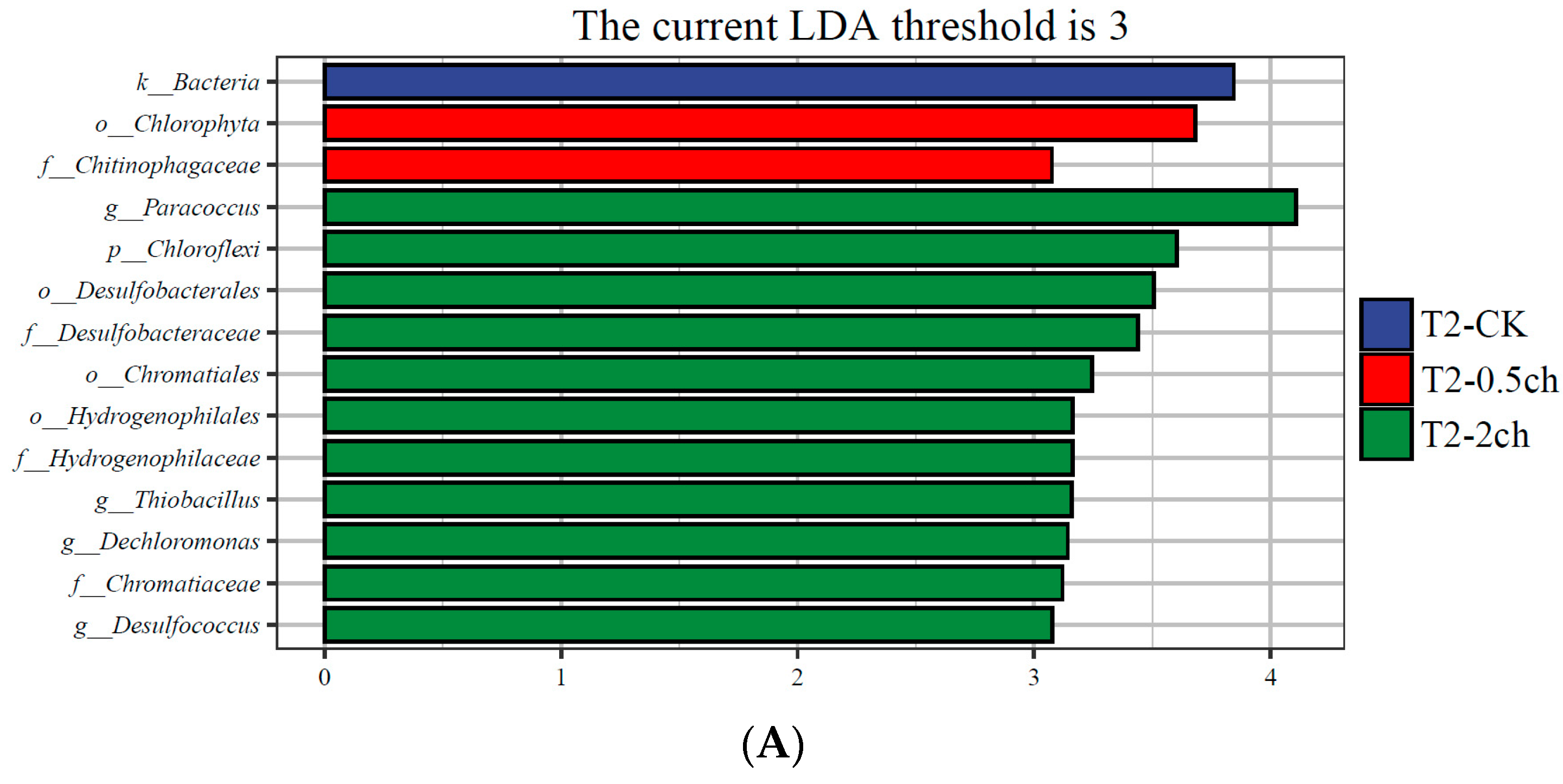
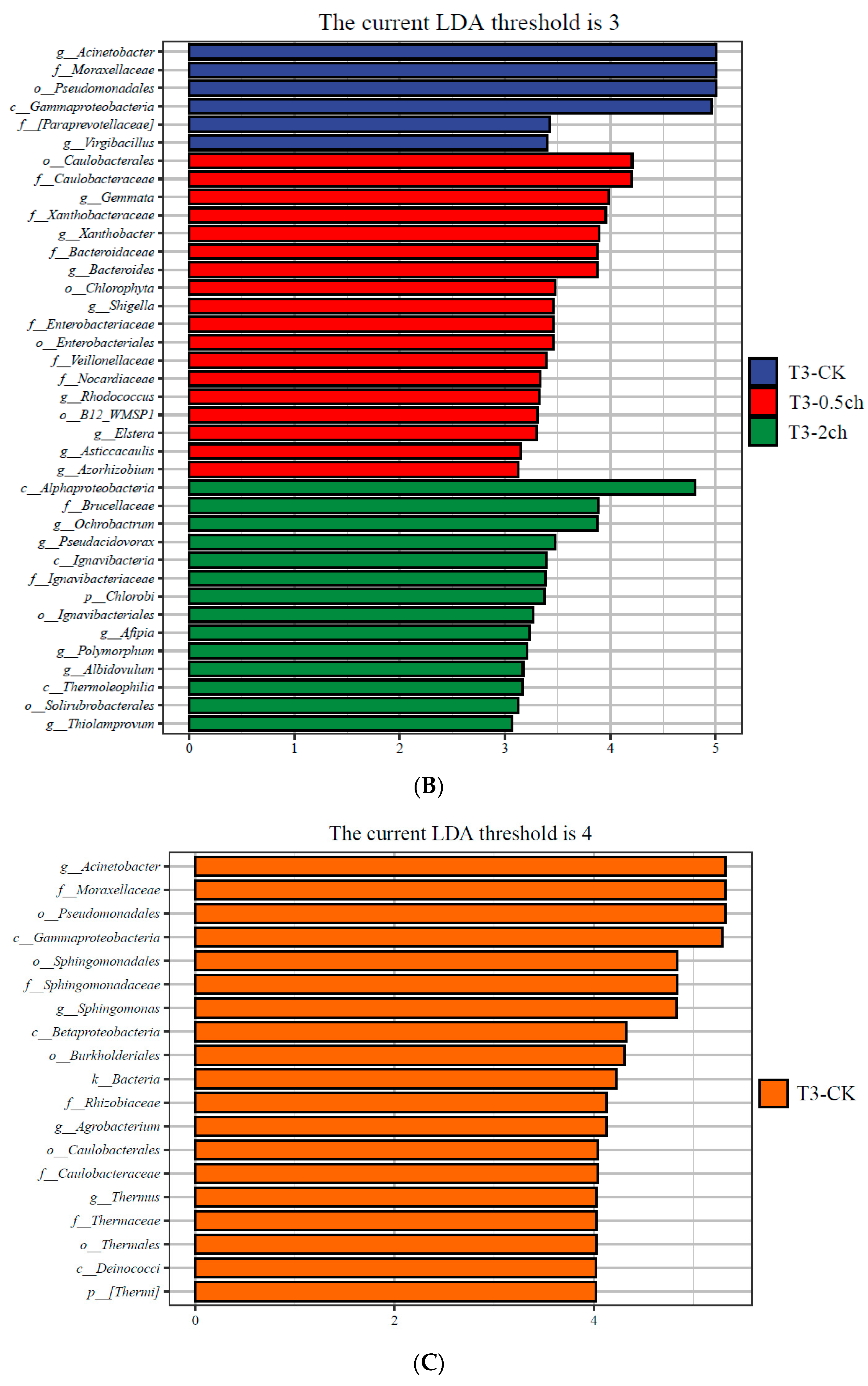
3.3. LEfSe Analysis
LEfSe analysis was utilized to identify discriminating taxa in the gut microbiota of Nile tilapia during different feeding times (p < 0.05, LDA > 4) and those associated with different amounts of C. vulgaris in diets (p < 0.05, LDA > 3) (Figure 3). During a 15-day feeding trial, the gut microbiota of fish fed 2% C. vulgaris was represented by biomarkers such as Paracoccus, Thiobacillus, Dechloromonas, and Desulfococcus. During a 30-day feeding trial, the gut microbiota of fish fed 2% C. vulgaris was represented by Afipia, Ochrobactrum, Polymorphum, Albidovulum, Pseudacidovorax, and Thiolamprovum; 0.5% C. vulgaris-enriched taxa included Rhodococcus, Bacteroides, Gemmata, Asticcacaulis, Azorhizobium, Xanthobacter, Elstera, and Shigella. During different life stages (0 days, 15 days, and 30 days), basal diet feeding for 30 days significantly stimulated the proliferation of Thermus, Agrobacterium, Sphingomonas, and Acinetobacter.
