Rumen Biogeographical Regions and Microbiome Variation
Abstract
1. Introduction
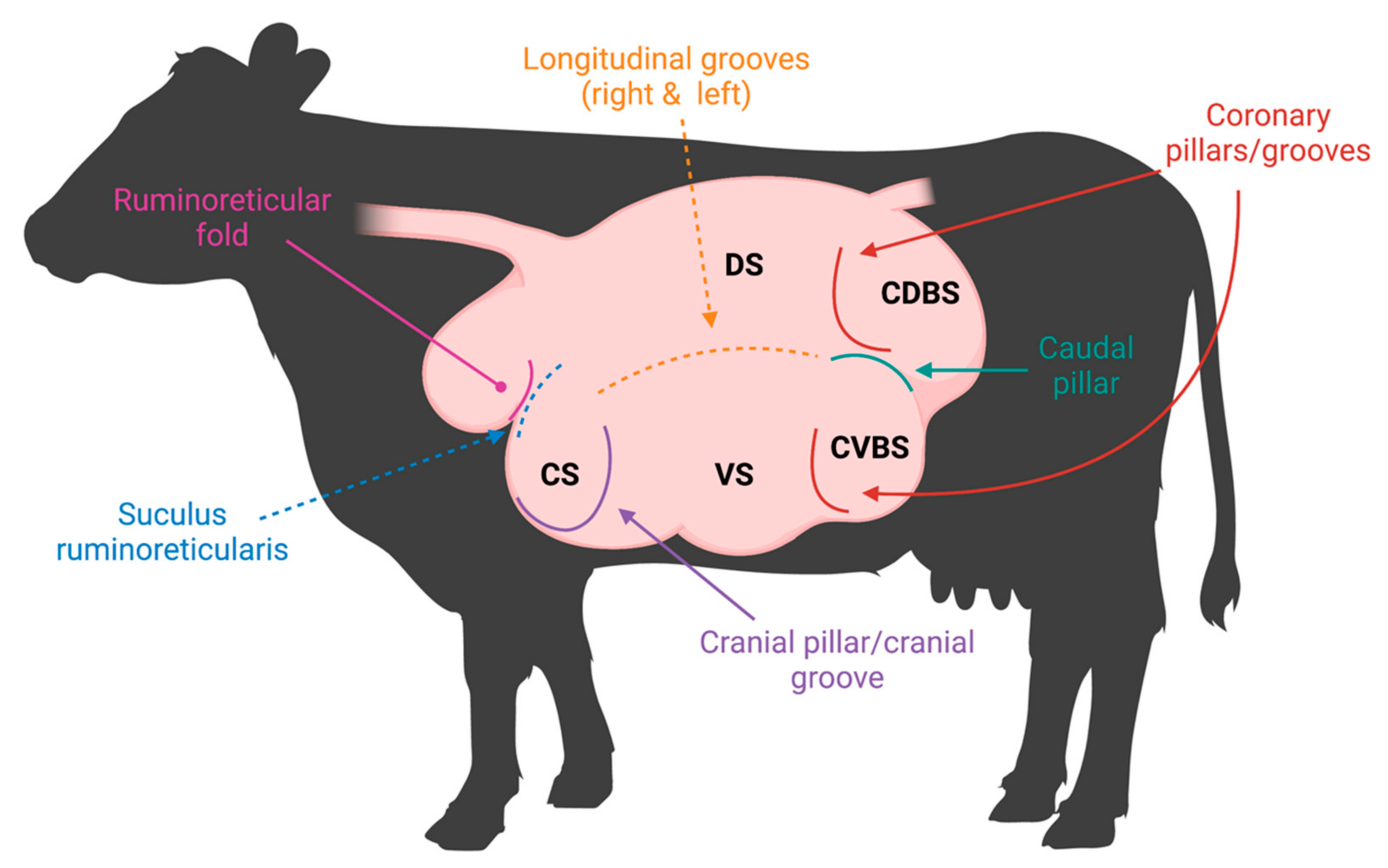
2. Anatomy of the Rumen Affecting the Microbiome
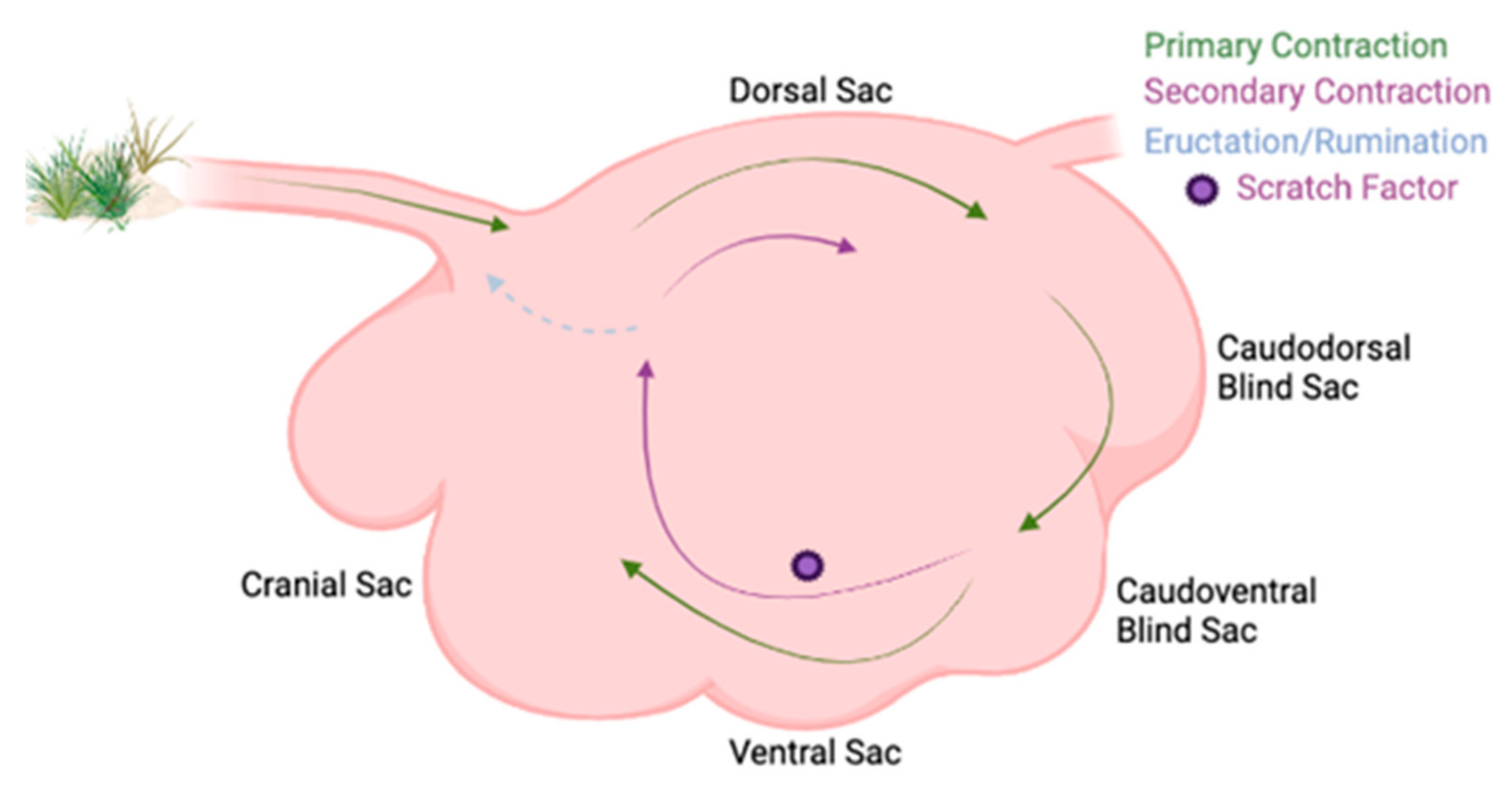
2.1. Ruminal Environment Variations and Their Impact on Microbial Community Variation
2.2. Ruminal Papillae Variation among the Different Sacs
3. Microbial Community Variation, Fractions, and Importance in the Ruminant
3.1. Epimural Microbial Community
3.2. Interactions among the Fiber-Adherent and Planktonic Microbial Communities
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Li, Z.; Wang, X.; Zhang, Y.; Yu, Z.; Zhang, T.; Dai, X.; Pan, X.; Jing, R.; Yan, Y.; Liu, Y.; et al. Genomic insights into the phylogeny and biomass-degrading enzymes of rumen ciliates. ISME J. 2022, 16, 2775–2787. [Google Scholar] [CrossRef] [PubMed]
- Stewart, R.D.; Auffret, M.D.; Warr, A.; Wiser, A.H.; Press, M.O.; Langford, K.W.; Liachko, I.; Snelling, T.J.; Dewhurst, R.J.; Walker, A.W.; et al. Assembly of 913 microbial genomes from metagenomic sequencing of the cow rumen. Nat. Commun. 2018, 9, 870. [Google Scholar] [CrossRef]
- Sbardellati, D.L.; Fischer, A.; Cox, M.S.; Li, W.; Kalscheur, K.F.; Suen, G. The bovine epimural microbiota displays compositional and structural heterogeneity across different ruminal locations. J. Dairy Sci. 2020, 103, 3636–3647. [Google Scholar] [CrossRef] [PubMed]
- Van Soest, P.J. Nutritional Ecology of the Ruminant; Cornell University Press: Ithaca, NY, USA, 1994. [Google Scholar]
- Orpin, C.G. Studies on the rumen flagellate Sphaeromonas communis. J. Gen. Microbiol. 1976, 94, 270–280. [Google Scholar] [CrossRef] [PubMed]
- Carballo, O.C.; Khan, M.A.; Knol, F.W.; Lewis, S.J.; Stevens, D.R.; Laven, R.A.; McCoard, S.A. Impact of weaning age on rumen development in artificially reared lambs1. J. Anim. Sci. 2019, 97, 3498–3510. [Google Scholar] [CrossRef]
- Schmitz-Esser, S. The Rumen Epithelial Microbiota: Possible Gatekeepers of the Rumen Epithelium and Its Potential Contributions to Epithelial Barrier Function and Animal Health and Performance. Meat Muscle Biol. 2021, 4, 1–11. [Google Scholar] [CrossRef]
- Mizrahi, I.; Jami, E. A method to the madness: Disentangling the individual forces that shape the rumen microbiome. EMBO Rep. 2021, 22, e52269. [Google Scholar] [CrossRef]
- Weimer, P.J. Redundancy, resilience, and host specificity of the ruminal microbiota: Implications for engineering improved ruminal fermentations. Front. Microbiol. 2015, 6, 296. [Google Scholar] [CrossRef]
