Influence of Extremely High Pressure and Oxygen on Hydrocarbon-Enriched Microbial Communities in Sediments from the Challenger Deep, Mariana Trench
Abstract
1. Introduction
2. Materials and Methods
2.1. Sediments Sampling
2.2. Enrichments of n-Alkanes Experiments under High-Pressure
2.3. DNA Extraction
2.4. PCR Amplification of 16S rRNA Gene
2.5. Illumina Miseq Sequencing and Data Processing
2.6. Statistical and Ecological Analyses
3. Results
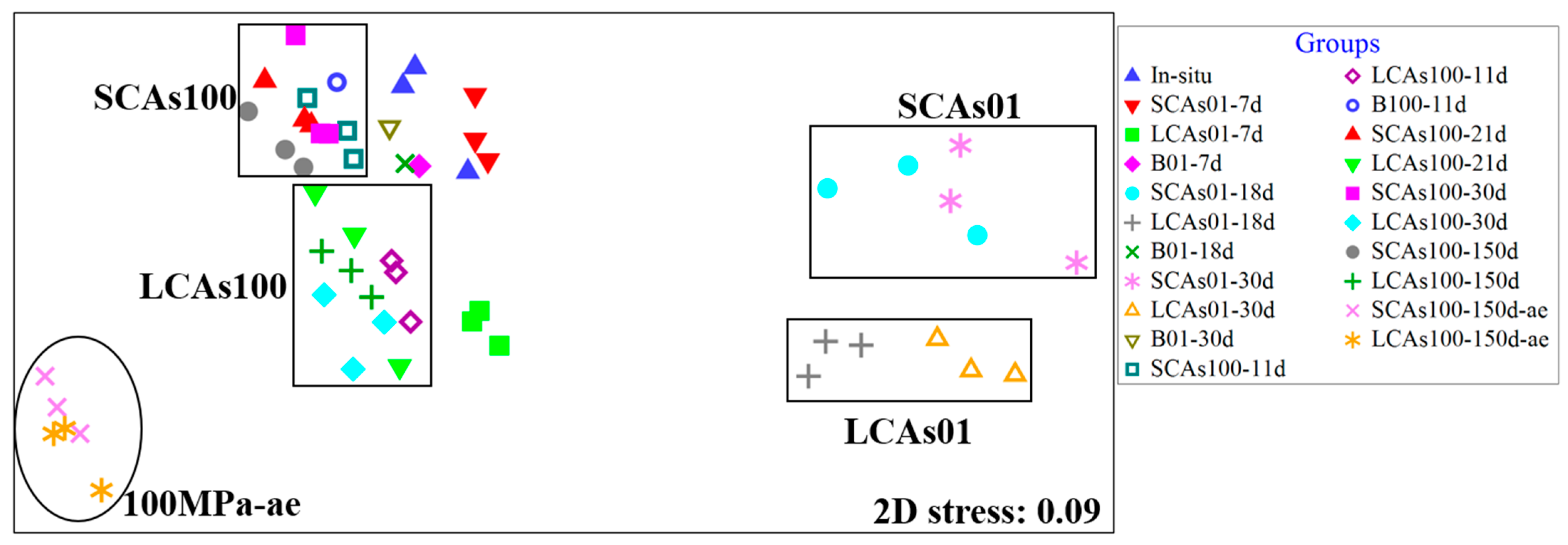
3.1. Analysis of Microbial Diversity
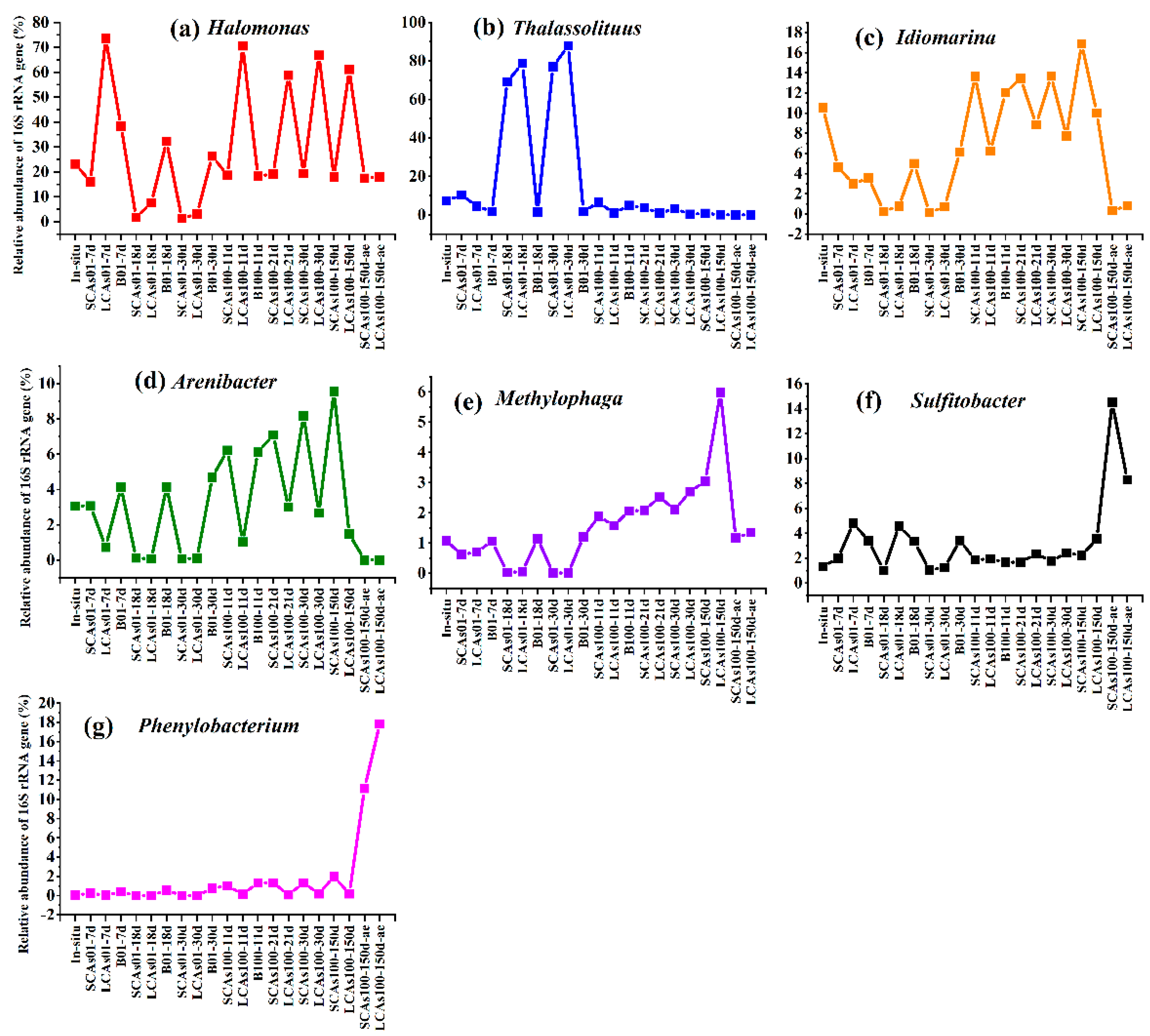
3.2. Microbial Community Structure
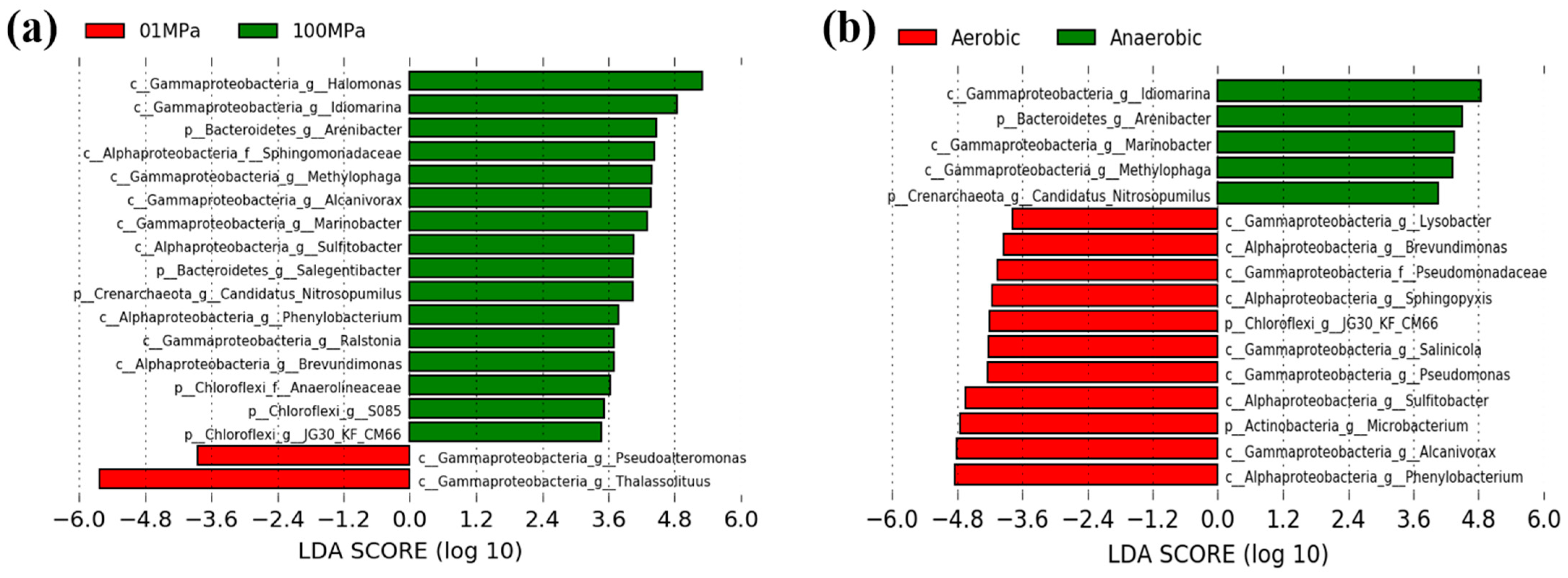
3.3. LDA Analysis
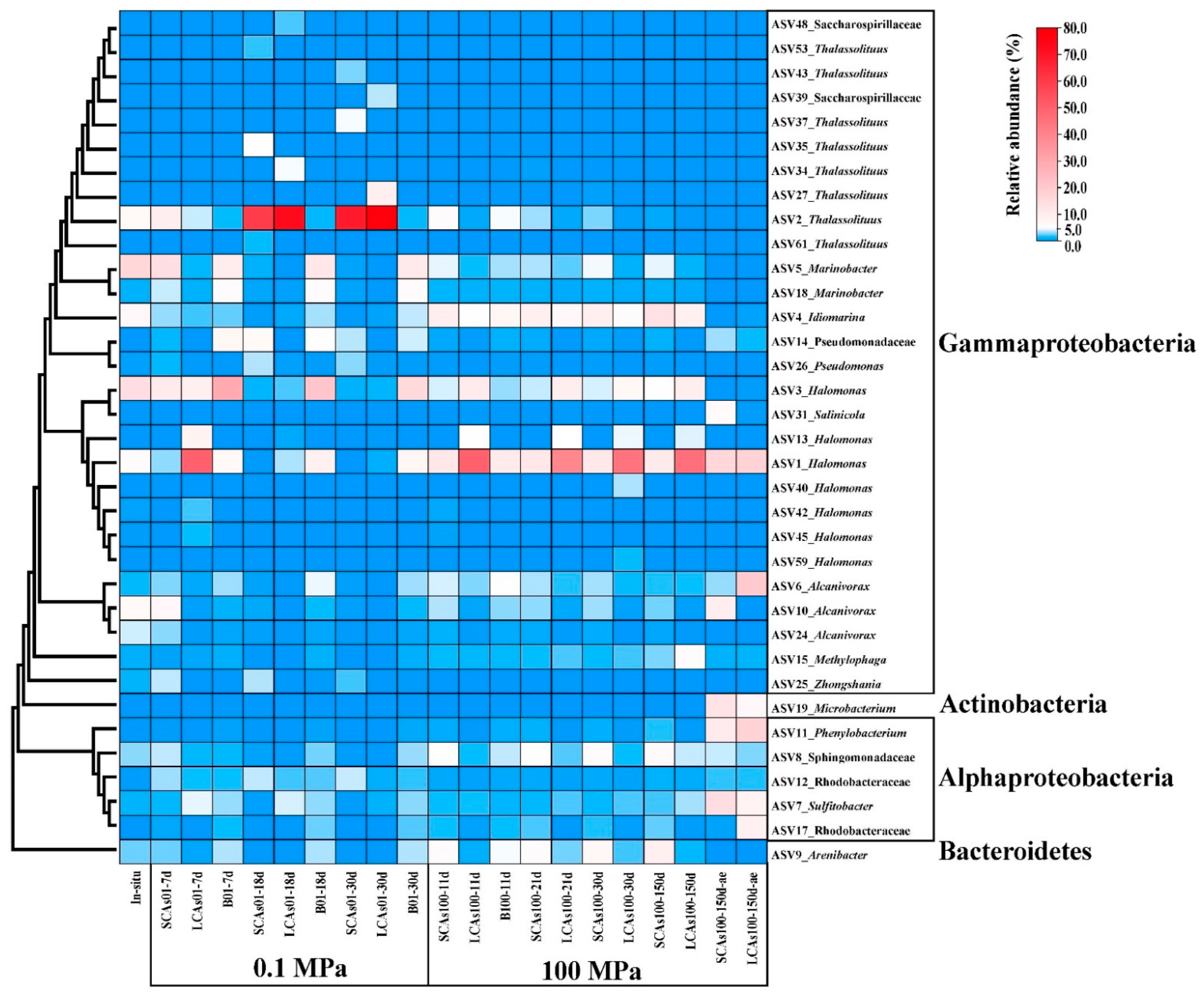
3.4. Phylogenetic Analysis
4. Discussion
4.1. Effect of Hydrostatic Pressure on Microbial Diversity and Community Composition in n-Alkane Enrichment Cultures
4.2. Effect of Different Hydrocarbon Components on Microbial Diversity and Community Structure
4.3. Effect of Oxygen Concentration on Microbial Community Structure of n-Alkanes Enrichment Cultures
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schirmer, A.; Rude, M.A.; Li, X.; Popova, E.; del Cardayre, S.B. Microbial Biosynthesis of Alkanes. Science 2010, 329, 559–562. [Google Scholar] [CrossRef] [PubMed]
- Lea-Smith, D.J.; Biller, S.J.; Davey, M.P.; Cotton, C.A.R.; Perez Sepulveda, B.M.; Turchyn, A.V.; Scanlan, D.J.; Smith, A.G.; Chisholm, S.W.; Howe, C.J. Contribution of cyanobacterial alkane production to the ocean hydrocarbon cycle. Proc. Natl. Acad. Sci. USA 2015, 112, 13591–13596. [Google Scholar] [CrossRef] [PubMed]
- Love, C.R.; Arrington, E.C.; Gosselin, K.M.; Reddy, C.M.; Van Mooy, B.A.S.; Nelson, R.K.; Valentine, D.L. Microbial production and consumption of hydrocarbons in the global ocean. Nat. Microbiol. 2021, 6, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Widdel, F.; Knittel, K.; Galushko, A. Anaerobic Hydrocarbon-Degrading Microorganisms: An Overview. In Handbook of Hydrocarbon and Lipid Microbiology; Timmis, K.N., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 1997–2021. [Google Scholar]
- Rojo, F. Degradation of alkanes by bacteria. Environ. Microbiol. 2009, 11, 2477–2490. [Google Scholar] [CrossRef]
- Valentine, D.L.; Reddy, C.M. Latent hydrocarbons from cyanobacteria. Proc. Natl. Acad. Sci. USA 2015, 112, 13434–13435. [Google Scholar] [CrossRef]
- Grossi, V.; Yakimov, M.M.; Al Ali, B.; Tapilatu, Y.; Cuny, P.; Goutx, M.; La Cono, V.; Giuliano, L.; Tamburini, C. Hydrostatic pressure affects membrane and storage lipid compositions of the piezotolerant hydrocarbon-degrading Marinobacter hydrocarbonoclasticus strain# 5. Environ. Microbiol. 2010, 12, 2020–2033. [Google Scholar]
- Xue, J.; Yu, Y.; Bai, Y.; Wang, L.; Wu, Y. Marine oil-degrading microorganisms and biodegradation process of petroleum hydrocarbon in marine environments: A review. Curr. Microbiol. 2015, 71, 220–228. [Google Scholar] [CrossRef]
- Hazen, T.C.; Prince, R.C.; Mahmoudi, N. Marine oil biodegradation. Environ. Sci. Technol. 2016, 50, 2121–2129. [Google Scholar] [CrossRef]
- Harayama, S.; Kasai, Y.; Hara, A. Microbial communities in oil-contaminated seawater. Curr. Opin. Biotechnol. 2004, 15, 205–214. [Google Scholar] [CrossRef]
- Coulon, F.; McKew, B.A.; Osborn, A.M.; McGenity, T.J.; Timmis, K.N. Effects of temperature and biostimulation on oil-degrading microbial communities in temperate estuarine waters. Environ. Microbiol. 2007, 9, 177–186. [Google Scholar] [CrossRef]
- Hazen, T.C.; Dubinsky, E.A.; DeSantis, T.Z.; Andersen, G.L.; Piceno, Y.M.; Singh, N.; Jansson, J.K.; Probst, A.; Borglin, S.E.; Fortney, J.L. Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science 2010, 330, 204–208. [Google Scholar] [CrossRef] [PubMed]
- Shao, Z.; Wang, W. Enzymes and genes involved in aerobic alkane degradation. Front. Microbiol. 2013, 4, 116. [Google Scholar] [CrossRef]
- Liu, J.; Zheng, Y.; Lin, H.; Wang, X.; Li, M.; Liu, Y.; Yu, M.; Zhao, M.; Pedentchouk, N.; Lea-Smith, D.J. Proliferation of hydrocarbon-degrading microbes at the bottom of the Mariana Trench. Microbiome 2019, 7, 47. [Google Scholar] [CrossRef] [PubMed]
- Yakimov, M.M.; Giuliano, L.; Gentile, G.; Crisafi, E.; Chernikova, T.N.; Abraham, W.-R.; Lünsdorf, H.; Timmis, K.N.; Golyshin, P.N. Oleispira antarctica gen. nov., sp. nov., a novel hydrocarbonoclastic marine bacterium isolated from Antarctic coastal sea water. Int. J. Syst. Evol. Microbiol. 2003, 53, 779–785. [Google Scholar] [CrossRef]
- Ron, E.Z.; Rosenberg, E. Biosurfactants and oil bioremediation. Curr. Opin. Biotechnol. 2002, 13, 249–252. [Google Scholar] [CrossRef] [PubMed]