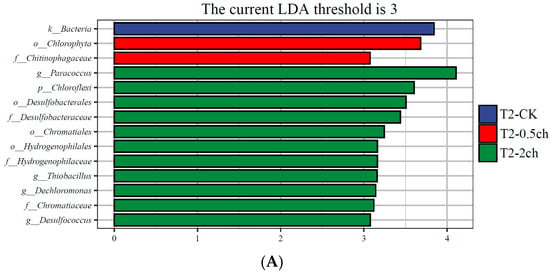
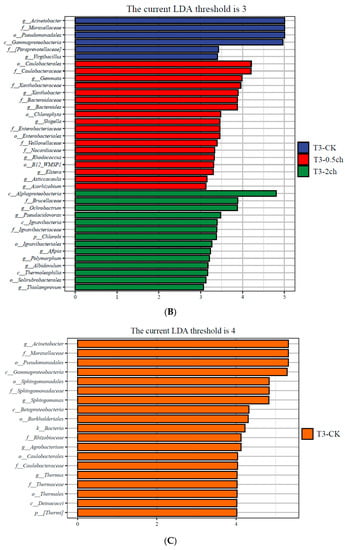
Figure 3.
Linear discriminant analysis effect size (LEfSe) analysis of gut microbiota in Nile tilapia. (A) The taxa with different abundances of gut microbiota in Nile tilapia fed 0.5% and 2% C. vulgaris for 15 days. T2-CK, control group; T2-0.5ch, 0.5% C. vulgaris-treated group; T2-2ch, 2% C. vulgaris-treated group. (B) The taxa with different abundances of gut microbiota in Nile tilapia fed 0.5% and 2% C. vulgaris for 30 days. T3-CK, control group; T3-0.5ch, 0.5% C. vulgaris-treated group; T3-2ch, 2% C. vulgaris-treated group. (C) The taxa with different abundances of gut microbiota in Nile tilapia during different feeding times. T1-CK, before feeding; T2-CK, fed for 15 days without C. vulgaris; T3-CK, fed for 30 days without C. vulgaris.
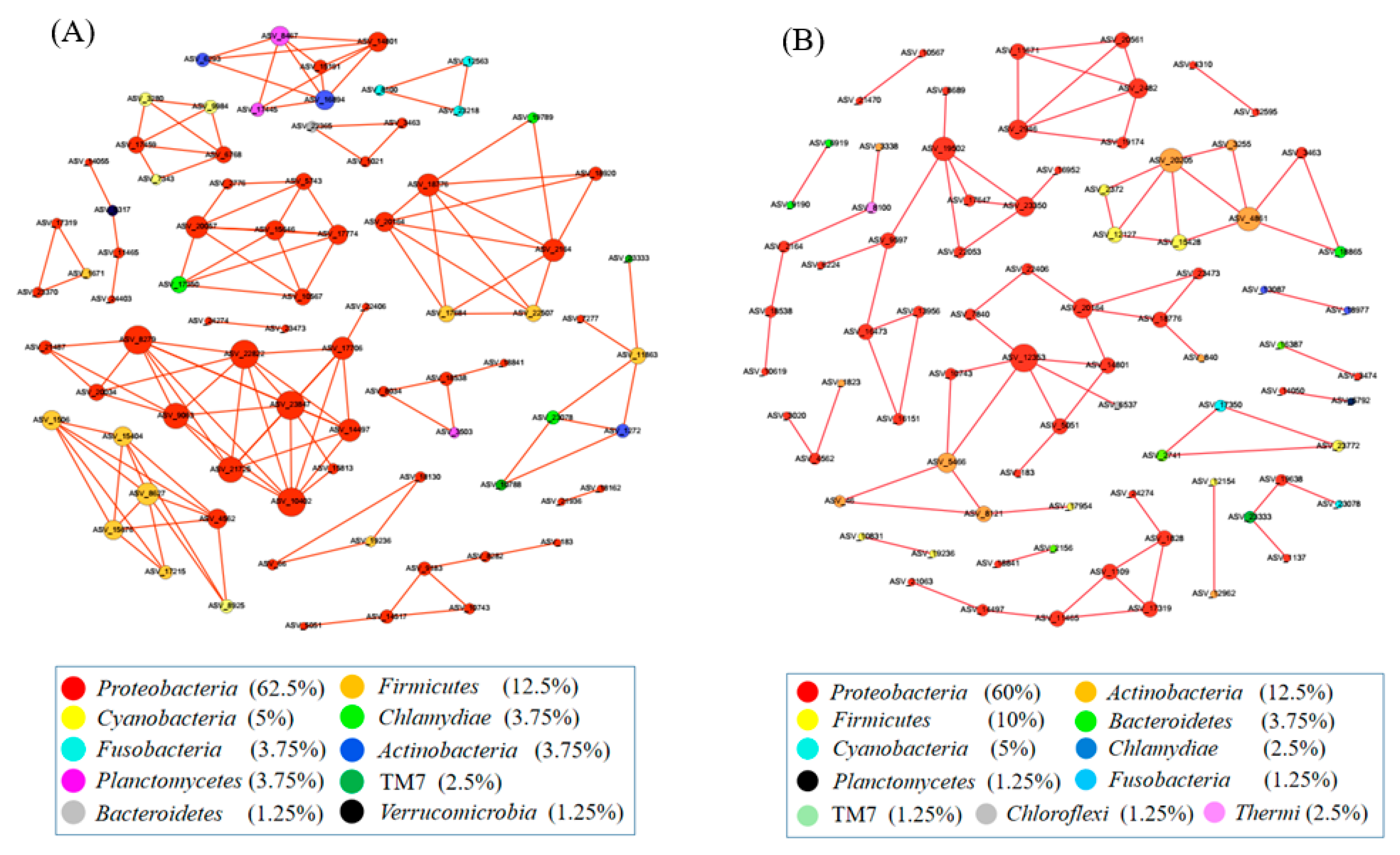
3.4. Network Analysis
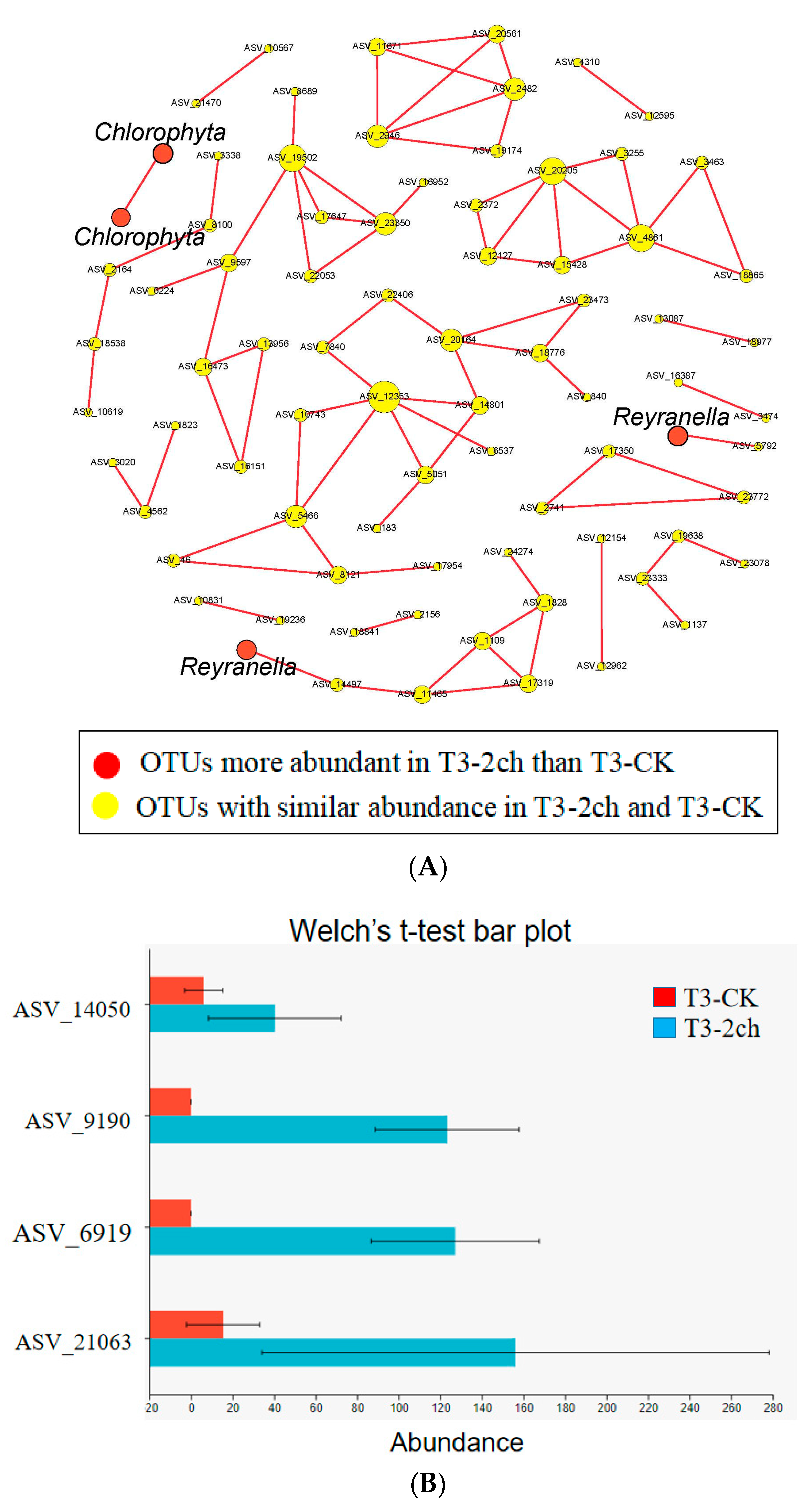
The co-occurrence network among ASVs was constructed using random matrix theory (RMT) (Figure 4). During the growth time of 15 days, a total of 80 ASVs (nodes) and 134 correlation links (edges) were rendered in the network of gut microbiota (R > 0.80, p-value > 0.05) (Netwoek15 days, Figure 4A). During the growth time of 30 days, a total of 80 ASVs (nodes) and 82 correlation links (edges) were rendered (Netwoek30 days, R > 0.80, p-value > 0.05) (Figure 4B). Eight and five large modules, which possessed more than five nodes in each module, were recovered from Network15 days and Network30 days, respectively. In both co-occurrence networks above, Proteobacteria was the most predominant (Figure 4). Four ASVs in the co-occurrence network, named ASV_9190, ASV_6919, ASV_21063, and ASV_14050, were more abundant in 2% C. vulgaris-treated Nile tilapia at a feeding time of 30 days (Welch’s t-test, p < 0.05) (Figure 5). ASV_9190 and ASV_6919 were annotated as unidentified_Chlorophyta, while ASV_21063 and ASV_14050 were annotated as Reyranella_massiliensis.
