HIV Tat Expression and Cocaine Exposure Lead to Sex- and Age-Specific Changes of the Microbiota Composition in the Gut
Abstract
1. Introduction
2. Materials and Methods
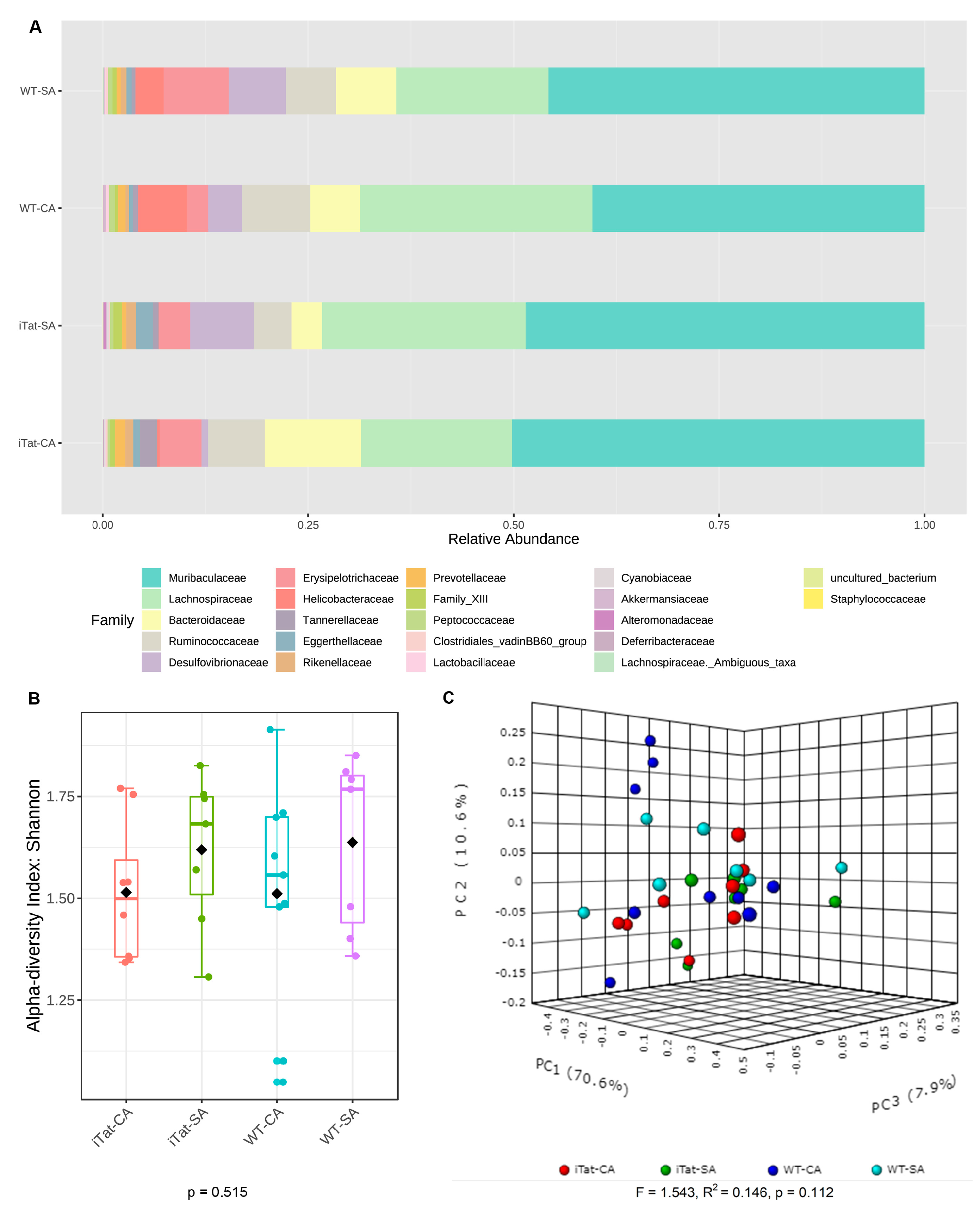
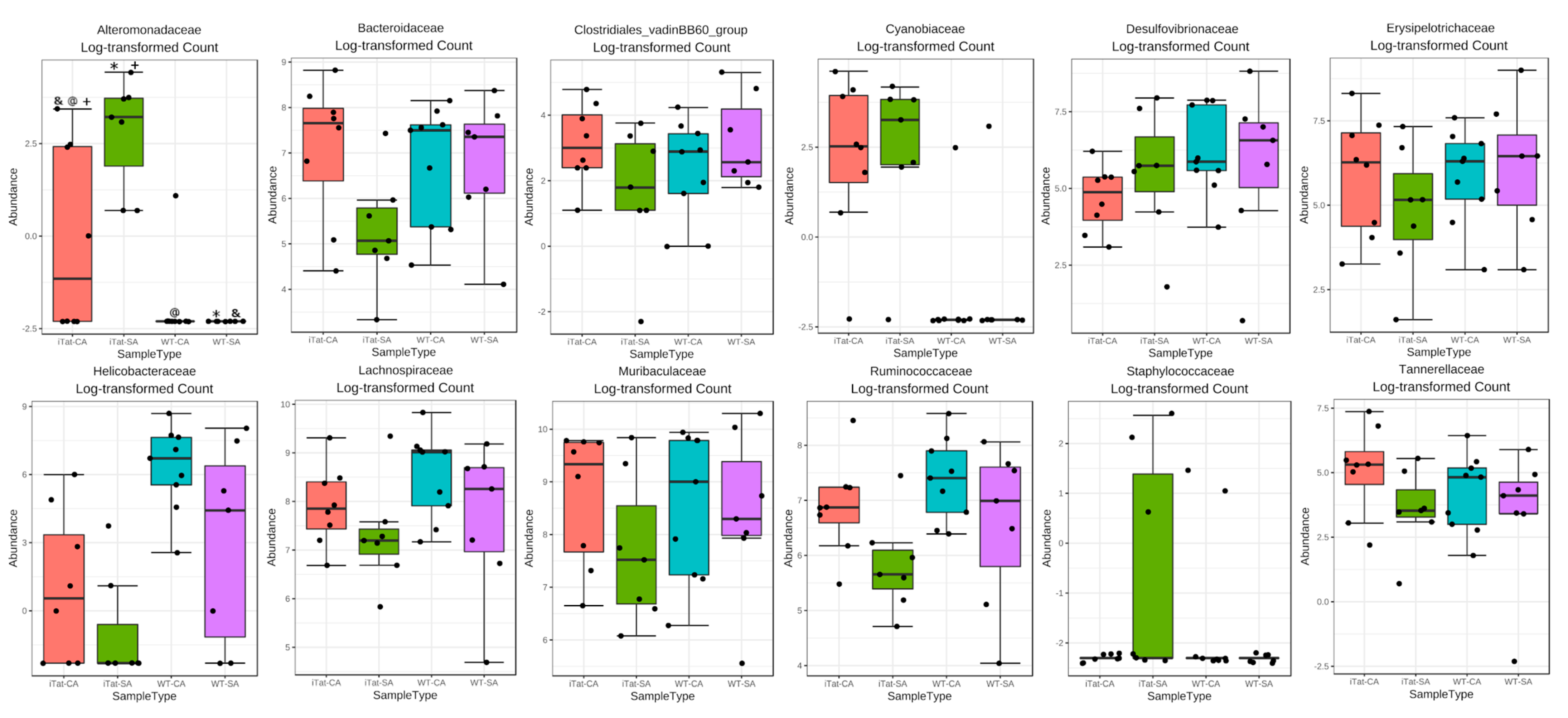
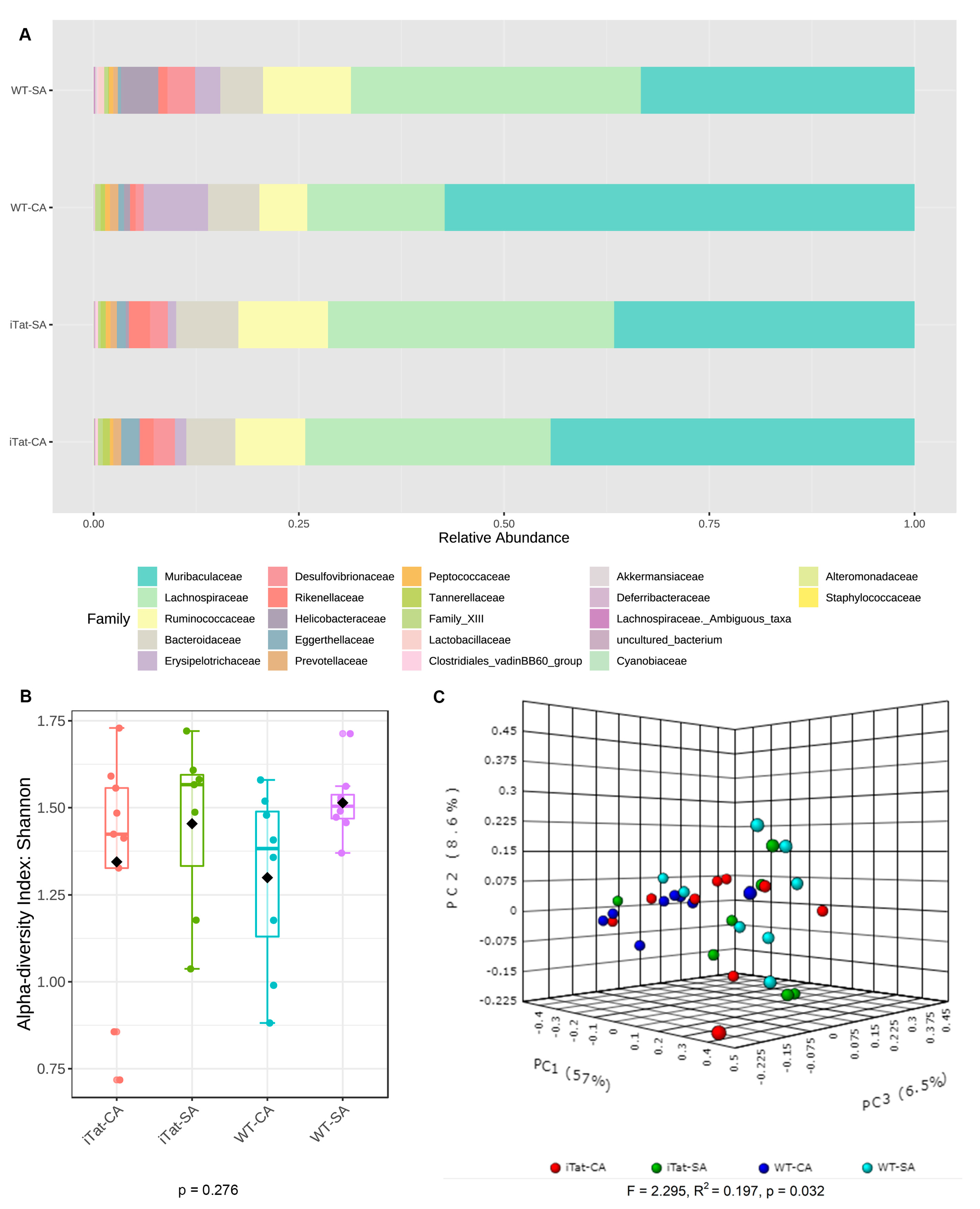
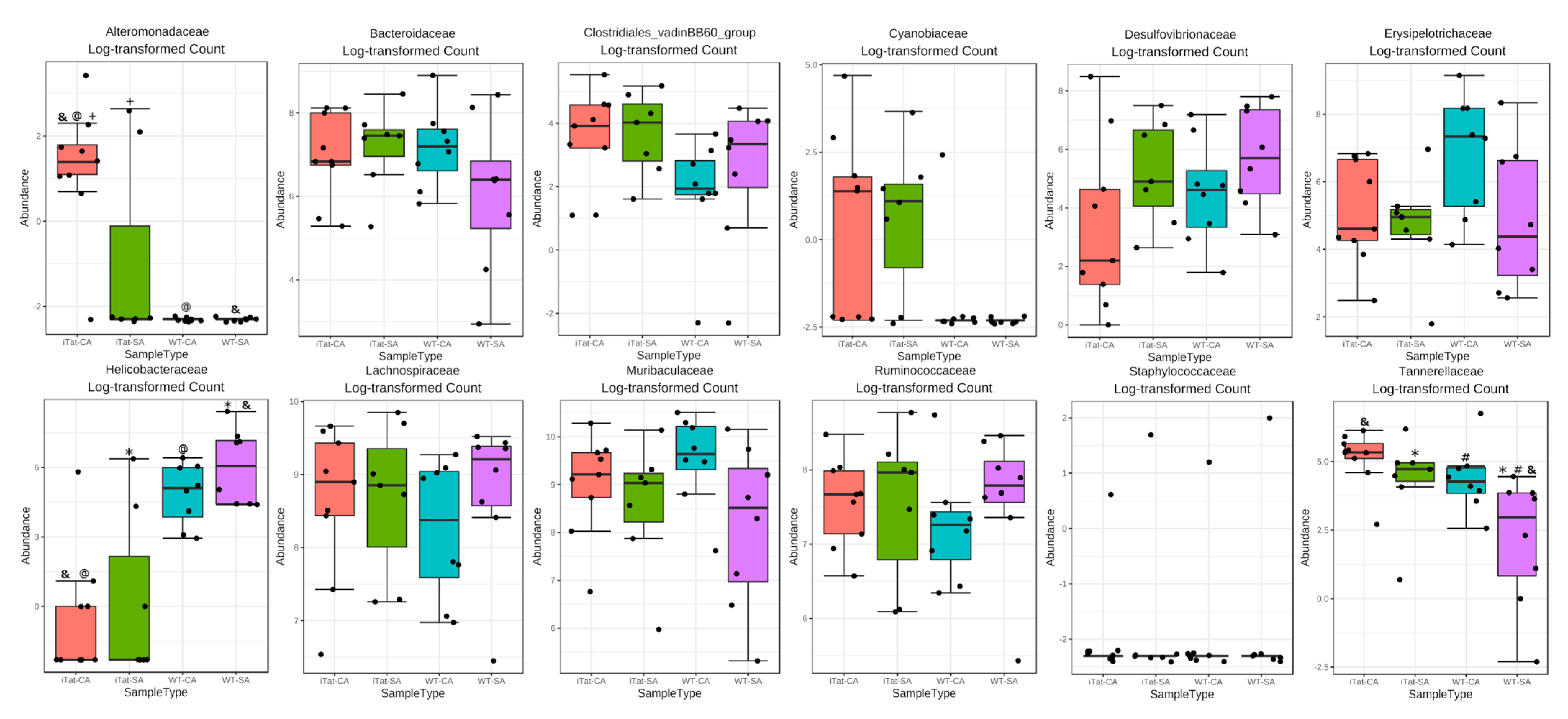
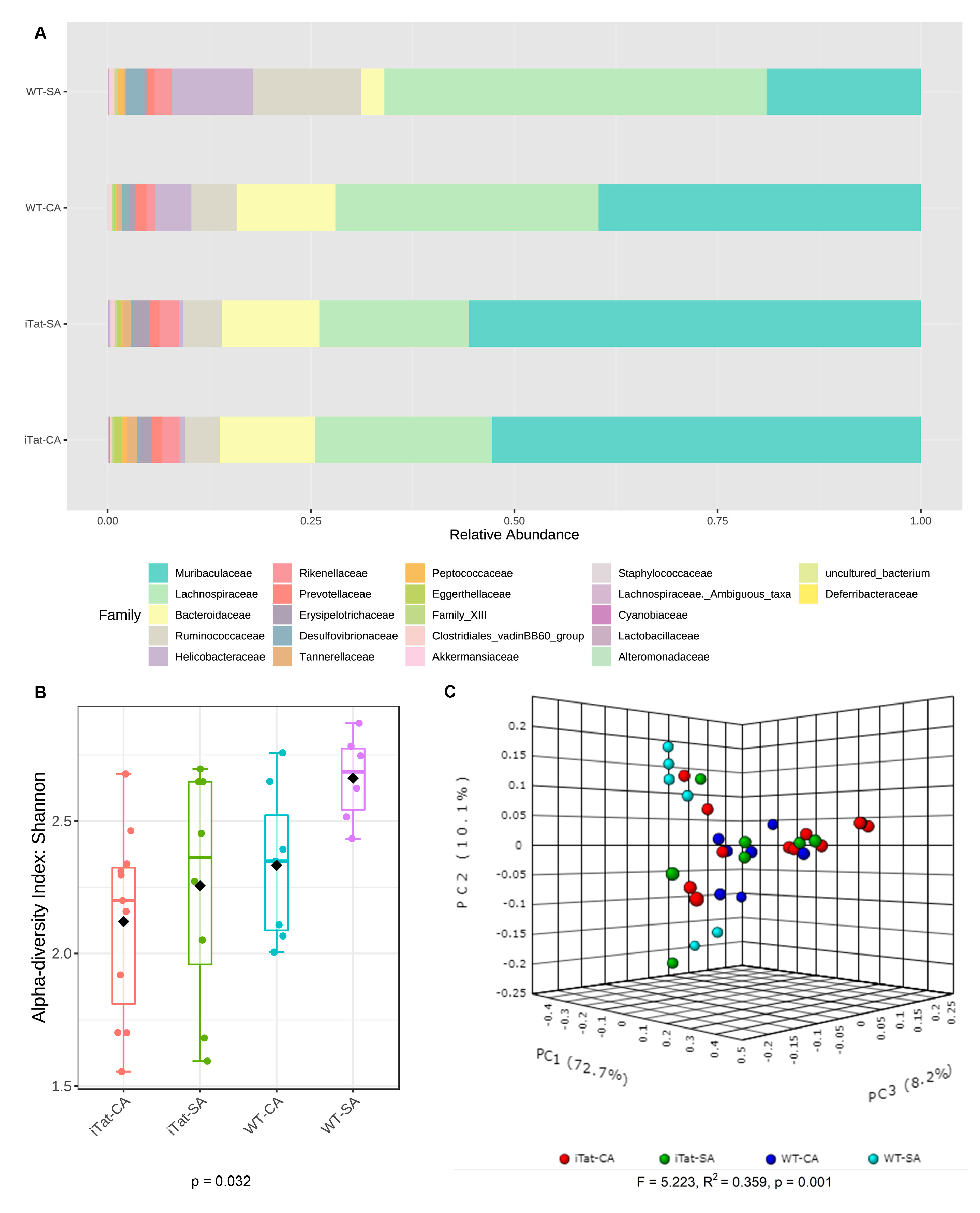
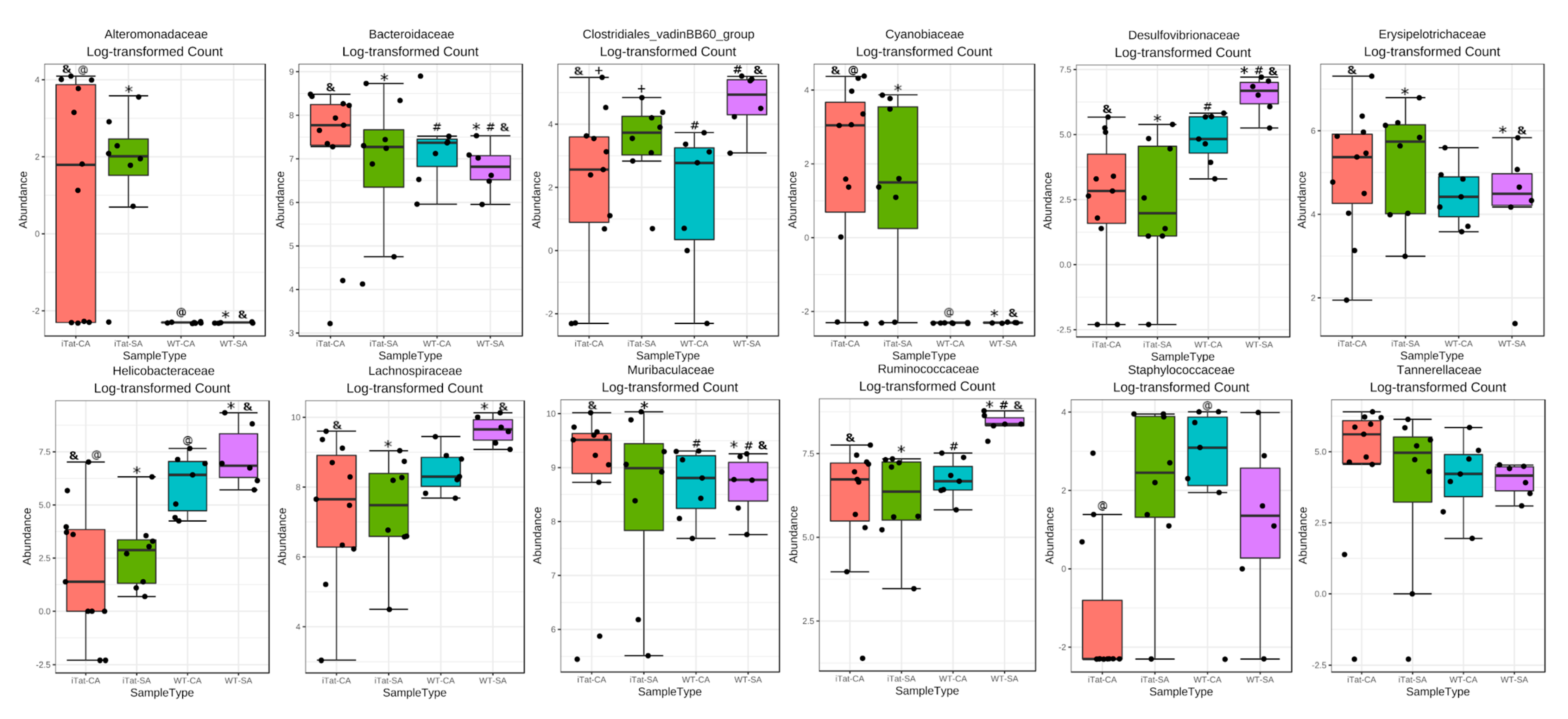
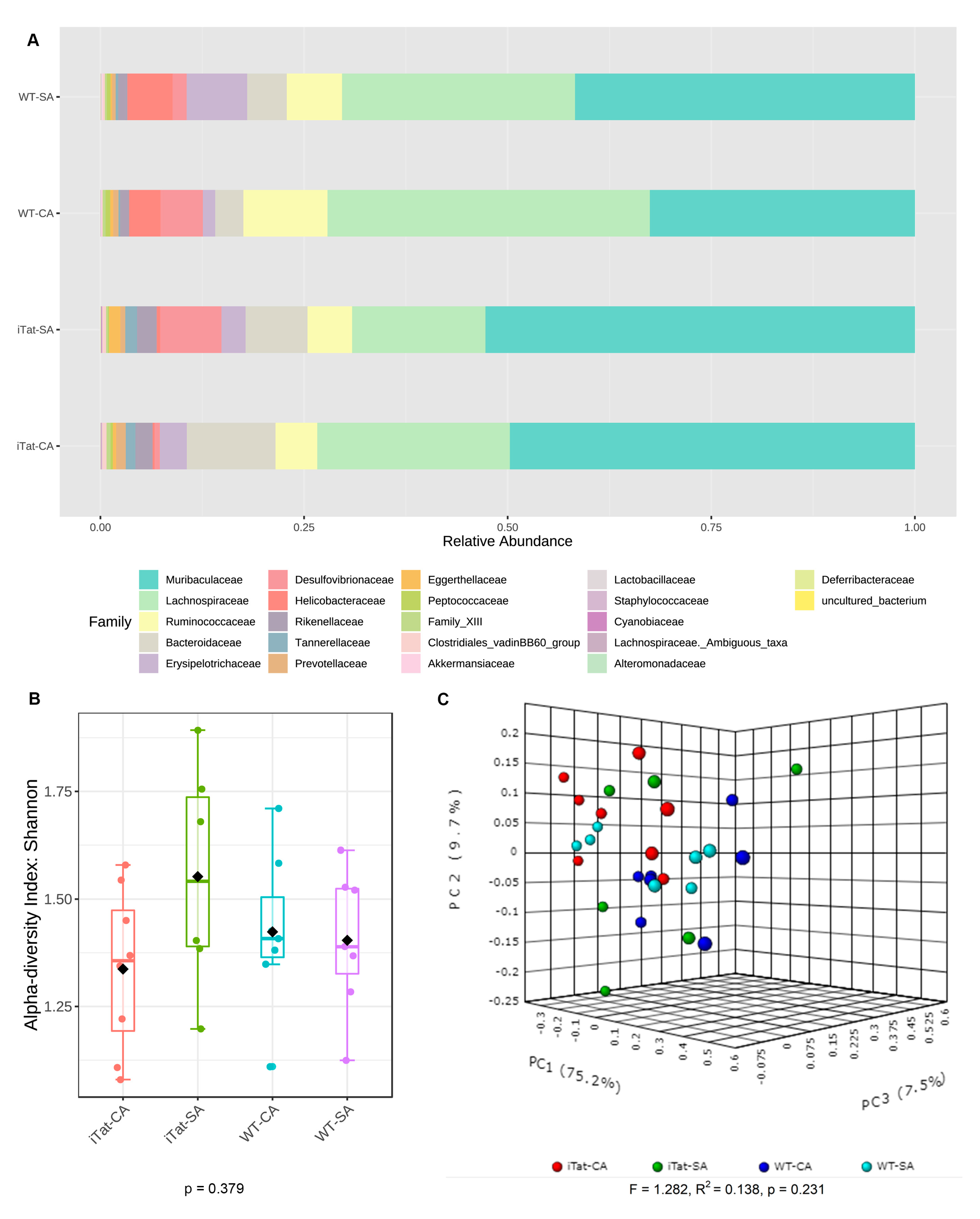
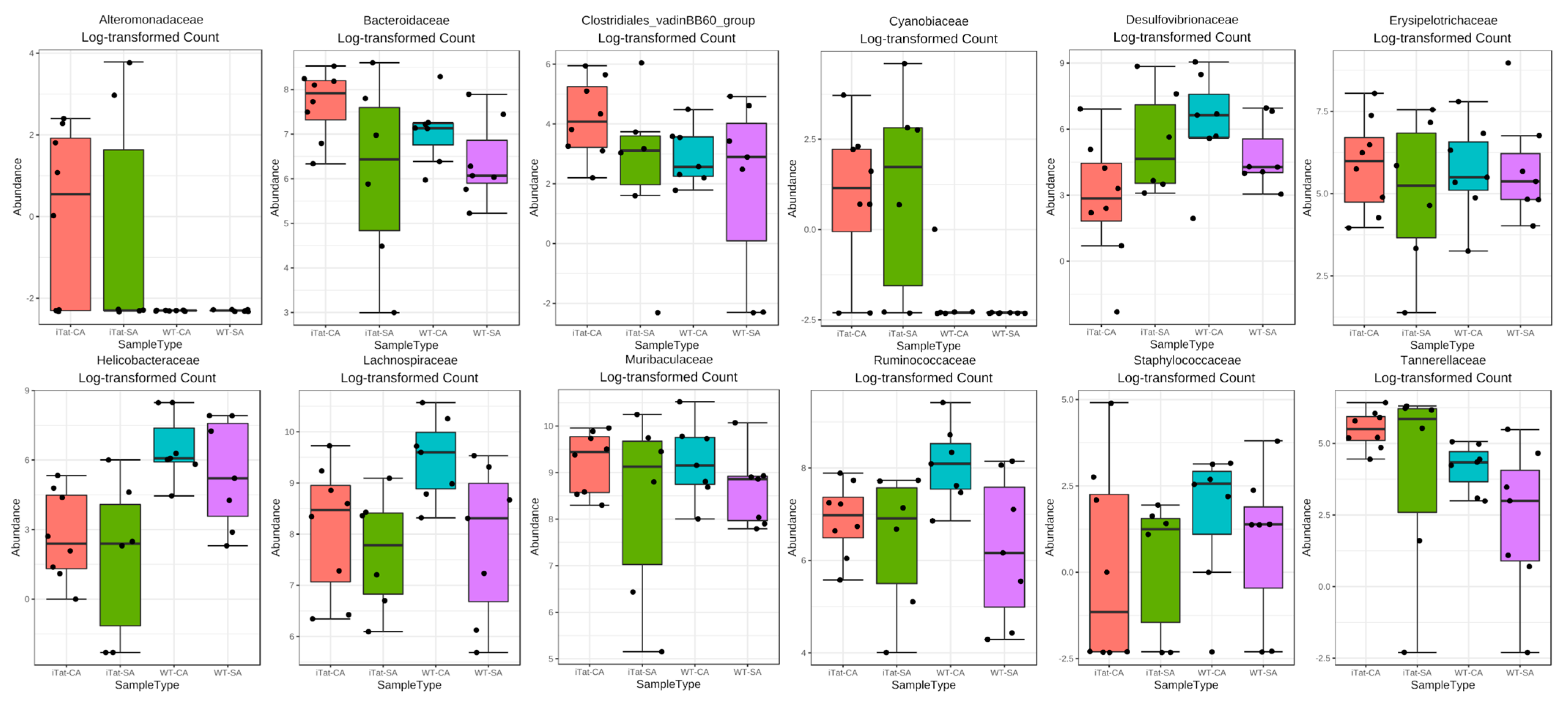
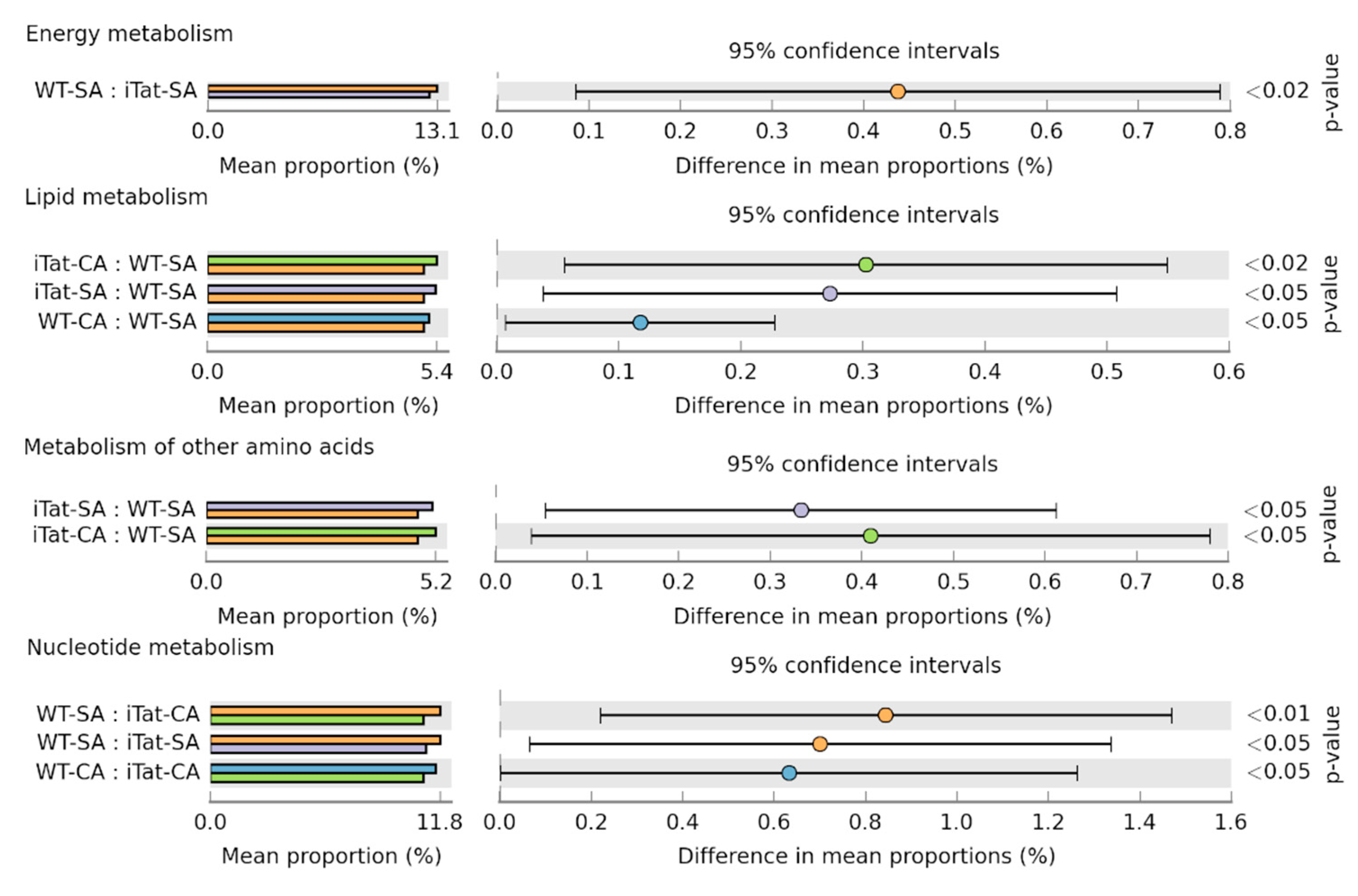
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bruzzese, E.; Volpicelli, M.; Squaglia, M.; Tartaglione, A.; Guarino, A. Impact of prebiotics on human health. Dig. Liver Dis. 2006, 38 (Suppl. S2), S283–S287. [Google Scholar] [CrossRef] [PubMed]
- Boulangé, C.L.; Neves, A.L.; Chilloux, J.; Nicholson, J.K.; Dumas, M.E. Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Med. 2016, 8, 42. [Google Scholar] [CrossRef] [PubMed]
- Hugon, P.; Dufour, J.C.; Colson, P.; Fournier, P.E.; Sallah, K.; Raoult, D. A comprehensive repertoire of prokaryotic species identified in human beings. Lancet Infect. Dis. 2015, 15, 1211–1219. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Jia, H.; Cai, X.; Zhong, H.; Feng, Q.; Sunagawa, S.; Arumugam, M.; Kultima, J.R.; Prifti, E.; Nielsen, T.; et al. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 2014, 32, 834–841. [Google Scholar] [CrossRef]
- Rodríguez, J.M.; Murphy, K.; Stanton, C.; Ross, R.P.; Kober, O.I.; Juge, N.; Avershina, E.; Rudi, K.; Narbad, A.; Jenmalm, M.C.; et al. The composition of the gut microbiota throughout life, with an emphasis on early life. Microb. Ecol. Health Dis. 2015, 26, 26050. [Google Scholar] [CrossRef]
- Bäckhed, F. Programming of host metabolism by the gut microbiota. Ann. Nutr. Metab. 2011, 58 (Suppl. S2), 44–52. [Google Scholar] [CrossRef]
- Palmer, C.; Bik, E.M.; DiGiulio, D.B.; Relman, D.A.; Brown, P.O. Development of the human infant intestinal microbiota. PLoS Biol. 2007, 5, e177. [Google Scholar] [CrossRef]
- Claesson, M.J.; Cusack, S.; O’Sullivan, O.; Greene-Diniz, R.; de Weerd, H.; Flannery, E.; Marchesi, J.R.; Falush, D.; Dinan, T.; Fitzgerald, G. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. S1), 4586–4591. [Google Scholar] [CrossRef]
- Biagi, E.; Nylund, L.; Candela, M.; Ostan, R.; Bucci, L.; Pini, E.; Nikkïla, J.; Monti, D.; Satokari, R.; Franceschi, C.; et al. Through ageing, and beyond: Gut microbiota and inflammatory status in seniors and centenarians. PLoS ONE 2010, 5, e10667. [Google Scholar] [CrossRef]
- Ley, R.E.; Peterson, D.A.; Gordon, J.I. Ecological and Evolutionary Forces Shaping Microbial Diversity in the Human Intestine. Cell 2006, 124, 837–848. [Google Scholar] [CrossRef]
- Claesson, M.J.; Jeffery, I.B.; Conde, S.; Power, S.E.; O’Connor, E.M.; Cusack, S.; Harris, H.M.; Coakley, M.; Lakshminarayanan, B.; O’Sullivan, O.; et al. Gut microbiota composition correlates with diet and health in the elderly. Nature 2012, 488, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Valdes, A.M.; Walter, J.; Segal, E.; Spector, T.D. Role of the gut microbiota in nutrition and health. BMJ 2018, 361, k2179. [Google Scholar] [CrossRef] [PubMed]