- Rathore, D.S.; Sheikh, M.; Singh, S.P. Marine Actinobacteria: New Horizons in Bioremediation. In Recent Developments in Microbial Technologies; Prasad, R., Kumar, V., Singh, J., Upadhyaya, C.P., Eds.; Springer Nature: Singapore, 2021; pp. 425–449. [Google Scholar]
- Margesin, R.; Schinner, F. Biodegradation and bioremediation of hydrocarbons in extreme environments. Appl. Microbiol. Biotechnol. 2001, 56, 650–663. [Google Scholar] [CrossRef]
- Scoma, A.; Yakimov, M.M.; Boon, N. Challenging Oil Bioremediation at Deep-Sea Hydrostatic Pressure. Front. Microbiol. 2016, 7, 1203. [Google Scholar] [CrossRef]
- Schwarz, J.R.; Walker, J.D.; Colwell, R.R. Deep-sea bacteria: Growth and utilization of n-hexadecane at in situ temperature and pressure. Can. J. Microbiol. 1975, 21, 682–687. [Google Scholar] [CrossRef]
- Head, I.M.; Jones, D.M.; Larter, S.R. Biological activity in the deep subsurface and the origin of heavy oil. Nature 2003, 426, 344–352. [Google Scholar] [CrossRef]
- Schedler, M.; Hiessl, R.; Valladares Juárez, A.G.; Gust, G.; Müller, R. Effect of high pressure on hydrocarbon-degrading bacteria. AMB Express 2014, 4, 77. [Google Scholar] [CrossRef]
- Prince, R.C.; Nash, G.W.; Hill, S.J. The biodegradation of crude oil in the deep ocean. Mar. Pollut. Bull. 2016, 111, 354–357. [Google Scholar] [CrossRef]
- Scoma, A.; Boon, N. Osmotic Stress Confers Enhanced Cell Integrity to Hydrostatic Pressure but Impairs Growth in Alcanivorax borkumensis SK2. Front. Microbiol. 2016, 7, 729. [Google Scholar] [CrossRef] [PubMed]
- Marietou, A.; Chastain, R.; Beulig, F.; Scoma, A.; Hazen, T.C.; Bartlett, D.H. The Effect of Hydrostatic Pressure on Enrichments of Hydrocarbon Degrading Microbes from the Gulf of Mexico Following the Deepwater Horizon Oil Spill. Front. Microbiol. 2018, 9, 808. [Google Scholar] [CrossRef] [PubMed]
- Hackbusch, S.; Noirungsee, N.; Viamonte, J.; Sun, X.; Bubenheim, P.; Kostka, J.E.; Müller, R.; Liese, A. Influence of pressure and dispersant on oil biodegradation by a newly isolated Rhodococcus strain from deep-sea sediments of the gulf of Mexico. Mar. Pollut. Bull. 2020, 150, 110683. [Google Scholar] [CrossRef] [PubMed]
- Scoma, A.; Barbato, M.; Borin, S.; Daffonchio, D.; Boon, N. An impaired metabolic response to hydrostatic pressure explains Alcanivorax borkumensis recorded distribution in the deep marine water column. Sci. Rep. 2016, 6, 31316. [Google Scholar] [CrossRef]
- Nguyen, U.T.; Lincoln, S.A.; Valladares Juárez, A.G.; Schedler, M.; Macalady, J.L.; Müller, R.; Freeman, K.H. The influence of pressure on crude oil biodegradation in shallow and deep Gulf of Mexico sediments. PLoS ONE 2018, 13, e0199784. [Google Scholar] [CrossRef] [PubMed]
- Hackbusch, S. The Influence of Elevated Pressure and Hydrocarbon Input on the Deep Sea Microbial Community of the Gulf of Mexico; Technische Universität Hamburg: Hamburg, Germany, 2019. [Google Scholar]
- Fasca, H.; de Castilho, L.V.A.; de Castilho, J.F.M.; Pasqualino, I.P.; Alvarez, V.M.; de Azevedo Jurelevicius, D.; Seldin, L. Response of marine bacteria to oil contamination and to high pressure and low temperature deep sea conditions. MicrobiologyOpen 2018, 7, e00550. [Google Scholar] [CrossRef]
- McKew, B.A.; Coulon, F.; Osborn, A.M.; Timmis, K.N.; McGenity, T.J. Determining the identity and roles of oil-metabolizing marine bacteria from the Thames estuary, UK. Environ. Microbiol. 2007, 9, 165–176. [Google Scholar] [CrossRef]
- Potts, L.D.; Perez Calderon, L.J.; Gontikaki, E.; Keith, L.; Gubry-Rangin, C.; Anderson, J.A.; Witte, U. Effect of spatial origin and hydrocarbon composition on bacterial consortia community structure and hydrocarbon biodegradation rates. FEMS Microbiol. Ecol. 2018, 94, fiy127. [Google Scholar] [CrossRef]
- Valentine, D.L.; Kessler, J.D.; Redmond, M.C.; Mendes, S.D.; Heintz, M.B.; Farwell, C.; Hu, L.; Kinnaman, F.S.; Yvon-Lewis, S.; Du, M.; et al. Propane Respiration Jump-Starts Microbial Response to a Deep Oil Spill. Science 2010, 330, 208–211. [Google Scholar] [CrossRef]
- Kessler, J.D.; Valentine, D.L.; Redmond, M.C.; Du, M.; Chan, E.W.; Mendes, S.D.; Quiroz, E.W.; Villanueva, C.J.; Shusta, S.S.; Werra, L.M.; et al. A Persistent Oxygen Anomaly Reveals the Fate of Spilled Methane in the Deep Gulf of Mexico. Science 2011, 331, 312–315. [Google Scholar] [CrossRef] [PubMed]
- Redmond, M.C.; Valentine, D.L. Natural gas and temperature structured a microbial community response to the Deepwater Horizon oil spill. Proc. Natl. Acad. Sci. USA 2012, 109, 20292–20297. [Google Scholar] [CrossRef]