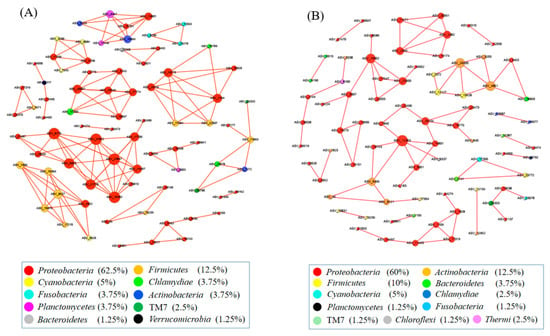
Figure 4.
The interactions of gut microbes in Nile tilapia are illustrated by co-occurrence network analysis. (A). The co-occurrence network of gut microbiota in Nile tilapia fed 0%, 0.5%, and 2% C. vulgaris for 15 days. (B). The co-occurrence network of gut microbiota in Nile tilapia fed 0%, 0.5%, and 2% C. vulgaris for 30 days. The different colored circles indicate some ASVs in the network annotated at the phylum level.
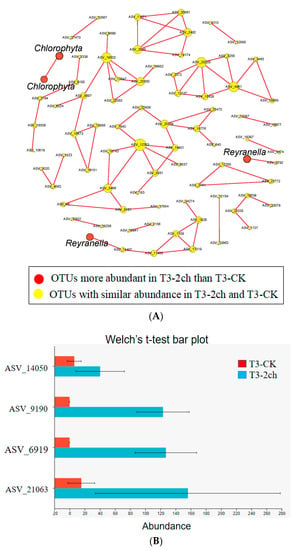
Figure 5.
Some ASVs in the co-occurrence network were especially enriched in 2% C. vulgaris-treated Nile tilapia at the life stage of 30 days. (A) The red nodes indicate ASVs enriched in 2% C. vulgaris-treated fish; (B) The ASVs with higher abundance in 2% C. vulgaris-treated fish were detected by Welch’s t-test (p < 0.05).
4. Discussion
Gut microbes in fish influence various host functions, including nutrition, digestion, development, disease resistance, and immunity [35]. Studies of the gut microbiota are beneficial for developing effective strategies to promote host health and improve growth [36]. Host phylogeny and diet may significantly influence gut microbial community diversity in fish [37]. Among them, the effect of diets on the microbial community present in the fish gut has recently attracted increasing attention [38]. Changes in dietary nutrients and ingredients have been applied to evaluate their distinct impact on the gut microbial community of fish [39,40,41]. The impact of different diets on the alpha and beta diversity of the gut microbiota in fish varies [38,42,43,44]. Moreover, the enzyme activity and physiological structure of the GI tract in fish may change during ontogenesis [45]. Therefore, when we evaluate the influence of diets on the gut microbial community in fish, we should consider the influence of different lifetimes. To date, we know little about this effect, and it is urgent to conduct in-depth investigations.
In this study, we found that the impact of C. vulgaris on gut microbiota diversity in fish is feeding-time dependent. First, the impact of C. vulgaris on the alpha diversity of the gut microbiota in Nile tilapia was correlated with feeding time. As the study illustrated, only by feeding for 30 days (not 15 days) did the addition of 2% C. vulgaris to diets significantly elevate the alpha diversity of gut microbiota. Second, the influence of C. vulgaris on the beta diversity of fish gut microbiota was feeding-time dependent as well. After feeding with C. vulgaris for 30 days, 0.5% and 2% of C. vulgaris exerted a significant effect on the beta diversity of the fish gut microbiota. However, the 15-day feeding trial did not significantly change the bacterial community structure from that observed during the initiation stage of feeding. The impact of C. vulgaris on the beta diversity of the gut microbiota in large fish was stronger than that in small fish. This is the first report that the impact of C. vulgaris on the diversity of gut microbiota is dependent on the growth time in fish.
Recently, many studies have reported that diet could strongly impact the GI microbiota in fish [46,47,48,49,50]. Chlorella has been widely used as a direct feed or an ingredient normally incorporated in diets for different stages of fish or prawn culturing [51]. The addition of C. vulgaris to fish diets significantly improved immunity [4,50], disease resistance [52], and growth rate [6,53,54]. However, the effect of C. vulgaris dietary supplementation on the fish intestinal microbiome has seldom been studied previously. Zhang et al. noted that 50% replacement of fishmeal with Chlorella did not influence species richness, Shannon diversity, or the community structure of the gut microbiota. However, high levels of fishmeal replacement (between 75 and 100%) significantly induced intestinal community disturbance and diversity loss in largemouth bass [55]. The effect of C. vulgaris on the gut microbiota depended on a simulated in vitro digestion process [56]. In summary, no study has been conducted on the impact of C. vulgaris on the fish gut microbiota during different feeding times.
The impact of C. vulgaris on the diversity of the gut microbiota in fish may correlate with the digestible feature of C. vulgaris in the GI tract of Nile tilapia. The cell wall of C. vulgaris is rigid, and it restricts the access of digestive enzymes in the GI tract to intracellular components for proper digestion and assimilation [1]. Moreover, the morphological and cellular characteristics of the GI tract in fish change during the first month of life [57]. Young larval fish cannot digest dietary components in a manner exactly analogous to that of juveniles [57]. A similar study found that the apparent digestibility of crude fiber increased with pig age, which was correlated with the levels of gut microbes (such as Anaeroplasma, Campylobacter, and Clostridium) in pigs at different ages [58]. However, the digestibility of C. vulgaris in Nile tilapia at different feeding times have seldom been investigated. We still know little about whether the changing gut microbiota during different feeding times of Nile tilapia is the driving force of the degradation and assimilation of C. vulgaris or whether a switch in intestinal physiology improves the degradation and digestion of C. vulgaris and then modulates gut microbes.