- Kho, Z.Y.; Lal, S.K. The Human Gut Microbiome—A Potential Controller of Wellness and Disease. Front. Microbiol. 2018, 9, 1835. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.A.; Fitzstevens, J.L.; Schmidt, V.T.; Enav, H.; Huus, K.E.; Mbong Ngwese, M.; Grieβhammer, A.; Pfleiderer, A.; Adegbite, B.R.; Zinsou, J.F.; et al. Codiversification of gut microbiota with humans. Science 2022, 377, 1328–1332. [Google Scholar] [CrossRef] [PubMed]
- Garrett, W.S. Cancer and the microbiota. Science 2015, 348, 80–86. [Google Scholar] [CrossRef]
- Dominguez-Diaz, C.; Garcia-Orozco, A.; Riera-Leal, A.; Padilla-Arellano, J.R.; Fafutis-Morris, M. Microbiota and Its Role on Viral Evasion: Is It With Us or Against Us? Front. Cell. Infect. Microbiol. 2019, 9, 256. [Google Scholar] [CrossRef]
- Nagao-Kitamoto, H.; Kitamoto, S.; Kuffa, P.; Kamada, N. Pathogenic role of the gut microbiota in gastrointestinal diseases. Intest. Res. 2016, 14, 127–138. [Google Scholar] [CrossRef]
- Laitinen, K.; Mokkala, K. Overall Dietary Quality Relates to Gut Microbiota Diversity and Abundance. Int. J. Mol. Sci. 2019, 20, 1835. [Google Scholar] [CrossRef]
- Durack, J.; Lynch, S.V. The gut microbiome: Relationships with disease and opportunities for therapy. J. Exp. Med. 2019, 216, 20–40. [Google Scholar] [CrossRef]
- Martin, A.M.; Sun, E.W.; Rogers, G.B.; Keating, D.J. The Influence of the Gut Microbiome on Host Metabolism through the Regulation of Gut Hormone Release. Front. Physiol. 2019, 10, 428. [Google Scholar] [CrossRef]
- Chun, T.W.; Nickle, D.C.; Justement, J.S.; Meyers, J.H.; Roby, G.; Hallahan, C.W.; Kottilil, S.; Moir, S.; Mican, J.M.; Mullins, J.I.; et al. Persistence of HIV in gut-associated lymphoid tissue despite long-term antiretroviral therapy. J. Infect. Dis. 2008, 197, 714–720. [Google Scholar] [CrossRef]
- Yukl, S.A.; Gianella, S.; Sinclair, E.; Epling, L.; Li, Q.; Duan, L.; Choi, A.L.; Girling, V.; Ho, T.; Li, P.; et al. Differences in HIV burden and immune activation within the gut of HIV-positive patients receiving suppressive antiretroviral therapy. J. Infect. Dis. 2010, 202, 1553–1561. [Google Scholar] [CrossRef]
- Estes, J.D.; Kityo, C.; Ssali, F.; Swainson, L.; Makamdop, K.N.; Del Prete, G.Q.; Deeks, S.G.; Luciw, P.A.; Chipman, J.G.; Beilman, G.J.; et al. Defining total-body AIDS-virus burden with implications for curative strategies. Nat. Med. 2017, 23, 1271–1276. [Google Scholar] [CrossRef]
- Raffatellu, M.; Santos, R.L.; Verhoeven, D.E.; George, M.D.; Wilson, R.P.; Winter, S.E.; Godinez, I.; Sankaran, S.; Paixao, T.A.; Gordon, M.A.; et al. Simian immunodeficiency virus-induced mucosal interleukin-17 deficiency promotes Salmonella dissemination from the gut. Nat. Med. 2008, 14, 421–428. [Google Scholar] [CrossRef]
- Epple, H.J.; Allers, K.; Tröger, H.; Kühl, A.; Erben, U.; Fromm, M.; Zeitz, M.; Loddenkemper, C.; Schulzke, J.D.; Schneider, T. Acute HIV infection induces mucosal infiltration with CD4+ and CD8+ T cells, epithelial apoptosis, and a mucosal barrier defect. Gastroenterology 2010, 139, 1289–1300. [Google Scholar] [CrossRef] [PubMed]