- Krehbiel, C. Invited review: Applied nutrition of ruminants: Fermentation and digestive physiology. Prof. Anim. Sci. 2014, 30, 129–139. [Google Scholar] [CrossRef]
- Shen, J.S.; Chai, Z.; Song, L.J.; Liu, J.X.; Wu, Y.M. Insertion depth of oral stomach tubes may affect the fermentation parameters of ruminal fluid collected in dairy cows. J. Dairy Sci. 2012, 95, 5978–5984. [Google Scholar] [CrossRef]
- Harlow, B.E.; Flythe, M.D.; Aiken, G.E. Effect of biochanin A on corn grain (Zea mays) fermentation by bovine rumen amylolytic bacteria. J. Appl. Microbiol. 2017, 122, 870–880. [Google Scholar] [CrossRef] [PubMed]
- Rieu, F.; Fonty, G.; Gaillard, B.; Gouet, P. Electron microscopy study of the bacteria adherent to the rumen wall in young conventional lambs. Can. J. Microbiol. 1990, 36, 140–144. [Google Scholar] [CrossRef]
- McCowan, R.; Cheng, K.; Bailey, C.; Costerton, J. Adhesion of bacteria to epithelial cell surfaces within the reticulo-rumen of cattle. Appl. Environ. Microbiol. 1978, 35, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Cheng, K.J.; McCowan, R.P.; Costerton, J.W. Adherent epithelial bacteria in ruminants and their roles in digestive tract function. Am. J. Clin. Nutr. 1979, 32, 139–148. [Google Scholar] [CrossRef]
- Bryant, A. Variations in the pH and volatile fatty acid concentration within the bovine reticulo-rumen. N. Z. J. Agric. Res. 1964, 7, 694–706. [Google Scholar] [CrossRef]
- Church, D.C. The Ruminant Animal: Digestive Physiology and Nutrition; Waveland Press: Long Grove, IL, USA, 1993. [Google Scholar]
- Tafaj, M.; Junck, B.; Maulbetsch, A.; Steingass, H.; Piepho, H.P.; Drochner, W. Digesta characteristics of dorsal, middle and ventral rumen of cows fed with different hay qualities and concentrate levels. Arch. Anim. Nutr. 2004, 58, 325–342. [Google Scholar] [CrossRef]
- Grünberg, W.; Constable, P.D. CHAPTER 6—Function and Dysfunction of the Ruminant Forestomach. In Food Animal Practice, 5th ed.; Anderson, D.E., Rings, D.M., Eds.; W.B. Saunders: Saint Louis, MO, USA, 2009; pp. 12–19. [Google Scholar]
- Millen, D.D.; De Beni Arrigoni, M.; Lauritano Pacheco, R.D. Rumenology, 1st ed.; Springer: Cham, Switzerland, 2016. [Google Scholar]
- Li, M.; Penner, G.B.; Hernandez-Sanabria, E.; Oba, M.; Guan, L.L. Effects of sampling location and time, and host animal on assessment of bacterial diversity and fermentation parameters in the bovine rumen. J. Appl. Microbiol. 2009, 107, 1924–1934. [Google Scholar] [CrossRef]