The use of precise control over dietary intake, especially nondigestible carbohydrates, to modulate gut microbes and improve health has recently attracted great attention [59]. The consumption of nondigestible or food with limited digestibility, especially nondigestible carbohydrates, could manipulate and modulate the gut microbial composition and is beneficial for human health [60,61]. In this work, LEfSe analysis showed that some biomarkers, such as Paracoccus, Thiobacillus, Dechloromonas, and Desulfococcus, were enriched in the gut microbiota of fish under 2% C. vulgaris treatment during a 15-day feeding trial. Paracoccus, Thiobacillus, Dechloromonas, and Desulfococcus are chemolithotrophic bacteria. Thiobacillus and Desulfococcus are believed to play major roles in sulfur oxidation and sulfate reduction, respectively. Paracoccus has been reported as the core microbiome in the gut of prawns [62] and shrimp [63]. Moreover, Paracoccus promoted growth and enhanced intestinal innate immunity in sea cucumbers [64]. Notably, the abundance of Paracoccus increased in the gut microbiota of fish under 2% C. vulgaris treatment during the 15-day feeding trial, which exhibited the beneficial effects of C. vulgaris on promoting the growth of probiotics in the fish gut. Traditional Chinese medicine (TCM) is believed to be beneficial for enhancing fish immune functions and improving intestinal health conditions [65]. Dechloromonas was found to be significantly more abundant in the intestinal microbiota of gibel carp fed TCM than in nontreated fish [66]. This finding further illustrates that C. vulgaris is able to promote the growth of gut bacteria beneficial for the health of fish. Some taxa, such as Polymorphum, were more abundant in 2% C. vulgaris-treated fish during the 30-day feeding trial. Polymorphum was detected in the gut of Litopenaeus vannamei fed the probiotic Lactobacillus pentosus but not in the control group [67]. However, the function of some other taxa, including Afipia, Ochrobactrum, Albidovulum, Pseudacidovorax, and Thiolamprovum, in the fish gut has not been reported previously. Therefore, a profound investigation should be conducted to illustrate their effect on the fish gut.
The co-occurrence network analysis showed some interesting results on how C. vulgaris impacted the gut microbiota of Nile tilapia. First, this finding indicated that the addition of C. vulgaris to diets did not significantly impact the interaction of microbial communities in the gut of Nile tilapia during the feeding time of 15 days. One reason for this may be that the rigidity of the cell wall restricts the utilization of C. vulgaris [1], and another reason could be that the abundant extracellular polysaccharides in C. vulgaris may restrict its absorption by fish [68]. However, Reyranella, which was more abundant in 0.5% or 2% C. vulgaris-treated Nile tilapia during the feeding time of 30 days, formed an interaction network with Rhodobacter, Rhizobiales, Hyphomicrobium, and Sphingomonas. This means that C. vulgaris may promote the interaction of gut microbiota by increasing the abundance of Reyranella. Reyranella, Rhodobacter, Hyphomicrobium, and Sphingomonas have been commonly found in Nile tilapia and other fish guts [69,70,71], and their functions in the gut have not been clearly illustrated until now. Second, in this work, more correlation links and modules were presented in the network during the feeding time of 15 days than at 30 days in the fish gut. This finding indicated that feeding time significantly influenced the interaction of microbes in the fish gut. More precisely, the microbes in the fish gut during the feeding time of 15 days interact more closely than those during the feeding time of 30 days. This finding could be explained by the fact that the digestive function of the fish gut during the growth time of 30 days was more developed than that of 15 days. Therefore, the digestion of food in the GI tract of fish during a feeding time of 15 days requires more cooperation of microbes, which collaborate and form a closer interaction network.
5. Conclusions
The impact of C. vulgaris on the gut microbiota of juvenile Nile tilapia is feeding time dependent. Feeding for 30 days, but not 15 days, with 2% C. vulgaris significantly elevated the alpha diversity of the gut microbiota, and treatment with 0.5% and 2% C. vulgaris significantly affected the beta diversity of the gut microbiota. During the 15-day feeding trial, Paracoccus, Thiobacillus, Dechloromonas, and Desulfococcus were enriched in the 2% C. vulgaris group. During the 30-day feeding trial, Afipia, Ochrobactrum, Polymorphum, Albidovulum, Pseudacidovorax, and Thiolamprovum were more abundant in 2% C. vulgaris-treated fish. The gut microbes during the feeding time of 15 days interacted more closely than those during the feeding time of 30 days. C. vulgaris promotes the interaction of gut microbiota in juvenile Nile tilapia by increasing the abundance of Reyranella and further promotes the interaction of Rhodobacter, Rhizobiales, Hyphomicrobium, and Sphingomonas. This work will be valuable for understanding how C. vulgaris in diets impacts the gut microbiota of fish.
Author Contributions
Z.H.: Carried out the experiments, writing-original draft preparation; J.G. and C.P.: data curation, carried out the experiments; J.S., Z.X., J.J., H.L. and S.Z.: Data analysis; B.G. and Y.L.: conceptualization, methodology, writing-original draft preparation, data analysis, Supervision. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the National Natural Science Foundation of China (31560727), Guangxi Natural Science Foundation (2020GXNSFDA238015), Marine Science First-Class Subject, Beibu Gulf University (DRA001, TRA003, TTB002, DTC002), and the funds of The Guangxi Key Laboratory of Beibu Gulf Marine Biodiversity Conservation (2021ZA01, 2019ZC01).
Institutional Review Board Statement