- Borgognone, A.; Noguera-Julian, M.; Oriol, B.; Noël-Romas, L.; Ruiz-Riol, M.; Guillén, Y.; Parera, M.; Casadellà, M.; Duran, C.; Puertas, M.C.; et al. Gut microbiome signatures linked to HIV-1 reservoir size and viremia control. Microbiome 2022, 10, 59. [Google Scholar] [CrossRef]
- Nowak, P.; Troseid, M.; Avershina, E.; Barqasho, B.; Neogi, U.; Holm, K.; Hov, J.R.; Noyan, K.; Vesterbacka, J.; Svärd, J.; et al. Gut microbiota diversity predicts immune status in HIV-1 infection. Aids 2015, 29, 2409–2418. [Google Scholar] [CrossRef]
- Dillon, S.M.; Lee, E.J.; Kotter, C.V.; Austin, G.L.; Dong, Z.; Hecht, D.K.; Gianella, S.; Siewe, B.; Smith, D.M.; Landay, A.L.; et al. An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol. 2014, 7, 983–994. [Google Scholar] [CrossRef] [PubMed]
- Vujkovic-Cvijin, I.; Dunham, R.M.; Iwai, S.; Maher, M.C.; Albright, R.G.; Broadhurst, M.J.; Hernandez, R.D.; Lederman, M.M.; Huang, Y.; Somsouk, M.; et al. Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci. Transl. Med. 2013, 5, 193ra91. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Lin, H.; Cole, M.; Morris, A.; Martinson, J.; McKay, H.; Mimiaga, M.; Margolick, J.; Fitch, A.; Methe, B.; et al. Signature changes in gut microbiome are associated with increased susceptibility to HIV-1 infection in MSM. Microbiome 2021, 9, 237. [Google Scholar] [CrossRef] [PubMed]
- Brenchley, J.M.; Price, D.A.; Schacker, T.W.; Asher, T.E.; Silvestri, G.; Rao, S.; Kazzaz, Z.; Bornstein, E.; Lambotte, O.; Altmann, D.; et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat. Med. 2006, 12, 1365–1371. [Google Scholar] [CrossRef]
- Timmons, T.; Shen, C.; Aldrovandi, G.; Rollie, A.; Gupta, S.K.; Stein, J.H.; Dubé, M.P. Microbial translocation and metabolic and body composition measures in treated and untreated HIV infection. AIDS Res. Hum. Retrovir. 2014, 30, 272–277. [Google Scholar] [CrossRef]
- Moreno-Pérez, Ó.; Giner, L.; Reus, S.; Boix, V.; Alfayate, R.; Frances, R.; Merino, E.; Pico, A.; Portilla, J. Impact of circulating bacterial DNA in long-term glucose homeostasis in non-diabetic patients with HIV infection: Cohort study. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Haissman, J.M.; Knudsen, A.; Hoel, H.; Kjær, A.; Kristoffersen, U.S.; Berge, R.K.; Katzenstein, T.L.; Svardal, A.; Ueland, T.; Aukrust, P.; et al. Microbiota-Dependent Marker TMAO Is Elevated in Silent Ischemia but Is Not Associated with First-Time Myocardial Infarction in HIV Infection. J. Acquir. Immune Defic. Syndr. 2016, 71, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Srinivasa, S.; Fitch, K.V.; Lo, J.; Kadar, H.; Knight, R.; Wong, K.; Abbara, S.; Gauguier, D.; Capeau, J.; Boccara, F.; et al. Plaque burden in HIV-infected patients is associated with serum intestinal microbiota-generated trimethylamine. Aids 2015, 29, 443–452. [Google Scholar] [CrossRef]
- Ancuta, P.; Kamat, A.; Kunstman, K.J.; Kim, E.Y.; Autissier, P.; Wurcel, A.; Zaman, T.; Stone, D.; Mefford, M.; Morgello, S.; et al. Microbial translocation is associated with increased monocyte activation and dementia in AIDS patients. PLoS ONE 2008, 3, e2516. [Google Scholar] [CrossRef]