- Rumsey, T.S.; Putnam, P.A.; Bond, J.; Oltjen, R.R. Influence of level and type of diet on ruminal pH and VFA, respiratory rate and EKG patterns of steers. J. Anim. Sci. 1970, 31, 608–616. [Google Scholar] [CrossRef]
- Adebayo Arowolo, M.; Zhang, X.M.; Wang, M.; Wang, R.; Wen, J.N.; Hao, L.Z.; He, J.H.; Shen, W.J.; Ma, Z.Y.; Tan, Z.L. Proper motility enhances rumen fermentation and microbial protein synthesis with decreased saturation of dissolved gases in rumen simulation technique. J. Dairy Sci. 2022, 105, 231–241. [Google Scholar] [CrossRef]
- Waghorn, G.; Reid, C. Rumen motility in sheep and cattle given different diets. N. Z. J. Agric. Res. 1983, 26, 289–295. [Google Scholar] [CrossRef]
- Beauchemin, K. Invited review: Current perspectives on eating and rumination activity in dairy cows. J. Dairy Sci. 2018, 101, 4762–4784. [Google Scholar] [CrossRef]
- Crichlow, E. Ruminal lactic acidosis: Forestomach epithelial receptor activation by undissociated volatile fatty acids and rumen fluids collected during loss of reticuloruminal motility. Res. Vet. Sci. 1988, 45, 364–368. [Google Scholar] [CrossRef] [PubMed]
- Storm, A.C.; Kristensen, N.B. Effects of particle size and dry matter content of a total mixed ration on intraruminal equilibration and net portal flux of volatile fatty acids in lactating dairy cows. J. Dairy Sci. 2010, 93, 4223–4238. [Google Scholar] [CrossRef] [PubMed]
- Ahn, G.; Ricconi, K.; Avila, S.; Klotz, J.L.; Harmon, D.L. Ruminal motility, reticuloruminal fill, and eating patterns in steers exposed to ergovaline. J. Anim. Sci. 2019, 98, skz374. [Google Scholar] [CrossRef]
- Melchior, E.A.; Smith, J.K.; Schneider, L.G.; Mulliniks, J.T.; Bates, G.E.; Flythe, M.D.; Klotz, J.L.; Ji, H.; Goodman, J.P.; Lee, A.R.; et al. Effects of endophyte-infected tall fescue seed and red clover isoflavones on rumen microbial populations and physiological parameters of beef cattle. Transl. Anim. Sci. 2019, 3, 315–328. [Google Scholar] [CrossRef]
- Melchior, E.A.; Myer, P.R. Fescue toxicosis and its influence on the rumen microbiome: Mitigation of production losses through clover isoflavones. J. Appl. Anim. Res. 2018, 46, 1280–1288. [Google Scholar] [CrossRef]
- Koester, L.R.; Poole, D.H.; Serão, N.V.; Schmitz-Esser, S. Beef cattle that respond differently to fescue toxicosis have distinct gastrointestinal tract microbiota. PLoS ONE 2020, 15, e0229192. [Google Scholar] [CrossRef] [PubMed]