The animal study protocol was approved by the Institutional Review Board (or Ethics Committee) of Beibu Gulf University (protocol code 597/04, 9 March 2021).
Data Availability Statement
The high-throughput sequencing data from this research has been uploaded to GenBank with the BioProject accession number PRJNA924963, and the data will be released in 24 November 2024.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ahmad, M.T.; Shariff, M.; Yusoff, F.M.; Goh, Y.M.; Banerjee, S. Applications of microalga Chlorella vulgaris in aquaculture. Rev. Aquac. 2020, 12, 328–346. [Google Scholar] [CrossRef]
- Maliwat, G.C.; Velasquez, S.; Robil, J.L.; Chan, M.; Traifalgar, R.F.; Tayamen, M.; Ragaza, J.A. Growth and immune response of giant freshwater prawn Macrobrachium rosenbergii (De Man) postlarvae fed diets containing Chlorella vulgaris (Beijerinck). Aquac. Res. 2017, 48, 1666–1676. [Google Scholar] [CrossRef]
- Mahmoud, E.A.; El-Sayed, B.M.; Mahsoub, Y.H.; Neamat-Allah, A.N. Effect of Chlorella vulgaris enriched diet on growth performance, hemato-immunological responses, antioxidant and transcriptomics profile disorders caused by deltamethrin toxicity in Nile tilapia (Oreochromis niloticus). Fish Shellfish Immun. 2020, 102, 422–429. [Google Scholar] [CrossRef] [PubMed]
- Galal, A.A.; Reda, R.M.; Mohamed, A.A.R. Influences of Chlorella vulgaris dietary supplementation on growth performance, hematology, immune response and disease resistance in Oreochromis niloticus exposed to sub-lethal concentrations of penoxsulam herbicide. Fish Shellfish Immun. 2018, 77, 445–456. [Google Scholar] [CrossRef]
- Gouveia, L.; Gomes, E.; Empis, J. Use of Chlorella vulgaris in rainbow trout, Oncorhynchus mykiss, diets to enhance muscle pigmentation. J. Appl. Aquac. 1997, 7, 61–70. [Google Scholar] [CrossRef]
- Enyidi, U.D. Chlorella vulgaris as protein source in the diets of African catfish Clarias gariepinus. Fishes 2017, 2, 17. [Google Scholar] [CrossRef]
- Pakravan, S.; Akbarzadeh, A.; Sajjadi, M.M.; Hajimoradloo, A.; Noori, F. Chlorella vulgaris meal improved growth performance, digestive enzyme activities, fatty acid composition and tolerance of hypoxia and ammonia stress in juvenile Pacific white shrimp Litopenaeus vannamei. Aquac. Nutr. 2018, 24, 594–604. [Google Scholar] [CrossRef]
- Radhakrishnan, S.; Saravana, B.P.; Seenivasan, C.; Muralisankar, T. Effect of dietary replacement of fishmeal with Chlorella vulgaris on growth performance, energy utilization and digestive enzymes in Macrobrachium rosenbergii postlarvae. Int. J. Fish. Aquac. 2015, 7, 62–70. [Google Scholar]
- Karapanagiotidis, I.T.; Metsoviti, M.N.; Gkalogianni, E.Z.; Psofakis, P.; Asimaki, A.; Katsoulas, N.; Zarkadas, I. The effects of replacing fishmeal by Chlorella vulgaris and fish oil by Schizochytrium sp. and Microchloropsis gaditana blend on growth performance, feed efficiency, muscle fatty acid composition and liver histology of gilthead seabream (Sparus aurata). Aquaculture 2022, 561, 738709. [Google Scholar] [CrossRef]
- Abdel-Tawwab, M.; Mousa, M.A.; Mamoon, A.; Abdelghany, M.F.; Abdel-Hamid, E.A.; Abdel-Razek, N.; Gewida, A.G. Dietary Chlorella vulgaris modulates the performance, antioxidant capacity, innate immunity, and disease resistance capability of Nile tilapia fingerlings fed on plant-based diets. Anim. Feed. Sci. Technol. 2022, 283, 115181. [Google Scholar] [CrossRef]
- Glencross, B.D.; Huyben, D.; Schrama, J.W. The application of single-cell ingredients in aquaculture feeds-a review. Fishes 2020, 5, 22. [Google Scholar] [CrossRef]
- Butt, R.L.; Volkoff, H. Gut microbiota and energy homeostasis in fish. Front. Endocrinol. 2019, 10, 9. [Google Scholar] [CrossRef]
- Banerjee, G.; Ray, A.K. Bacterial symbiosis in the fish gut and its role in health and metabolism. Symbiosis 2017, 72, 1–11. [Google Scholar] [CrossRef]
- Cui, K.; Fu, Z.; Cheng, D.; Yang, Q.; Ma, Z.; Qin, J.G.; Hu, J. Development of immune functionality in larval and juvenile crimson snapper Lutjanus erythropterus (Bloch 1790). Aquac. Rep. 2018, 10, 1–7. [Google Scholar] [CrossRef]
- Vargas-Albores, F.; Martínez-Córdova, L.R.; Hernández-Mendoza, A.; Cicala, F.; Lago-Lestón, A.; Martínez-Porchas, M. Therapeutic modulation of fish gut microbiota, a feasible strategy for aquaculture? Aquaculture 2021, 544, 737050. [Google Scholar] [CrossRef]
- Peredo-Lovillo, A.; Romero-Luna, H.E.; Jiménez-Fernández, M. Health promoting microbial metabolites produced by gut microbiota after prebiotics metabolism. Food Res. Int. 2020, 136, 109473. [Google Scholar] [CrossRef]
- Fan, Y.; Pedersen, O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 2021, 19, 55–71. [Google Scholar] [CrossRef]
- Li, W.; Zhou, Z.; Li, H.; Wang, S.; Ren, L.; Hu, J.; Liu, S. Successional Changes of Microbial Communities and Host-Microbiota Interactions Contribute to Dietary Adaptation in Allodiploid Hybrid Fish. Microb. Ecol. 2022, 1–12. [Google Scholar] [CrossRef]
- Safari, O.; Paolucci, M.; Motlagh, H.A. Dietary supplementation of Chlorella vulgaris improved growth performance, immunity, intestinal microbiota and stress resistance of juvenile narrow clawed crayfish, Pontastacus leptodactylus Eschscholtz, 1823. Aquaculture 2022, 554, 738138. [Google Scholar] [CrossRef]
- De Souza, F.P.; de Lima, E.C.S.; Urrea-Rojas, A.M.; Suphoronski, S.A.; Facimoto, C.T.; Bezerra Júnior, J.D.S.; Lopera-Barrero, N.M. Effects of dietary supplementation with a microalga (Schizochytrium sp.) on the hemato-immunological, and intestinal histological parameters and gut microbiota of Nile tilapia in net cages. PLoS ONE 2020, 15, e0226977. [Google Scholar] [CrossRef]
- Gong, B.; Cao, H.; Peng, C.; Perčulija, V.; Tong, G.; Fang, H.; Ouyang, S. High-throughput sequencing and analysis of microbial communities in the mangrove swamps along the coast of Beibu Gulf in Guangxi, China. Sci. Rep. 2019, 9, 9377. [Google Scholar] [CrossRef]