- Lyons, J.L.; Uno, H.; Ancuta, P.; Kamat, A.; Moore, D.J.; Singer, E.J.; Morgello, S.; Gabuzda, D. Plasma sCD14 is a biomarker associated with impaired neurocognitive test performance in attention and learning domains in HIV infection. J. Acquir. Immune Defic. Syndr. 2011, 57, 371–379. [Google Scholar] [CrossRef]
- Chen, M.F.; Gill, A.J.; Kolson, D.L. Neuropathogenesis of HIV-associated neurocognitive disorders: Roles for immune activation, HIV blipping and viral tropism. Curr. Opin. HIV AIDS 2014, 9, 559–564. [Google Scholar] [CrossRef]
- Heaton, R.K.; Franklin, D.R.; Ellis, R.J.; McCutchan, J.A.; Letendre, S.L.; Leblanc, S.; Corkran, S.H.; Duarte, N.A.; Clifford, D.B.; Woods, S.P.; et al. HIV-associated neurocognitive disorders before and during the era of combination antiretroviral therapy: Differences in rates, nature, and predictors. J. Neurovirol. 2011, 17, 3–16. [Google Scholar] [CrossRef]
- Sacktor, N.; Skolasky, R.L.; Seaberg, E.; Munro, C.; Becker, J.T.; Martin, E.; Ragin, A.; Levine, A.; Miller, E. Prevalence of HIV-associated neurocognitive disorders in the Multicenter AIDS Cohort Study. Neurology 2016, 86, 334–340. [Google Scholar] [CrossRef] [PubMed]
- Kesby, J.P.; Markou, A.; Semenova, S. The effects of HIV-1 regulatory TAT protein expression on brain reward function, response to psychostimulants and delay-dependent memory in mice. Neuropharmacology 2016, 109, 205–215. [Google Scholar] [CrossRef]
- Kesby, J.P.; Markou, A.; Semenova, S.; Group, T. Effects of HIV/TAT protein expression and chronic selegiline treatment on spatial memory, reversal learning and neurotransmitter levels in mice. Behav. Brain Res. 2016, 311, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.O.; Liu, Y.; Ruan, Y.; Xu, Z.C.; Schantz, L.; He, J.J. Neuropathologies in transgenic mice expressing human immunodeficiency virus type 1 Tat protein under the regulation of the astrocyte-specific glial fibrillary acidic protein promoter and doxycycline. Am. J. Pathol. 2003, 162, 1693–1707. [Google Scholar] [CrossRef] [PubMed]
- Raybuck, J.D.; Hargus, N.J.; Thayer, S.A. A GluN2B-Selective NMDAR Antagonist Reverses Synapse Loss and Cognitive Impairment Produced by the HIV-1 Protein Tat. J. Neurosci. 2017, 37, 7837–7847. [Google Scholar] [CrossRef] [PubMed]
- Carey, A.N.; Liu, X.; Mintzopoulos, D.; Paris, J.J.; Muschamp, J.W.; McLaughlin, J.P.; Kaufman, M.J. Conditional Tat protein expression in the GT-tg bigenic mouse brain induces gray matter density reductions. Prog. Neuropsychopharmacol. Biol. Psychiatry 2013, 43, 49–54. [Google Scholar] [CrossRef]
- Carey, A.N.; Sypek, E.I.; Singh, H.D.; Kaufman, M.J.; McLaughlin, J.P. Expression of HIV-Tat protein is associated with learning and memory deficits in the mouse. Behav. Brain Res. 2012, 229, 48–56. [Google Scholar] [CrossRef]
- Li, S.T.; Matsushita, M.; Moriwaki, A.; Saheki, Y.; Lu, Y.F.; Tomizawa, K.; Wu, H.Y.; Terada, H.; Matsui, H. HIV-1 Tat inhibits long-term potentiation and attenuates spatial learning [corrected]. Ann. Neurol. 2004, 55, 362–371. [Google Scholar] [CrossRef]
- Nookala, A.R.; Schwartz, D.C.; Chaudhari, N.S.; Glazyrin, A.; Stephens, E.B.; Berman, N.E.J.; Kumar, A. Methamphetamine augment HIV-1 Tat mediated memory deficits by altering the expression of synaptic proteins and neurotrophic factors. Brain Behav. Immun. 2018, 71, 37–51. [Google Scholar] [CrossRef]