- Mason, O.U.; Hazen, T.C.; Borglin, S.; Chain, P.S.G.; Dubinsky, E.A.; Fortney, J.L.; Han, J.; Holman, H.-Y.N.; Hultman, J.; Lamendella, R.; et al. Metagenome, metatranscriptome and single-cell sequencing reveal microbial response to Deepwater Horizon oil spill. ISME J. 2012, 6, 1715–1727. [Google Scholar] [CrossRef]
- Head, I.M.; Gray, N.D.; Larter, S.R. Life in the slow lane; biogeochemistry of biodegraded petroleum containing reservoirs and implications for energy recovery and carbon management. Front. Microbiol. 2014, 5, 566. [Google Scholar] [CrossRef] [PubMed]
- Prince, R.C.; Walters, C.C. 19—Biodegradation of oil hydrocarbons and its implications for source identification. In Standard Handbook Oil Spill Environmental Forensics, 2nd ed.; Stout, S.A., Wang, Z., Eds.; Academic Press: Boston, MA, USA, 2016; pp. 869–916. [Google Scholar]
- Prince, R.C.; McFarlin, K.M.; Butler, J.D.; Febbo, E.J.; Wang, F.C.Y.; Nedwed, T.J. The primary biodegradation of dispersed crude oil in the sea. Chemosphere 2013, 90, 521–526. [Google Scholar] [CrossRef]
- Netzer, F.v.; Pilloni, G.; Kleindienst, S.; Krüger, M.; Knittel, K.; Gründger, F.; Lueders, T. Enhanced Gene Detection Assays for Fumarate-Adding Enzymes Allow Uncovering of Anaerobic Hydrocarbon Degraders in Terrestrial and Marine Systems. Appl. Environ. Microbiol. 2013, 79, 543–552. [Google Scholar] [CrossRef] [PubMed]
- Stagars, M.H.; Ruff, S.E.; Amann, R.; Knittel, K. High Diversity of Anaerobic Alkane-Degrading Microbial Communities in Marine Seep Sediments Based on (1-methylalkyl) succinate Synthase Genes. Front. Microbiol. 2016, 6, 1511. [Google Scholar] [CrossRef]
- van Beilen, J.B.; Funhoff, E.G. Alkane hydroxylases involved in microbial alkane degradation. Appl. Microbiol. Biotechnol. 2007, 74, 13–21. [Google Scholar] [CrossRef]
- Austin, R.N.; Groves, J.T. Alkane-oxidizing metalloenzymes in the carbon cycle. Metallomics 2011, 3, 775–787. [Google Scholar] [CrossRef]
- Throne-Holst, M.; Wentzel, A.; Ellingsen, T.E.; Kotlar, H.-K.; Zotchev, S.B. Identification of Novel Genes Involved in Long-Chain n-Alkane Degradation by Acinetobacter sp. Strain DSM 17874. Appl. Environ. Microbiol. 2007, 73, 3327–3332. [Google Scholar] [CrossRef]
- Feng, L.; Wang, W.; Cheng, J.; Ren, Y.; Zhao, G.; Gao, C.; Tang, Y.; Liu, X.; Han, W.; Peng, X.; et al. Genome and proteome of long-chain alkane degrading Geobacillus thermodenitrificans NG80-2 isolated from a deep-subsurface oil reservoir. Proc. Natl. Acad. Sci. USA 2007, 104, 5602–5607. [Google Scholar] [CrossRef]
- Foght, J. Anaerobic Biodegradation of Aromatic Hydrocarbons: Pathways and Prospects. Microb. Physiol. 2008, 15, 93–120. [Google Scholar] [CrossRef]
- Heider, J.; Schühle, K. Anaerobic Biodegradation of Hydrocarbons Including Methane; Springer: Berlin/Heidelberg, Germany, 2006; pp. 605–634. [Google Scholar]
- Gray, N.D.; Sherry, A.; Hubert, C.; Dolfing, J.; Head, I.M. Chapter 5—Methanogenic Degradation of Petroleum Hydrocarbons in Subsurface Environments: Remediation, Heavy Oil Formation, and Energy Recovery. In Advances in Applied Microbiology; Laskin, A.I., Sariaslani, S., Gadd, G.M., Eds.; Academic Press: Cambridge, MA, USA, 2010; Volume 72, pp. 137–161. [Google Scholar]
- Khelifi, N.; Amin Ali, O.; Roche, P.; Grossi, V.; Brochier-Armanet, C.; Valette, O.; Ollivier, B.; Dolla, A.; Hirschler-Réa, A. Anaerobic oxidation of long-chain n-alkanes by the hyperthermophilic sulfate-reducing archaeon, Archaeoglobus fulgidus. ISME J. 2014, 8, 2153–2166. [Google Scholar] [CrossRef]
- Acosta-González, A.; Rosselló-Móra, R.; Marqués, S. Characterization of the anaerobic microbial community in oil-polluted subtidal sediments: Aromatic biodegradation potential after the Prestige oil spill. Environ. Microbiol. 2013, 15, 77–92. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.M.; Wawrik, B.; Isom, C.; Boling, W.B.; Callaghan, A.V. Interrogation of Chesapeake Bay sediment microbial communities for intrinsic alkane-utilizing potential under anaerobic conditions. FEMS Microbiol Ecol 2015, 91, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Jamieson, A.J.; Fujii, T.; Mayor, D.J.; Solan, M.; Priede, I.G. Hadal trenches: The ecology of the deepest places on Earth. Trends Ecol. Evol. 2010, 25, 190–197. [Google Scholar] [CrossRef]
- Nunoura, T.; Takaki, Y.; Hirai, M.; Shimamura, S.; Makabe, A.; Koide, O.; Kikuchi, T.; Miyazaki, J.; Koba, K.; Yoshida, N. Hadal biosphere: Insight into the microbial ecosystem in the deepest ocean on Earth. Proc. Natl. Acad. Sci. USA 2015, 112, E1230–E1236. [Google Scholar] [CrossRef]
- Liu, R.; Wang, L.; Wei, Y.; Fang, J. The hadal biosphere: Recent insights and new directions. Deep Sea Res. Part II Top. Stud. Oceanogr. 2018, 155, 11–18. [Google Scholar] [CrossRef]