- Holm, J.B.; Humphrys, M.S.; Robinson, C.K.; Settles, M.L.; Ott, S.; Fu, L.; Ravel, J. Ultrahigh-throughput multiplexing and sequencing of >500-base-pair amplicon regions on the Illumina HiSeq 2500 platform. MSystems 2019, 4, e00029-19. [Google Scholar] [CrossRef] [PubMed]
- Bacci, G.; Bazzicalupo, M.; Benedetti, A.; Mengoni, A. StreamingTrim 1.0: A Java software for dynamic trimming of 16S rRNA sequence data from metagenetic studies. Mol. Ecol. Resour. 2014, 14, 426–434. [Google Scholar] [CrossRef] [PubMed]
- Parikh, H.I.; Koparde, V.N.; Bradley, S.P.; Buck, G.A.; Sheth, N.U. MeFiT: Merging and filtering tool for illumina paired-end reads for 16S rRNA amplicon sequencing. BMC Bioinform. 2016, 17, 491. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Env. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef]
- García-López, R.; Cornejo-Granados, F.; Lopez-Zavala, A.A.; Cota-Huízar, A.; Sotelo-Mundo, R.R.; Gómez-Gil, B.; Ochoa-Leyva, A. OTUs and ASVs produce comparable taxonomic and diversity from shrimp microbiota 16S profiles using tailored abundance filters. Genes 2021, 124, 564. [Google Scholar] [CrossRef]
- Edgar, R.C. SINTAX: A simple non-Bayesian taxonomy classifier for 16S and ITS sequences. bioRxiv 2016. [Google Scholar] [CrossRef]
- Glöckner, F.O. The SILVA database project: An ELIXIR core data resource for high-quality ribosomal RNA sequences. Biodivers. Inf. Sci. Stand. 2019, 3, e36125. [Google Scholar] [CrossRef]
- McMURDIE, P.J.; Holmes, S. Phyloseq: A bioconductor package for handling and analysis of high-throughput phylogenetic sequence data. Biocomputing 2012, 235–246. [Google Scholar] [CrossRef]
- Ricotta, C.; Podani, J. On some properties of the Bray-Curtis dissimilarity and their ecological meaning. Ecol. Complex 2017, 31, 201–205. [Google Scholar] [CrossRef]
- Mercier, L.; Estrada, R.V.; Hernández, L. Beta-diversity of macroinvertebrates associated to Pocillopora corals along the Mexican Pacific coast. Reg. Stud. Mar. Sci. 2022, 53, 102387. [Google Scholar] [CrossRef]
- Chang, F.; He, S.; Dang, C. Assisted Selection of Biomarkers by Linear Discriminant Analysis Effect Size (LEfSe) in Microbiome Data. JOVE J. Vis. Exp. 2022, 183, e61715. [Google Scholar] [CrossRef]
- Ren, Y.; Yu, G.; Shi, C.; Liu, L.; Guo, Q.; Han, C.; Huang, H. Majorbio Cloud: A one-stop, comprehensive bioinformatic platform for multiomics analyses. iMeta 2022, 1, e12. [Google Scholar] [CrossRef]
- Field, A. Discovering Statistics Using IBM SPSS Statistics; Sage: Washington, DC, USA, 2013. [Google Scholar]
- Ghanbari, M.; Kneifel, W.; Domig, K.J. A new view of the fish gut microbiome: Advances from next-generation sequencing. Aquaculture 2015, 448, 464–475. [Google Scholar] [CrossRef]
- Romero, J.; Ringø, E.; Merrifield, D.L. The gut microbiota of fish. In Aquaculture Nutrition: Gut Health, Probiotics and Prebiotics; Wiley: Hoboken, NJ, USA, 2014; pp. 75–100. [Google Scholar]
- Pan, B.; Han, X.; Yu, K.; Sun, H.; Mu, R.; Lian, C.A. Geographical distance, host evolutionary history, and diet drive gut microbiome diversity of fish across the Yellow River. Mol. Ecol. 2022, 32, 1183–1196. [Google Scholar] [CrossRef] [PubMed]
- Egerton, S.; Culloty, S.; Whooley, J.; Stanton, C.; Ross, R.P. The gut microbiota of marine fish. Front. Microbiol. 2018, 9, 873. [Google Scholar] [CrossRef] [PubMed]
- Bruce, T.J.; Neiger, R.D.; Brown, M.L. Gut histology, immunology and the intestinal microbiota of rainbow trout, Oncorhynchus mykiss (Walbaum), fed process variants of soybean meal. Aquac. Res. 2018, 49, 492–504. [Google Scholar] [CrossRef]
- Yang, P.; Hu, H.; Liu, Y.; Li, Y.; Ai, Q.; Xu, W.; Mai, K. Dietary stachyose altered the intestinal microbiota profile and improved the intestinal mucosal barrier function of juvenile turbot, Scophthalmus maximus L. Aquaculture 2018, 486, 98–106. [Google Scholar] [CrossRef]
- Huyben, D.; Roehe, B.K.; Bekaert, M.; Ruyter, B.; Glencross, B. Dietary lipid: Protein ratio and n-3 long-chain polyunsaturated fatty acids alters the gut microbiome of Atlantic salmon under hypoxic and normoxic conditions. Front. Microbiol. 2020, 11, 589898. [Google Scholar] [CrossRef]
- Smith, C.C.; Snowberg, L.K.; Gregory Caporaso, J.; Knight, R.; Bolnick, D.I. Dietary input of microbes and host genetic variation shape among-population differences in stickleback gut microbiota. ISME J. 2015, 9, 2515–2526. [Google Scholar] [CrossRef]
- Long, S.; You, Y.; Dong, X.; Tan, B.; Zhang, S.; Chi, S.; Zhang, H. Effect of dietary oxidized fish oil on growth performance, physiological homeostasis and intestinal microbiome in hybrid grouper (♀Epi-nephelus fuscoguttatus×♂ Epinephelus lanceolatus). Aquac. Rep. 2022, 24, 101130. [Google Scholar] [CrossRef]
- Gaudioso, G.; Marzorati, G.; Faccenda, F.; Weil, T.; Lunelli, F.; Cardinaletti, G.; Fava, F. Processed animal proteins from insect and poultry by-products in a fish meal-free diet for rainbow trout: Impact on intestinal microbiota and inflammatory markers. Int. J. Mol. Sci. 2021, 22, 5454. [Google Scholar] [CrossRef] [PubMed]
- Kolkovski, S. Digestive enzymes in fish larvae and juveniles-implications and applications to formulated diets. Aquaculture 2001, 200, 181–201. [Google Scholar] [CrossRef]
- Miyake, S.; Ngugi, D.K.; Stingl, U. Diet strongly influences the gut microbiota of surgeonfishes. Mol. Ecol. 2015, 24, 656–672. [Google Scholar] [CrossRef] [PubMed]
- Baldo, L.; Riera, J.L.; Tooming-Klunderud, A.; Albà, M.M.; Salzburger, W. Gut microbiota dynamics during dietary shift in eastern African cichlid fishes. PLoS ONE 2015, 10, e0127462. [Google Scholar] [CrossRef]
- Ringø, E.; Zhou, Z.; Vecino, J.G.; Wadsworth, S.; Romero, J.; Krogdahl, Å.; Merrifield, D.L. Effect of dietary components on the gut microbiota of aquatic animals. A never-ending story? Aquac. Nutr. 2016, 22, 219–282. [Google Scholar] [CrossRef]