- Fu, X.; Lawson, M.A.; Kelley, K.W.; Dantzer, R. HIV-1 Tat activates indoleamine 2,3 dioxygenase in murine organotypic hippocampal slice cultures in a p38 mitogen-activated protein kinase-dependent manner. J. Neuroinflamm. 2011, 8, 88. [Google Scholar] [CrossRef]
- Hahn, Y.K.; Masvekar, R.R.; Xu, R.; Hauser, K.F.; Knapp, P.E. Chronic HIV-1 Tat and HIV reduce Rbfox3/NeuN: Evidence for sex-related effects. Curr. HIV Res. 2015, 13, 10–20. [Google Scholar] [CrossRef]
- Hahn, Y.K.; Podhaizer, E.M.; Farris, S.P.; Miles, M.F.; Hauser, K.F.; Knapp, P.E. Effects of chronic HIV-1 Tat exposure in the CNS: Heightened vulnerability of males versus females to changes in cell numbers, synaptic integrity, and behavior. Brain Struct. Funct. 2015, 220, 605–623. [Google Scholar] [CrossRef] [PubMed]
- Moran, L.M.; Fitting, S.; Booze, R.M.; Webb, K.M.; Mactutus, C.F. Neonatal intrahippocampal HIV-1 protein Tat(1-86) injection: Neurobehavioral alterations in the absence of increased inflammatory cytokine activation. Int. J. Dev. Neurosci. 2014, 38, 195–203. [Google Scholar] [CrossRef]
- Zou, W.; Kim, B.O.; Zhou, B.Y.; Liu, Y.; Messing, A.; He, J.J. Protection against human immunodeficiency virus type 1 Tat neurotoxicity by Ginkgo biloba extract EGb 761 involving glial fibrillary acidic protein. Am. J. Pathol. 2007, 171, 1923–1935. [Google Scholar] [CrossRef] [PubMed]
- Henderson, L.J.; Johnson, T.P.; Smith, B.R.; Reoma, L.B.; Santamaria, U.A.; Bachani, M.; Demarino, C.; Barclay, R.A.; Snow, J.; Sacktor, N.; et al. Presence of Tat and transactivation response element in spinal fluid despite antiretroviral therapy. Aids 2019, 33 (Suppl. S2), S145–S157. [Google Scholar] [CrossRef] [PubMed]
- Hudson, L.; Liu, J.; Nath, A.; Jones, M.; Raghavan, R.; Narayan, O.; Male, D.; Everall, I. Detection of the human immunodeficiency virus regulatory protein tat in CNS tissues. J. Neurovirol. 2000, 6, 145–155. [Google Scholar] [CrossRef]
- Johnson, T.P.; Patel, K.; Johnson, K.R.; Maric, D.; Calabresi, P.A.; Hasbun, R.; Nath, A. Induction of IL-17 and nonclassical T-cell activation by HIV-Tat protein. Proc. Natl. Acad. Sci. USA 2013, 110, 13588–13593. [Google Scholar] [CrossRef]
- Vassallo, M.; Dunais, B.; Durant, J.; Carsenti-Dellamonica, H.; Harvey-Langton, A.; Cottalorda, J.; Ticchioni, M.; Laffon, M.; Lebrun-Frenay, C.; Dellamonica, P.; et al. Relevance of lipopolysaccharide levels in HIV-associated neurocognitive impairment: The Neuradapt study. J. Neurovirol. 2013, 19, 376–382. [Google Scholar] [CrossRef] [PubMed]
- Ramendra, R.; Isnard, S.; Mehraj, V.; Chen, J.; Zhang, Y.; Finkelman, M.; Routy, J.P. Circulating LPS and (1→3)-β-D-Glucan: A Folie à Deux Contributing to HIV-Associated Immune Activation. Front. Immunol. 2019, 10, 465. [Google Scholar] [CrossRef] [PubMed]
- Ceccarelli, G.; Brenchley, J.M.; Cavallari, E.N.; Scheri, G.C.; Fratino, M.; Pinacchio, C.; Schietroma, I.; Fard, S.N.; Scagnolari, C.; Mezzaroma, I.; et al. Impact of High-Dose Multi-Strain Probiotic Supplementation on Neurocognitive Performance and Central Nervous System Immune Activation of HIV-1 Infected Individuals. Nutrients 2017, 9, 1269. [Google Scholar] [CrossRef]