- Guan, H.; Chen, L.; Luo, M.; Liu, L.; Mao, S.; Ge, H.; Zhang, M.; Fang, J.; Chen, D. Composition and origin of lipid biomarkers in the surface sediments from the southern Challenger Deep, Mariana Trench. Geosci. Front. 2019, 10, 351–360. [Google Scholar] [CrossRef]
- Liu, C.; Shao, Z. Alcanivorax dieselolei sp. nov., a novel alkane-degrading bacterium isolated from sea water and deep-sea sediment. Int. J. Syst. Evol. Microbiol. 2005, 55, 1181–1186. [Google Scholar] [CrossRef]
- Cui, Z.; Lai, Q.; Dong, C.; Shao, Z. Biodiversity of polycyclic aromatic hydrocarbon-degrading bacteria from deep sea sediments of the Middle Atlantic Ridge. Environ. Microbiol. 2008, 10, 2138–2149. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Lai, Q.; Cui, Z.; Tan, T.; Shao, Z. A pyrene-degrading consortium from deep-sea sediment of the West Pacific and its key member Cycloclasticus sp. P1. Environ. Microbiol. 2008, 10, 1948–1963. [Google Scholar] [CrossRef]
- Tapilatu, Y.; Acquaviva, M.; Guigue, C.; Miralles, G.; Bertrand, J.-C.; Cuny, P. Isolation of alkane-degrading bacteria from deep-sea Mediterranean sediments. Lett. Appl. Microbiol. 2010, 50, 234–236. [Google Scholar] [CrossRef] [PubMed]
- Lai, Q.; Wang, J.; Gu, L.; Zheng, T.; Shao, Z. Alcanivorax marinus sp. nov., isolated from deep-sea water. Int. J. Syst. Evol. Microbiol. 2013, 63, 4428–4432. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Wei, X.; Song, W.; Wang, L.; Cao, J.; Wu, J.; Thomas, T.; Jin, T.; Wang, Z.; Wei, W.; et al. Novel Chloroflexi genomes from the deepest ocean reveal metabolic strategies for the adaptation to deep-sea habitats. Microbiome 2022, 10, 75. [Google Scholar] [CrossRef]
- Liu, Y.; Zeng, M.; Xie, Z.; Ning, D.; Zhou, J.; Yu, X.; Liu, R.; Zhang, L.; Fang, J. Microbial Community Structure and Ecological Networks during Simulation of Diatom Sinking. Microorganisms 2022, 10, 639. [Google Scholar] [CrossRef]
- Walters, W.; Hyde, E.R.; Berg-Lyons, D.; Ackermann, G.; Humphrey, G.; Parada, A.; Gilbert, J.A.; Jansson, J.K.; Caporaso, J.G.; Fuhrman, J.A.; et al. Improved Bacterial 16S rRNA Gene (V4 and V4-5) and Fungal Internal Transcribed Spacer Marker Gene Primers for Microbial Community Surveys. Msystems 2016, 1, e00009–e00015. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F. Reproducible, Interactive, Scalable, and Extensible Microbiome Data Science using QIIME 2. Nat. Biotechnol. 2019, 852–857. [Google Scholar] [CrossRef]
- Clarke, K.; Gorley, R. Getting started with PRIMER v7. PRIMER-E Plymouth Plymouth Mar. Lab. 2015, 20. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Kleindienst, S.; Grim, S.; Sogin, M.; Bracco, A.; Crespo-Medina, M.; Joye, S.B. Diverse, rare microbial taxa responded to the Deepwater Horizon deep-sea hydrocarbon plume. ISME J. 2016, 10, 400–415. [Google Scholar] [CrossRef] [PubMed]
- Jurelevicius, D.; Alvarez, V.M.; Marques, J.M.; Lima, L.R.F.D.S.; Dias, F.D.A.; Seldin, L. Bacterial Community Response to Petroleum Hydrocarbon Amendments in Freshwater, Marine, and Hypersaline Water-Containing Microcosms. Appl. Environ. Microbiol. 2013, 79, 5927–5935. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Liu, J. Evaluating bacterial community structures in oil collected from the sea surface and sediment in the northern Gulf of Mexico after the Deepwater Horizon oil spill. MicrobiologyOpen 2013, 2, 492–504. [Google Scholar] [CrossRef]
- Yakimov, M.M.; Timmis, K.N.; Golyshin, P.N. Obligate oil-degrading marine bacteria. Curr. Opin. Biotechnol. 2007, 18, 257–266. [Google Scholar] [CrossRef]
- Jung, S.W.; Park, J.S.; Kown, O.Y.; Kang, J.-H.; Shim, W.J.; Kim, Y.-O. Effects of crude oil on marine microbial communities in short term outdoor microcosms. J. Microbiol. 2010, 48, 594–600. [Google Scholar] [CrossRef]
- Dubinsky, E.A.; Conrad, M.E.; Chakraborty, R.; Bill, M.; Borglin, S.E.; Hollibaugh, J.T.; Mason, O.U.; Piceno, Y.M.; Reid, F.C.; Stringfellow, W.T.; et al. Succession of Hydrocarbon-Degrading Bacteria in the Aftermath of the Deepwater Horizon Oil Spill in the Gulf of Mexico. Environ. Sci. Technol. 2013, 47, 10860–10867. [Google Scholar] [CrossRef]
- Gutierrez, T.; Singleton, D.R.; Berry, D.; Yang, T.; Aitken, M.D.; Teske, A. Hydrocarbon-degrading bacteria enriched by the Deepwater Horizon oil spill identified by cultivation and DNA-SIP. ISME J. 2013, 7, 2091–2104. [Google Scholar] [CrossRef]
- Meng, L.; Liu, H.; Bao, M.; Sun, P. Microbial community structure shifts are associated with temperature, dispersants and nutrients in crude oil-contaminated seawaters. Mar. Pollut. Bull. 2016, 111, 203–212. [Google Scholar] [CrossRef]