- Bergman, E.N. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol. Rev. 1990, 70, 567–590. [Google Scholar] [CrossRef]
- Steele, M.A.; Croom, J.; Kahler, M.; AlZahal, O.; Hook, S.E.; Plaizier, K.; McBride, B.W. Bovine rumen epithelium undergoes rapid structural adaptations during grain-induced subacute ruminal acidosis. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2011, 300, R1515–R1523. [Google Scholar] [CrossRef]
- Wang, B.; Wang, D.; Wu, X.; Cai, J.; Liu, M.; Huang, X.; Wu, J.; Liu, J.; Guan, L. Effects of dietary physical or nutritional factors on morphology of rumen papillae and transcriptome changes in lactating dairy cows based on three different forage-based diets. BMC Genom. 2017, 18, 353. [Google Scholar] [CrossRef]
- Dehority, B.A. Gastrointestinal tracts of herbivores, particularly the ruminant: Anatomy, physiology and microbial digestion of plants. J. Appl. Anim. Res. 2002, 21, 145–160. [Google Scholar] [CrossRef]
- Niwińska, B.; Hanczakowska, E.; Arciszewski, M.; Klebaniuk, R. Exogenous butyrate: Implications for the functional development of ruminal epithelium and calf performance. Animal 2017, 11, 1522–1530. [Google Scholar] [CrossRef] [PubMed]
- Mach, N.; Devant, M.; Bach, A. Rumen fermentation parameters and rumen papillae characteristics in finishing bulls as affected by nonfibrous carbohydrate level and lipid source of the diet. J. Anim. Vet. Adv. 2006, 5, 220–225. [Google Scholar]
- Aschenbach, J.R.; Penner, G.B.; Stumpff, F.; Gabel, G. Ruminant Nutrition Symposium: Role of fermentation acid absorption in the regulation of ruminal pH. J. Anim. Sci. 2011, 89, 1092–1107. [Google Scholar] [CrossRef] [PubMed]
- Dieho, K.; Bannink, A.; Geurts, I.A.L.; Schonewille, J.T.; Gort, G.; Dijkstra, J. Morphological adaptation of rumen papillae during the dry period and early lactation as affected by rate of increase of concentrate allowance. J. Dairy Sci. 2016, 99, 2339–2352. [Google Scholar] [CrossRef]
- Dijkstra, J.; Ellis, J.; Kebreab, E.; Strathe, A.; López, S.; France, J.; Bannink, A. Ruminal pH regulation and nutritional consequences of low pH. Anim. Feed Sci. Technol. 2012, 172, 22–33. [Google Scholar] [CrossRef]
- Penner, G.B.; Aschenbach, J.R.; Gabel, G.; Rackwitz, R.; Oba, M. Epithelial capacity for apical uptake of short chain fatty acids is a key determinant for intraruminal pH and the susceptibility to subacute ruminal acidosis in sheep. J. Nutr. 2009, 139, 1714–1720. [Google Scholar] [CrossRef]