- Ceccarelli, G.; Fratino, M.; Selvaggi, C.; Giustini, N.; Serafino, S.; Schietroma, I.; Corano Scheri, G.; Pavone, P.; Passavanti, G.; Alunni Fegatelli, D.; et al. A pilot study on the effects of probiotic supplementation on neuropsychological performance and microRNA-29a-c levels in antiretroviral-treated HIV-1-infected patients. Brain Behav. 2017, 7, e00756. [Google Scholar] [CrossRef]
- Byrd, D.A.; Fellows, R.P.; Morgello, S.; Franklin, D.; Heaton, R.K.; Deutsch, R.; Atkinson, J.H.; Clifford, D.B.; Collier, A.C.; Marra, C.M.; et al. Neurocognitive impact of substance use in HIV infection. J. Acquir. Immune Defic. Syndr. 2011, 58, 154–162. [Google Scholar] [CrossRef]
- Chilunda, V.; Calderon, T.M.; Martinez-Aguado, P.; Berman, J.W. The impact of substance abuse on HIV-mediated neuropathogenesis in the current ART era. Brain Res. 2019, 1724, 146426. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhang, F.; Kandel, S.R.; Brau, F.; He, J.J. HIV Tat and cocaine interactively alter genome-wide DNA methylation and gene expression and exacerbate learning and memory impairments. Cell Rep. 2022, 39, 110765. [Google Scholar] [CrossRef] [PubMed]
- Fulcher, J.A.; Hussain, S.K.; Cook, R.; Li, F.; Tobin, N.H.; Ragsdale, A.; Shoptaw, S.; Gorbach, P.M.; Aldrovandi, G.M. Effects of Substance Use and Sex Practices on the Intestinal Microbiome During HIV-1 Infection. J. Infect. Dis. 2018, 218, 1560–1570. [Google Scholar] [CrossRef] [PubMed]
- Arimatsu, K.; Yamada, H.; Miyazawa, H.; Minagawa, T.; Nakajima, M.; Ryder, M.I.; Gotoh, K.; Motooka, D.; Nakamura, S.; Iida, T.; et al. Oral pathobiont induces systemic inflammation and metabolic changes associated with alteration of gut microbiota. Sci. Rep. 2014, 4, 4828. [Google Scholar] [CrossRef]
- Shao, T.; Zhao, C.; Li, F.; Gu, Z.; Liu, L.; Zhang, L.; Wang, Y.; He, L.; Liu, Y.; Liu, Q.; et al. Intestinal HIF-1alpha Deletion Exacerbates Alcoholic Liver Disease through Inducing Intestinal Dysbiosis and Barrier Dysfunction. J. Hepatol. 2018, 69, 886–895. [Google Scholar] [CrossRef]
- Hernandez, J.; Tamargo, J.A.; Sales Martinez, S.; Martin, H.R.; Campa, A.; Sékaly, R.P.; Bordi, R.; Sherman, K.E.; Rouster, S.D.; Meeds, H.L.; et al. Cocaine use associated gut permeability and microbial translocation in people living with HIV in the Miami Adult Study on HIV (MASH) cohort. PLoS ONE 2022, 17, e0275675. [Google Scholar] [CrossRef]
- Ul-Hasan, S.; Bowers, R.M.; Figueroa-Montiel, A.; Licea-Navarro, A.F.; Beman, J.M.; Woyke, T.; Nobile, C.J. Community ecology across bacteria, archaea and microbial eukaryotes in the sediment and seawater of coastal Puerto Nuevo, Baja California. PLoS ONE 2019, 14, e0212355. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Dhariwal, A.; Chong, J.; Habib, S.; King, I.L.; Agellon, L.B.; Xia, J. MicrobiomeAnalyst: A web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017, 45, W180–W188. [Google Scholar] [CrossRef]
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef] [PubMed]