- Koyama, S.; Kobayashi, H.; Inoue, A.; Miwa, T.; Aizawa, M. Effects of the piezo-tolerance of cultured deep-sea eel cells on survival rates, cell proliferation, and cytoskeletal structures. Extremophiles 2005, 9, 449–460. [Google Scholar] [CrossRef] [PubMed]
- Yakimov, M.M.; Giuliano, L.; Denaro, R.; Crisafi, E.; Chernikova, T.N.; Abraham, W.-R.; Luensdorf, H.; Timmis, K.N.; Golyshin, P.N. Thalassolituus oleivorans gen. nov., sp. nov., a novel marine bacterium that obligately utilizes hydrocarbons. Int. J. Syst. Evol. Microbiol. 2004, 54, 141–148. [Google Scholar] [CrossRef]
- Gregson, B.H.; Metodieva, G.; Metodiev, M.V.; Golyshin, P.N.; McKew, B.A. Differential Protein Expression during Growth on Medium Versus Long-Chain Alkanes in the Obligate Marine Hydrocarbon-Degrading Bacterium Thalassolituus oleivorans MIL-1. Front. Microbiol. 2018, 9, 3130. [Google Scholar] [CrossRef]
- Yakimov, M.M.; Denaro, R.; Genovese, M.; Cappello, S.; D’Auria, G.; Chernikova, T.N.; Timmis, K.N.; Golyshin, P.N.; Giluliano, L. Natural microbial diversity in superficial sediments of Milazzo Harbor (Sicily) and community successions during microcosm enrichment with various hydrocarbons. Environ. Microbiol. 2005, 7, 1426–1441. [Google Scholar] [CrossRef] [PubMed]
- McKew, B.A.; Coulon, F.; Yakimov, M.M.; Denaro, R.; Genovese, M.; Smith, C.J.; Osborn, A.M.; Timmis, K.N.; McGenity, T.J. Efficacy of intervention strategies for bioremediation of crude oil in marine systems and effects on indigenous hydrocarbonoclastic bacteria. Environ. Microbiol. 2007, 9, 1562–1571. [Google Scholar] [CrossRef] [PubMed]
- Camilli, R.; Reddy, C.M.; Yoerger, D.R.; Van Mooy, B.A.S.; Jakuba, M.V.; Kinsey, J.C.; McIntyre, C.P.; Sylva, S.P.; Maloney, J.V. Tracking Hydrocarbon Plume Transport and Biodegradation at Deepwater Horizon. Science 2010, 330, 201–204. [Google Scholar] [CrossRef]
- Kryachko, Y.; Dong, X.; Sensen, C.W.; Voordouw, G. Compositions of microbial communities associated with oil and water in a mesothermic oil field. Antonie Van Leeuwenhoek 2012, 101, 493–506. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, W.; Lai, Q.; Shao, Z. Gene diversity of CYP153A and AlkB alkane hydroxylases in oil-degrading bacteria isolated from the Atlantic Ocean. Environ. Microbiol. 2010, 12, 1230–1242. [Google Scholar] [CrossRef]
- Shao, Z.; Yuan, J.; Lai, Q.; Zheng, T. The Diversity of PAH-degrading bacteria in a deep-sea water column above the Southwest Indian Ridge. Front. Microbiol. 2015, 6, 853. [Google Scholar] [CrossRef]
- Hassanshahian, M.; Boroujeni, N.A. Enrichment and identification of naphthalene-degrading bacteria from the Persian Gulf. Mar. Pollut. Bull. 2016, 107, 59–65. [Google Scholar] [CrossRef]
- Gomes, M.B.; Gonzales-Limache, E.E.; Sousa, S.T.P.; Dellagnezze, B.M.; Sartoratto, A.; Silva, L.C.F.; Gieg, L.M.; Valoni, E.; Souza, R.S.; Torres, A.P.R.; et al. Exploring the potential of halophilic bacteria from oil terminal environments for biosurfactant production and hydrocarbon degradation under high-salinity conditions. Int. Biodeterior. Biodegrad. 2018, 126, 231–242. [Google Scholar] [CrossRef]
- Nzila, A.; Jung, B.K.; Kim, M.-C.; Ibal, J.C.; Budiyanto, F.; Musa, M.M.; Thukair, A.; Kim, S.-J.; Shin, J.-H. Complete genome sequence of the polycyclic aromatic hydrocarbons biodegrading bacterium Idiomarina piscisalsi strain 10PY1A isolated from oil-contaminated soil. Korean J. Microbiol. 2018, 54, 289–292. [Google Scholar] [CrossRef]
- Fakhrzadegan, I.; Hassanshahian, M.; Askari Hesni, M.; Saadatfar, A. A study of crude oil-degrading bacteria from mangrove forests in the Persian Gulf. Mar. Ecol. 2019, 40, e12544. [Google Scholar] [CrossRef]
- Rizzo, C.; Papale, M.; Lo Giudice, A. Idiomarina sp. Isolates from Cold and Temperate Environments as Biosurfactant Producers. J. Mar. Sci. Eng. 2022, 10, 1135. [Google Scholar] [CrossRef]
- Kwon, K.; Kwon, Y.M.; Kim, S.-J. Aerobic Hydrocarbon-Degrading Bacteroidetes. In Taxonomy, Genomics and Ecophysiology of Hydrocarbon-Degrading Microbes; McGenity, T.J., Ed.; Springer International Publishing: Cham, Switzerland, 2019; pp. 73–91. [Google Scholar]
- Gutierrez, T.; Rhodes, G.; Mishamandani, S.; Berry, D.; Whitman, W.B.; Nichols, P.D.; Semple, K.T.; Aitken, M.D. Polycyclic Aromatic Hydrocarbon Degradation of Phytoplankton-Associated Arenibacter spp. and Description of Arenibacter algicola sp. nov., an Aromatic Hydrocarbon-Degrading Bacterium. Appl. Environ. Microbiol. 2014, 80, 618–628. [Google Scholar] [CrossRef]