- Hofmann, R.R. Evolutionary steps of ecophysiological adaptation and diversification of ruminants: A comparative view of their digestive system. Oecologia 1989, 78, 443–457. [Google Scholar] [CrossRef]
- Eslamizad, M.; Albrecht, D.; Kuhla, B. The effect of chronic, mild heat stress on metabolic changes of nutrition and adaptations in rumen papillae of lactating dairy cows. J. Dairy Sci. 2020, 103, 8601–8614. [Google Scholar] [CrossRef]
- Petri, R.M.; Kleefisch, M.T.; Metzler-Zebeli, B.U.; Zebeli, Q.; Klevenhusen, F. Changes in the Rumen Epithelial Microbiota of Cattle and Host Gene Expression in Response to Alterations in Dietary Carbohydrate Composition. Appl. Environ. Microbiol. 2018, 84, e00384-18. [Google Scholar] [CrossRef]
- Chen, Y.; Oba, M.; Guan, L.L. Variation of bacterial communities and expression of Toll-like receptor genes in the rumen of steers differing in susceptibility to subacute ruminal acidosis. Vet. Microbiol. 2012, 159, 451–459. [Google Scholar] [CrossRef]
- Liu, J.H.; Bian, G.R.; Zhu, W.Y.; Mao, S.Y. High-grain feeding causes strong shifts in ruminal epithelial bacterial community and expression of Toll-like receptor genes in goats. Front. Microbiol. 2015, 6, 167. [Google Scholar] [CrossRef] [PubMed]
- Kern, R.J.; Lindholm-Perry, A.K.; Freetly, H.C.; Snelling, W.M.; Kern, J.W.; Keele, J.W.; Miles, J.R.; Foote, A.P.; Oliver, W.T.; Kuehn, L.A.; et al. Transcriptome differences in the rumen of beef steers with variation in feed intake and gain. Gene 2016, 586, 12–26. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Wang, X.; Xiao, J.; Chen, K.; Zhou, H.; Shen, D.; Li, H.; Tang, Q. Loss of regulator of G protein signaling 5 exacerbates obesity, hepatic steatosis, inflammation and insulin resistance. PLoS ONE 2012, 7, e30256. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Kozasa, T.; Bondjers, C.; Betsholtz, C.; Kehrl, J.H. Pericyte-specific expression of Rgs5: Implications for PDGF and EDG receptor signaling during vascular maturation. FASEB J. 2003, 17, 440–442. [Google Scholar] [CrossRef]
- Czerkawski, J.; Cheng, K. Compartmentation in the rumen. In The Rumen Microbe Ecosystem; Springer: Dordrecht, The Netherlands, 1988. [Google Scholar] [CrossRef]
- Czerkawski, J.W. An Introduction to Rumen Studies; Elsevier: Amsterdam, The Netherlands, 2013. [Google Scholar]