- Aβhauer, K.P.; Wemheuer, B.; Daniel, R.; Meinicke, P. Tax4Fun: Predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics 2015, 31, 2882–2884. [Google Scholar]
- Volpe, G.E.; Ward, H.; Mwamburi, M.; Dinh, D.; Bhalchandra, S.; Wanke, C.; Kane, A.V. Associations of cocaine use and HIV infection with the intestinal microbiota, microbial translocation, and inflammation. J. Stud. Alcohol Drugs 2014, 75, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Chivero, E.T.; Ahmad, R.; Thangaraj, A.; Periyasamy, P.; Kumar, B.; Kroeger, E.; Feng, D.; Guo, M.L.; Roy, S.; Dhawan, P.; et al. Cocaine Induces Inflammatory Gut Milieu by Compromising the Mucosal Barrier Integrity and Altering the Gut Microbiota Colonization. Sci. Rep. 2019, 9, 12187. [Google Scholar] [CrossRef] [PubMed]
- Scorza, C.; Piccini, C.; Martinez Busi, M.; Abin Carriquiry, J.A.; Zunino, P. Alterations in the Gut Microbiota of Rats Chronically Exposed to Volatilized Cocaine and Its Active Adulterants Caffeine and Phenacetin. Neurotox. Res. 2019, 35, 111–121. [Google Scholar] [CrossRef]
- An, R.; Wilms, E.; Masclee, A.A.M.; Smidt, H.; Zoetendal, E.G.; Jonkers, D. Age-dependent changes in GI physiology and microbiota: Time to reconsider? Gut 2018, 67, 2213–2222. [Google Scholar] [CrossRef]
- O’Toole, P.W.; Jeffery, I.B. Gut microbiota and aging. Science 2015, 350, 1214–1215. [Google Scholar] [CrossRef]
- Org, E.; Mehrabian, M.; Parks, B.W.; Shipkova, P.; Liu, X.; Drake, T.A.; Lusis, A.J. Sex differences and hormonal effects on gut microbiota composition in mice. Gut Microbes 2016, 7, 313–322. [Google Scholar] [CrossRef]
- Baker, J.M.; Al-Nakkash, L.; Herbst-Kralovetz, M.M. Estrogen-gut microbiome axis: Physiological and clinical implications. Maturitas 2017, 103, 45–53. [Google Scholar] [CrossRef]
- Kiraly, D.D.; Walker, D.M.; Calipari, E.S.; Labonte, B.; Issler, O.; Pena, C.J.; Ribeiro, E.A.; Russo, S.J.; Nestler, E.J. Alterations of the Host Microbiome Affect Behavioral Responses to Cocaine. Sci. Rep. 2016, 6, 35455. [Google Scholar] [CrossRef]
- Jandhyala, S.M.; Talukdar, R.; Subramanyam, C.; Vuyyuru, H.; Sasikala, M.; Nageshwar Reddy, D. Role of the normal gut microbiota. World J. Gastroenterol. 2015, 21, 8787–8803. [Google Scholar] [CrossRef]
- Vernocchi, P.; Del Chierico, F.; Putignani, L. Gut Microbiota Metabolism and Interaction with Food Components. Int. J. Mol. Sci. 2020, 21, 3688. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Q.; Zhang, A.H.; Miao, J.H.; Sun, H.; Yan, G.L.; Wu, F.F.; Wang, X.J. Gut microbiota as important modulator of metabolism in health and disease. RSC Adv. 2018, 8, 42380–42389. [Google Scholar] [CrossRef] [PubMed]
- Hui, L.; Chen, X.; Bhatt, D.; Geiger, N.H.; Rosenberger, T.A.; Haughey, N.J.; Masino, S.A.; Geiger, J.D. Ketone bodies protection against HIV-1 Tat-induced neurotoxicity. J. Neurochem. 2012, 122, 382–391. [Google Scholar] [CrossRef] [PubMed]
- Mohseni Ahooyi, T.; Shekarabi, M.; Torkzaban, B.; Langford, T.D.; Burdo, T.H.; Gordon, J.; Datta, P.K.; Amini, S.; Khalili, K. Dysregulation of Neuronal Cholesterol Homeostasis upon Exposure to HIV-1 Tat and Cocaine Revealed by RNA-Sequencing. Sci. Rep. 2018, 8, 16300. [Google Scholar] [CrossRef]