- Gutierrez, T.; Whitman, W.B.; Huntemann, M.; Copeland, A.; Chen, A.; Kyrpides, N.; Markowitz, V.; Pillay, M.; Ivanova, N.; Mikhailova, N.; et al. Genome Sequence of Arenibacter algicola Strain TG409, a Hydrocarbon-Degrading Bacterium Associated with Marine Eukaryotic Phytoplankton. Genome Announc. 2016, 4, e00765-16. [Google Scholar] [CrossRef]
- Mishamandani, S.; Gutierrez, T.; Berry, D.; Aitken, M.D. Response of the bacterial community associated with a cosmopolitan marine diatom to crude oil shows a preference for the biodegradation of aromatic hydrocarbons. Environ. Microbiol. 2016, 18, 1817–1833. [Google Scholar] [CrossRef]
- Bagby, S.C.; Reddy, C.M.; Aeppli, C.; Fisher, G.B.; Valentine, D.L. Persistence and biodegradation of oil at the ocean floor following Deepwater Horizon. Proc. Natl. Acad. Sci. USA 2017, 114, E9–E18. [Google Scholar] [CrossRef]
- Hu, P.; Dubinsky, E.A.; Probst, A.J.; Wang, J.; Sieber, C.M.K.; Tom, L.M.; Gardinali, P.R.; Banfield, J.F.; Atlas, R.M.; Andersen, G.L. Simulation of Deepwater Horizon oil plume reveals substrate specialization within a complex community of hydrocarbon degraders. Proc. Natl. Acad. Sci. USA 2017, 114, 7432–7437. [Google Scholar] [CrossRef]
- Viggor, S.; Juhanson, J.; Jõesaar, M.; Mitt, M.; Truu, J.; Vedler, E.; Heinaru, A. Dynamic changes in the structure of microbial communities in Baltic Sea coastal seawater microcosms modified by crude oil, shale oil or diesel fuel. Microbiol. Res. 2013, 168, 415–427. [Google Scholar] [CrossRef]
- Kostka, J.E.; Prakash, O.; Overholt, W.A.; Green, S.J.; Freyer, G.; Canion, A.; Delgardio, J.; Norton, N.; Hazen, T.C.; Huettel, M. Hydrocarbon-Degrading Bacteria and the Bacterial Community Response in Gulf of Mexico Beach Sands Impacted by the Deepwater Horizon Oil Spill. Appl. Environ. Microbiol. 2011, 77, 7962–7974. [Google Scholar] [CrossRef] [PubMed]
- Mnif, S.; Chamkha, M.; Sayadi, S. Isolation and characterization of Halomonas sp. strain C2SS100, a hydrocarbon-degrading bacterium under hypersaline conditions. J. Appl. Microbiol. 2009, 107, 785–794. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, T.; Whitman, W.B.; Huntemann, M.; Copeland, A.; Chen, A.; Kyrpides, N.; Markowitz, V.; Pillay, M.; Ivanova, N.; Mikhailova, N.; et al. Genome Sequence of Halomonas sp. Strain MCTG39a, a Hydrocarbon-Degrading and Exopolymeric Substance-Producing Bacterium. Genome Announc. 2015, 3, e00793-15. [Google Scholar] [CrossRef]
- Abed, R.M.M.; Al-Sabahi, J.; Al-Maqrashi, F.; Al-Habsi, A.; Al-Hinai, M. Characterization of hydrocarbon-degrading bacteria isolated from oil-contaminated sediments in the Sultanate of Oman and evaluation of bioaugmentation and biostimulation approaches in microcosm experiments. Int. Biodeterior. Biodegrad. 2014, 89, 58–66. [Google Scholar] [CrossRef]
- Melcher, R.J.; Apitz, S.E.; Hemmingsen, B.B. Impact of Irradiation and Polycyclic Aromatic Hydrocarbon Spiking on Microbial Populations in Marine Sediment for Future Aging and Biodegradability Studies. Appl. Environ. Microbiol. 2002, 68, 2858–2868. [Google Scholar] [CrossRef]
- García, M.T.; Mellado, E.; Ostos, J.C.; Ventosa, A. Halomonas organivorans sp. nov., a moderate halophile able to degrade aromatic compounds. Int. J. Syst. Evol. Microbiol. 2004, 54, 1723–1728. [Google Scholar] [CrossRef]
- Wright, M.H.; Bentley, S.R.; Greene, A.C. Draft Genome Sequence of Halomonas sp. Strain ML-15, a Haloalkaliphilic, Polycyclic Aromatic Hydrocarbon-Degrading Bacterium. Microbiol. Resour. Announc. 2020, 9, e01175-20. [Google Scholar] [CrossRef] [PubMed]
- Pepi, M.; Cesàro, A.; Liut, G.; Baldi, F. An antarctic psychrotrophic bacterium Halomonas sp. ANT-3b, growing on n-hexadecane, produces a new emulsyfying glycolipid. FEMS Microbiol. Ecol. 2005, 53, 157–166. [Google Scholar] [CrossRef]
- Yan, F.; Fang, J.; Cao, J.; Wei, Y.; Liu, R.; Wang, L.; Xie, Z. Halomonas piezotolerans sp. nov., a multiple-stress-tolerant bacterium isolated from a deep-sea sediment sample of the New Britain Trench. Int. J. Syst. Evol. Microbiol. 2020, 70, 2560–2568. [Google Scholar] [CrossRef]
- Janvier, M.; Grimont, P.A.D. The genus Methylophaga, a new line of descent within phylogenetic branch γ of proteobacteria. Res. Microbiol. 1995, 146, 543–550. [Google Scholar] [CrossRef]
- Vila, J.; Nieto, J.M.; Mertens, J.; Springael, D.; Grifoll, M. Microbial community structure of a heavy fuel oil-degrading marine consortium: Linking microbial dynamics with polycyclic aromatic hydrocarbon utilization. FEMS Microbiol. Ecol. 2010, 73, 349–362. [Google Scholar] [CrossRef]