- Cho, S.-J.; Cho, K.-M.; Shin, E.-C.; Lim, W.-J.; Hong, S.-Y.; Choi, B.-R.; Kang, J.-M.; Lee, S.-M.; Kim, Y.-H.; Kim, H. 16S rDNA analysis of bacterial diversity in three fractions of cow rumen. J. Microbiol. Biotechnol. 2006, 16, 92–101. [Google Scholar]
- Tajima, K.; Aminov, R.I.; Nagamine, T.; Ogata, K.; Nakamura, M.; Matsui, H.; Benno, Y. Rumen bacterial diversity as determined by sequence analysis of 16S rDNA libraries. FEMS Microbiol. Ecol. 1999, 29, 159–169. [Google Scholar] [CrossRef]
- Tajima, K.; Arai, S.; Ogata, K.; Nagamine, T.; Matsui, H.; Nakamura, M.; Aminov, R.I.; Benno, Y. Rumen Bacterial Community Transition During Adaptation to High-grain Diet. Anaerobe 2000, 6, 273–284. [Google Scholar] [CrossRef]
- Whitford, M.F.; Forster, R.J.; Beard, C.E.; Gong, J.; Teather, R.M. Phylogenetic analysis of rumen bacteria by comparative sequence analysis of cloned 16S rRNA genes. Anaerobe 1998, 4, 153–163. [Google Scholar] [CrossRef]
- Fay, J.; Cheng, K.; Costerton, J. Production of alkaline phosphatase by epithelial cells and adherent bacteria of the bovine rumen and abomasum. Can. J. Microbiol. 1979, 25, 932–936. [Google Scholar] [CrossRef]
- Mao, S.; Zhang, M.; Liu, J.; Zhu, W. Characterising the bacterial microbiota across the gastrointestinal tracts of dairy cattle: Membership and potential function. Sci. Rep. 2015, 5, 16116. [Google Scholar] [CrossRef]
- Wallace, R.; Cheng, K.-J.; Dinsdale, D.; Ørskov, E. An independent microbial flora of the epithelium and its role in the ecomicrobiology of the rumen. Nature 1979, 279, 424–426. [Google Scholar] [CrossRef] [PubMed]
- Wetzels, S.U.; Mann, E.; Pourazad, P.; Qumar, M.; Pinior, B.; Metzler-Zebeli, B.U.; Wagner, M.; Schmitz-Esser, S.; Zebeli, Q. Epimural bacterial community structure in the rumen of Holstein cows with different responses to a long-term subacute ruminal acidosis diet challenge. J. Dairy Sci. 2017, 100, 1829–1844. [Google Scholar] [CrossRef] [PubMed]
- Schären, M.; Kiri, K.; Riede, S.; Gardener, M.; Meyer, U.; Hummel, J.; Urich, T.; Breves, G.; Dänicke, S. Alterations in the Rumen Liquid-, Particle- and Epithelium-Associated Microbiota of Dairy Cows during the Transition from a Silage- and Concentrate-Based Ration to Pasture in Spring. Front. Microbiol. 2017, 8, 744. [Google Scholar] [CrossRef] [PubMed]
- Petri, R.M.; Schwaiger, T.; Penner, G.B.; Beauchemin, K.A.; Forster, R.J.; McKinnon, J.J.; McAllister, T.A. Changes in the Rumen Epimural Bacterial Diversity of Beef Cattle as Affected by Diet and Induced Ruminal Acidosis. Appl. Environ. Microbiol. 2013, 79, 3744–3755. [Google Scholar] [CrossRef] [PubMed]