- Fontinha, F.; Magalhães, R.; Moutinho, S.; Santos, R.; Campos, P.; Serra, C.R.; Aires, T.; Oliva-Teles, A.; Peres, H. Effect of dietary poultry meal and oil on growth, digestive capacity, and gut microbiota of gilthead seabream (Sparus aurata) juveniles. Aquaculture 2021, 530, 735879. [Google Scholar] [CrossRef]
- Li, L.; Liu, X.; Wang, Y.; Huang, Y.; Wang, C. Effects of alternate feeding between fish meal and novel protein diets on the intestinal health of juvenile largemouth bass (Micropterus salmoides). Aquac. Rep. 2022, 23, 101023. [Google Scholar] [CrossRef]
- Maliwat, G.C.F.; Velasquez, S.F.; Buluran, S.M.D.; Tayamen, M.M.; Ragaza, J.A. Growth and immune response of pond-reared giant freshwater prawn Macrobrachium rosenbergii post larvae fed diets containing Chlorella vulgaris. Aquac. Fish 2021, 6, 465–470. [Google Scholar] [CrossRef]
- Chen, W.; Gao, S.; Huang, Y.; Chang, K.; Zhao, X. Addition of Chlorella sorokiniana meal in the diet of juvenile rainbow trout (Oncorhynchus mykiss): Influence on fish growth, gut histology, oxidative stress, immune response, and disease resistance against Aeromonas salmonicida. Fish Shellfish Immun. 2022, 129, 243–250. [Google Scholar] [CrossRef]
- Kandathil Radhakrishnan, D.; Velayudhannair, K.; Schmidt, B.V. Effects of bio-flocculated algae on the growth, digestive enzyme activity and microflora of freshwater fish Catla catla (Hamilton 1922). Aquac. Res. 2020, 51, 4533–4540. [Google Scholar] [CrossRef]
- Xi, L.; Lu, Q.; Liu, Y.; Su, J.; Chen, W.; Gong, Y.; Xie, S. Effects of fish meal replacement with Chlorella meal on growth performance, pigmentation, and liver health of largemouth bass (Micropterus salmoides). Anim. Nutr. 2022, 10, 26–40. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Xi, L.; Liu, H.; Jin, J.; Yang, Y.; Zhu, X.; Xie, S. High replacement of fishmeal by Chlorella meal affects intestinal microbiota and the potential metabolic function in largemouth bass (Micropterus salmoides). Front. Microbiol. 2022, 13, 3772. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.B.; Cha, J.W.; Shin, I.S.; Jeon, J.Y.; An, H.S.; Cha, K.H.; Pan, C.H. Effect of Chlorella vulgaris on gut microbiota through a simulated in vitro digestion process. J. Appl. Biol. Chem. 2021, 64, 49–55. [Google Scholar] [CrossRef]
- Infante, J.Z.; Cahu, C.L. Ontogeny of the gastrointestinal tract of marine fish larvae. Comp. Biochem. Phys. C 2001, 130, 477–487. [Google Scholar]
- Niu, Q.; Li, P.; Hao, S.; Zhang, Y.; Kim, S.W.; Li, H.; Huang, R. Dynamic distribution of the gut microbiota and the relationship with apparent crude fiber digestibility and growth stages in pigs. Sci. Rep. 2015, 5, 9938. [Google Scholar] [CrossRef]
- Davis, C.D. The gut microbiome and its role in obesity. Nutr. Today 2016, 51, 167. [Google Scholar] [CrossRef]
- Doré, J.; Blottière, H. The influence of diet on the gut microbiota and its consequences for health. Curr. Opin. Biotechnol. 2015, 32, 195–199. [Google Scholar] [CrossRef]
- Chung, W.S.F.; Walker, A.W.; Louis, P.; Parkhill, J.; Vermeiren, J.; Bosscher, D.; Flint, H.J. Modulation of the human gut microbiota by dietary fibres occurs at the species level. BMC Biol. 2016, 14, 3. [Google Scholar] [CrossRef]
- Tzeng, T.D.; Pao, Y.Y.; Chen, P.C.; Weng, F.C.H.; Jean, W.D.; Wang, D. Effects of host phylogeny and habitats on gut microbiomes of oriental river prawn (Macrobrachium nipponense). PLoS ONE 2015, 10, e0132860. [Google Scholar] [CrossRef]
- Deng, Y.; Xu, X.; Yin, X.; Lu, H.; Chen, G.; Yu, J.; Ruan, Y. Effect of stock density on the microbial community in biofloc water and Pacific white shrimp (Litopenaeus vannamei) gut microbiota. Appl. Microbiol. Biotechnol. 2019, 103, 4241–4252. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Tian, X.; Dong, S.; Peng, M.; Wang, D. Effects of dietary Bacillus cereus G19, B. cereus BC-01, and Paracoccus marcusii DB11 supplementation on the growth, immune response, and expression of immune-related genes in coelomocytes and intestine of the sea cucumber (Apostichopus japonicus Selenka). Fish Shellfish Immun. 2015, 45, 800–807. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Mu, G.; Lu, S.; Wang, D.; Jiang, H.; Sun, X.; Liu, Y. Effects of traditional Chinese medicines on immunity and culturable gut microflora to Oncorhynchus masou. Fish Shellfish Immun. 2019, 93, 322–327. [Google Scholar]
- Wu, Z.B.; Gatesoupe, F.J.; Li, T.T.; Wang, X.H.; Zhang, Q.Q.; Feng, D.Y.; Li, A.H. Significant improvement of intestinal microbiota of gibel carp (Carassius auratus gibelio) after traditional Chinese medicine feeding. J. Appl. Microbiol. 2018, 124, 829–841. [Google Scholar] [CrossRef] [PubMed]
- Sha, Y.; Liu, M.; Wang, B.; Jiang, K.; Qi, C.; Wang, L. Bacterial population in intestines of Litopenaeus vannamei fed different probiotics or probiotic supernatant S. J. Microbiol. Biotechnol. 2016, 26, 1736–1745. [Google Scholar] [CrossRef] [PubMed]
- Raji, A.A.; Jimoh, W.A.; Bakar, N.H.; Taufek, N.H.; Muin, H.; Alias, Z.; Razak, S.A. Dietary use of Spirulina (Arthrospira) and Chlorella instead of fish meal on growth and digestibility of nutrients, amino acids and fatty acids by African catfish. J. Appl. Phys. 2020, 32, 1763–1770. [Google Scholar] [CrossRef]
- Mondal, H.K.; Maji, U.J.; Mohanty, S.; Sahoo, P.K.; Maiti, N.K. Alteration of gut microbiota composition and function of Indian major carp, rohu (Labeo rohita) infected with Argulus siamensis. Microb. Pathog. 2022, 164, 105420. [Google Scholar] [CrossRef]
- Cao, H.; Zhang, S.; An, J.; Diao, J.; Xu, L.; Gai, C. Rhodobacter azotoformans supplementation improves defense ability of Chinese mitten crab Eriocheir sinensis against citrobacteriosis. Fish Shellfish Immun. 2022, 131, 991–998. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Q.; Lian, S.; Yue, Y.; Gao, Q.; Peng, S. Dietary jellyfish affect digestive enzyme activities and gut microbiota of Pampus argenteus. Comp. Biochem. Phys. D 2021, 40, 100923. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).