- Muangchinda, C.; Rungsihiranrut, A.; Prombutara, P.; Soonglerdsongpha, S.; Pinyakong, O. 16S metagenomic analysis reveals adaptability of a mixed-PAH-degrading consortium isolated from crude oil-contaminated seawater to changing environmental conditions. J. Hazard. Mater. 2018, 357, 119–127. [Google Scholar] [CrossRef]
- Mishamandani, S.; Gutierrez, T.; Aitken, M. DNA-based stable isotope probing coupled with cultivation methods implicates Methylophaga in hydrocarbon degradation. Front. Microbiol. 2014, 5, 76. [Google Scholar] [CrossRef]
- Gutierrez, T.; Aitken, M.D. Role of methylotrophs in the degradation of hydrocarbons during the Deepwater Horizon oil spill. ISME J. 2014, 8, 2543–2545. [Google Scholar] [CrossRef]
- Olajuyigbe, F.M.; Ehiosun, K.I. Assessment of crude oil degradation efficiency of newly isolated actinobacteria reveals untapped bioremediation potentials. Bioremediat. J. 2016, 20, 133–143. [Google Scholar] [CrossRef]
- Margesin, R.; Moertelmaier, C.; Mair, J. Low-temperature biodegradation of petroleum hydrocarbons (n-alkanes, phenol, anthracene, pyrene) by four actinobacterial strains. Int. Biodeterior. Biodegrad. 2013, 84, 185–191. [Google Scholar] [CrossRef]
- Isaac, P.; Martínez, F.L.; Bourguignon, N.; Sánchez, L.A.; Ferrero, M.A. Improved PAHs removal performance by a defined bacterial consortium of indigenous Pseudomonas and actinobacteria from Patagonia, Argentina. Int. Biodeterior. Biodegrad. 2015, 101, 23–31. [Google Scholar] [CrossRef]
- Chaturvedi, S.; Khurana, S.M.P. Importance of Actinobacteria for Bioremediation. In Plant Biotechnology: Progress in Genomic Era; Khurana, S.M.P., Gaur, R.K., Eds.; Springer: Singapore, 2019; pp. 277–307. [Google Scholar]
- De Pasquale, C.; Palazzolo, E.; Piccolo, L.L.; Quatrini, P. Degradation of long-chain n-alkanes in soil microcosms by two actinobacteria. J. Environ. Sci. Health Part A 2012, 47, 374–381. [Google Scholar] [CrossRef]
- Lumactud, R.; Fulthorpe, R.; Sentchilo, V.; Meer, J.R.v.d. Draft Genome Sequence of Microbacterium foliorum Strain 122 Isolated from a Plant Growing in a Chronically Hydrocarbon-Contaminated Site. Genome Announc. 2017, 5, e00434-17. [Google Scholar] [CrossRef]
- Cruz-Hernández, M.A.; Reyes-Peralta, J.; Mendoza-Herrera, A.; Rivera, G.; Bocanegra-García, V. Characterization of a Microbacterium sp. strain isolated from soils contaminated with hydrocarbons in the burgos basin, Mexico. Rev. Int. De Contam. Ambient. 2021, 37. [Google Scholar] [CrossRef]
- Schippers, A.; Bosecker, K.; Spröer, C.; Schumann, P. Microbacterium oleivorans sp. nov. and Microbacterium hydrocarbonoxydans sp. nov., novel crude-oil-degrading Gram-positive bacteria. Int. J. Syst. Evol. Microbiol. 2005, 55, 655–660. [Google Scholar] [CrossRef]
- Sheng, X.F.; He, L.Y.; Zhou, L.; Shen, Y.Y. Characterization of Microbacterium sp. F10a and its role in polycyclic aromatic hydrocarbon removal in low-temperature soil. Can. J. Microbiol. 2009, 55, 529–535. [Google Scholar] [CrossRef]
- Salam, L.B.; Obayori, O.S.; Olatoye, N.O. Biodegradation of anthracene by a novel actinomycete, Microbacterium sp. isolated from tropical hydrocarbon-contaminated soil. World J. Microbiol. Biotechnol. 2014, 30, 335–341. [Google Scholar] [CrossRef]
- Salam, L.; Obayori, O.; Campbell, C.; Ilori, M.; Amund, O. Pyrene biodegradation potentials of an actinomycete, Microbacterium esteraromaticum isolated from tropical hydrocarbon-contaminated soil. J. Microbiol. Biotechnol. Food Sci. 2017, 6, 995–1000. [Google Scholar] [CrossRef]
- Wongbunmak, A.; Khiawjan, S.; Suphantharika, M.; Pongtharangkul, T. BTEX- and naphthalene-degrading bacterium Microbacterium esteraromaticum strain SBS1-7 isolated from estuarine sediment. J. Hazard. Mater. 2017, 339, 82–90. [Google Scholar] [CrossRef]
- Logeshwaran, P.; Subashchandrabose, S.R.; Krishnan, K.; Sivaram, A.K.; Annamalai, P.; Naidu, R.; Megharaj, M. Polycyclic aromatic hydrocarbons biodegradation by fenamiphos degrading Microbacterium esteraromaticum MM1. Environ. Technol. Innov. 2022, 27, 102465. [Google Scholar] [CrossRef]
- Brakstad, O.G.; Lødeng, A.G.G. Microbial Diversity during Biodegradation of Crude Oil in Seawater from the North Sea. Microb. Ecol. 2005, 49, 94–103. [Google Scholar] [CrossRef]
- Størdal, I.F.; Olsen, A.J.; Jenssen, B.M.; Netzer, R.; Hansen, B.H.; Altin, D.; Brakstad, O.G. Concentrations of viable oil-degrading microorganisms are increased in feces from Calanus finmarchicus feeding in petroleum oil dispersions. Mar. Pollut. Bull. 2015, 98, 69–77. [Google Scholar] [CrossRef]
- Liu, J.; Bacosa, H.P.; Liu, Z. Potential Environmental Factors Affecting Oil-Degrading Bacterial Populations in Deep and Surface Waters of the Northern Gulf of Mexico. Front. Microbiol. 2017, 7, 2131. [Google Scholar] [CrossRef]