- Sadet-Bourgeteau, S.; Martin, C.; Morgavi, D.P. Bacterial diversity dynamics in rumen epithelium of wethers fed forage and mixed concentrate forage diets. Vet. Microbiol. 2010, 146, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Soltis, M.P.; Henniger, M.T.; Egert-Mclean, A.M.; Voy, B.H.; Moorey, S.E.; Schneider, L.G.; Shepherd, E.A.; Christopher, C.; Campagna, S.R.; Smith, J.S.; et al. Rumen biogeographical regions and their impact on microbial and metabolome varition. Front. Anim. Sci. 2022, accepted. [Google Scholar]
- Welkie, D.G.; Stevenson, D.M.; Weimer, P.J. ARISA analysis of ruminal bacterial community dynamics in lactating dairy cows during the feeding cycle. Anaerobe 2010, 16, 94–100. [Google Scholar] [CrossRef]
- De Mulder, T.; Goossens, K.; Peiren, N.; Vandaele, L.; Haegeman, A.; De Tender, C.; Ruttink, T.; de Wiele, T.V.; De Campeneere, S. Exploring the methanogen and bacterial communities of rumen environments: Solid adherent, fluid and epimural. FEMS Microbiol. Ecol. 2017, 93, fiw251. [Google Scholar] [CrossRef]
- Mizrahi, I. Rumen Symbioses. In The Prokaryotes: Prokaryotic Biology and Symbiotic Associations; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 533–544. [Google Scholar]
- Mizrahi, I.; Wallace, R.J.; Moraïs, S. The rumen microbiome: Balancing food security and environmental impacts. Nat. Rev. Microbiol. 2021, 19, 553–566. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soltis, M.P.; Moorey, S.E.; Egert-McLean, A.M.; Voy, B.H.; Shepherd, E.A.; Myer, P.R. Rumen Biogeographical Regions and Microbiome Variation. Microorganisms 2023, 11, 747. https://doi.org/10.3390/microorganisms11030747
Soltis MP, Moorey SE, Egert-McLean AM, Voy BH, Shepherd EA, Myer PR. Rumen Biogeographical Regions and Microbiome Variation. Microorganisms. 2023; 11(3):747. https://doi.org/10.3390/microorganisms11030747
Chicago/Turabian StyleSoltis, Macey P., Sarah E. Moorey, Amanda M. Egert-McLean, Brynn H. Voy, Elizabeth A. Shepherd, and Phillip R. Myer. 2023. "Rumen Biogeographical Regions and Microbiome Variation" Microorganisms 11, no. 3: 747. https://doi.org/10.3390/microorganisms11030747
APA StyleSoltis, M. P., Moorey, S. E., Egert-McLean, A. M., Voy, B. H., Shepherd, E. A., & Myer, P. R. (2023). Rumen Biogeographical Regions and Microbiome Variation. Microorganisms, 11(3), 747. https://doi.org/10.3390/microorganisms11030747