- Mayer, E.A.; Tillisch, K.; Gupta, A. Gut/brain axis and the microbiota. J. Clin. Investig. 2015, 125, 926–938. [Google Scholar] [CrossRef]
- Bagasra, O.; Lavi, E.; Bobroski, L.; Khalili, K.; Pestaner, J.P.; Tawadros, R.; Pomerantz, R.J. Cellular reservoirs of HIV-1 in the central nervous system of infected individuals: Identification by the combination of in situ polymerase chain reaction and immunohistochemistry. Aids 1996, 10, 573–585. [Google Scholar] [CrossRef]
- Brack-Werner, R. Astrocytes: HIV cellular reservoirs and important participants in neuropathogenesis. Aids 1999, 13, 1–22. [Google Scholar] [CrossRef]
- Frankel, A.D.; Pabo, C.O. Cellular uptake of the tat protein from human immunodeficiency virus. Cell 1988, 55, 1189–1193. [Google Scholar] [CrossRef]
- Liu, Y.; Jones, M.; Hingtgen, C.M.; Bu, G.; Laribee, N.; Tanzi, R.E.; Moir, R.D.; Nath, A.; He, J.J. Uptake of HIV-1 tat protein mediated by low-density lipoprotein receptor-related protein disrupts the neuronal metabolic balance of the receptor ligands. Nat. Med. 2000, 6, 1380–1387. [Google Scholar] [CrossRef]
- Wallet, C.; De Rovere, M.; Van Assche, J.; Daouad, F.; De Wit, S.; Gautier, V.; Mallon, P.W.G.; Marcello, A.; Van Lint, C.; Rohr, O.; et al. Microglial Cells: The Main HIV-1 Reservoir in the Brain. Front. Cell. Infect. Microbiol. 2019, 9, 362. [Google Scholar] [CrossRef] [PubMed]
- Green, M.; Loewenstein, P.M. Autonomous functional domains of chemically synthesized human immunodeficiency virus tat trans-activator protein. Cell 1988, 55, 1179–1188. [Google Scholar] [CrossRef] [PubMed]
- Esposito, G.; Capoccia, E.; Gigli, S.; Pesce, M.; Bruzzese, E.; D’Alessandro, A.; Cirillo, C.; di Cerbo, A.; Cuomo, R.; Seguella, L.; et al. HIV-1 Tat-induced diarrhea evokes an enteric glia-dependent neuroinflammatory response in the central nervous system. Sci. Rep. 2017, 7, 7735. [Google Scholar] [CrossRef]
- Sarnelli, G.; Seguella, L.; Pesce, M.; Lu, J.; Gigli, S.; Bruzzese, E.; Lattanzi, R.; D’Alessandro, A.; Cuomo, R.; Steardo, L.; et al. HIV-1 Tat-induced diarrhea is improved by the PPARalpha agonist, palmitoylethanolamide, by suppressing the activation of enteric glia. J. Neuroinflamm. 2018, 15, 94. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.; Zhao, X.; He, J.J. HIV Tat Expression and Cocaine Exposure Lead to Sex- and Age-Specific Changes of the Microbiota Composition in the Gut. Microorganisms 2023, 11, 799. https://doi.org/10.3390/microorganisms11030799
Li L, Zhao X, He JJ. HIV Tat Expression and Cocaine Exposure Lead to Sex- and Age-Specific Changes of the Microbiota Composition in the Gut. Microorganisms. 2023; 11(3):799. https://doi.org/10.3390/microorganisms11030799
Chicago/Turabian StyleLi, Lu, Xiaojie Zhao, and Johnny J. He. 2023. "HIV Tat Expression and Cocaine Exposure Lead to Sex- and Age-Specific Changes of the Microbiota Composition in the Gut" Microorganisms 11, no. 3: 799. https://doi.org/10.3390/microorganisms11030799
APA StyleLi, L., Zhao, X., & He, J. J. (2023). HIV Tat Expression and Cocaine Exposure Lead to Sex- and Age-Specific Changes of the Microbiota Composition in the Gut. Microorganisms, 11(3), 799. https://doi.org/10.3390/microorganisms11030799