- Gontikaki, E.; Potts, L.D.; Anderson, J.A.; Witte, U. Hydrocarbon-degrading bacteria in deep-water subarctic sediments (Faroe-Shetland Channel). J. Appl. Microbiol. 2018, 125, 1040–1053. [Google Scholar] [CrossRef]
- Ryther, C.M.; Ortmann, A.C.; Wohlgeschaffen, G.; Robinson, B.J. Temperate Coastal Microbial Communities Rapidly Respond to Low Concentrations of Partially Weathered Diesel. Microb. Ecol. 2022, 84, 1122–1132. [Google Scholar] [CrossRef] [PubMed]
- Prabagaran, S.R.; Manorama, R.; Delille, D.; Shivaji, S. Predominance of Roseobacter, Sulfitobacter, Glaciecola and Psychrobacter in seawater collected off Ushuaia, Argentina, Sub-Antarctica. FEMS Microbiol. Ecol. 2007, 59, 342–355. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, Y.; Yang, L.; Kong, Q.; Zhang, H. Microbial Degradation Mechanisms of Surface Petroleum Contaminated Seawater in a Typical Oil Trading Port. SSRN Electron. J. [CrossRef]
- Mas-Lladó, M.; Piña-Villalonga, J.M.; Brunet-Galmés, I.; Nogales, B.; Bosch, R. Draft Genome Sequences of Two Isolates of the Roseobacter Group, Sulfitobacter sp. Strains 3SOLIMAR09 and 1FIGIMAR09, from Harbors of Mallorca Island (Mediterranean Sea). Genome Announc. 2014, 2, e00350-14. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Pan, H.; Wang, Q.; Ge, Y.; Liu, W.; Christie, P. Enrichment of the soil microbial community in the bioremediation of a petroleum-contaminated soil amended with rice straw or sawdust. Chemosphere 2019, 224, 265–271. [Google Scholar] [CrossRef]
- Kuri, M.L.; Kumari, V.; Roy, S. Phenylobacterium Korensee Best Indigenous Petroleum Hydrocarbon Degrading Bacteria Isolated from Contaminated Soil of Bahror, Alwar Region, India. Int. J. Contemp. Res. Rev. 2019, 10, 20203–20211. [Google Scholar] [CrossRef]
- Kuri, M.; Kumari, V.; Roy, S. Biodegradable Capability of the Indigenous Micrococcus sp. Oil Degrading Bacteria Isolated from Oil Contaminated Soil, Motor Workshop Area of Bahror, Alwar, Rajasthan, India. Int. J. Adv. Eng. Nano Technol. 2021, 4, 1–4. [Google Scholar] [CrossRef]
- Yang, S.; Wen, X.; Zhao, L.; Shi, Y.; Jin, H. Crude oil treatment leads to shift of bacterial communities in soils from the deep active layer and upper permafrost along the China-Russia Crude Oil Pipeline route. PLoS ONE 2014, 9, e96552. [Google Scholar] [CrossRef]
- Rodgers-Vieira, E.A.; Zhang, Z.; Adrion, A.C.; Gold, A.; Aitken, M.D. Identification of Anthraquinone-Degrading Bacteria in Soil Contaminated with Polycyclic Aromatic Hydrocarbons. Appl. Environ. Microbiol. 2015, 81, 3775–3781. [Google Scholar] [CrossRef]
- Singleton, D.R.; Adrion, A.C.; Aitken, M.D. Surfactant-induced bacterial community changes correlated with increased polycyclic aromatic hydrocarbon degradation in contaminated soil. Appl. Microbiol. Biotechnol. 2016, 100, 10165–10177. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Uribe, M.L.; Peña-Cabriales, J.J.; Rivera-Cruz, M.D.C.; Délano-Frier, J.P. Native bacteria isolated from weathered petroleum oil-contaminated soils in Tabasco, Mexico, accelerate the degradation petroleum hydrocarbons in saline soil microcosms. Environ. Technol. Innov. 2021, 23, 101781. [Google Scholar] [CrossRef]
- Wang, B.; Teng, Y.; Yao, H.; Christie, P. Detection of functional microorganisms in benzene [a] pyrene-contaminated soils using DNA-SIP technology. J. Hazard. Mater. 2021, 407, 124788. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Chen, S.; Xie, Z.; Zhang, L.; Wang, J.; Fang, J. Influence of Extremely High Pressure and Oxygen on Hydrocarbon-Enriched Microbial Communities in Sediments from the Challenger Deep, Mariana Trench. Microorganisms 2023, 11, 630. https://doi.org/10.3390/microorganisms11030630
Liu Y, Chen S, Xie Z, Zhang L, Wang J, Fang J. Influence of Extremely High Pressure and Oxygen on Hydrocarbon-Enriched Microbial Communities in Sediments from the Challenger Deep, Mariana Trench. Microorganisms. 2023; 11(3):630. https://doi.org/10.3390/microorganisms11030630
Chicago/Turabian StyleLiu, Ying, Songze Chen, Zhe Xie, Li Zhang, Jiahua Wang, and Jiasong Fang. 2023. "Influence of Extremely High Pressure and Oxygen on Hydrocarbon-Enriched Microbial Communities in Sediments from the Challenger Deep, Mariana Trench" Microorganisms 11, no. 3: 630. https://doi.org/10.3390/microorganisms11030630
APA StyleLiu, Y., Chen, S., Xie, Z., Zhang, L., Wang, J., & Fang, J. (2023). Influence of Extremely High Pressure and Oxygen on Hydrocarbon-Enriched Microbial Communities in Sediments from the Challenger Deep, Mariana Trench. Microorganisms, 11(3), 630. https://doi.org/10.3390/microorganisms11030630




