Development of an Extraction Method to Detect Hepatitis A Virus, Hepatitis E Virus, and Noroviruses in Fish Products
Abstract
1. Introduction
2. Materials and Methods
2.1. Viruses
2.2. Artificial Contamination of Fish Products
2.3. Sample Processing for Recovery of Viruses
2.4. Viral RNA Extraction and RT-qPCR Conditions
2.5. Calculation and Interpretation of Results
2.6. Statistical Analysis
3. Results
3.1. Comparison of Four Methods to Recover MNV-1 from Artificially Contaminated Fish Products
3.2. Validation of the Selected Method for the Detection of HAV, HEV, and Norovirus in Samples
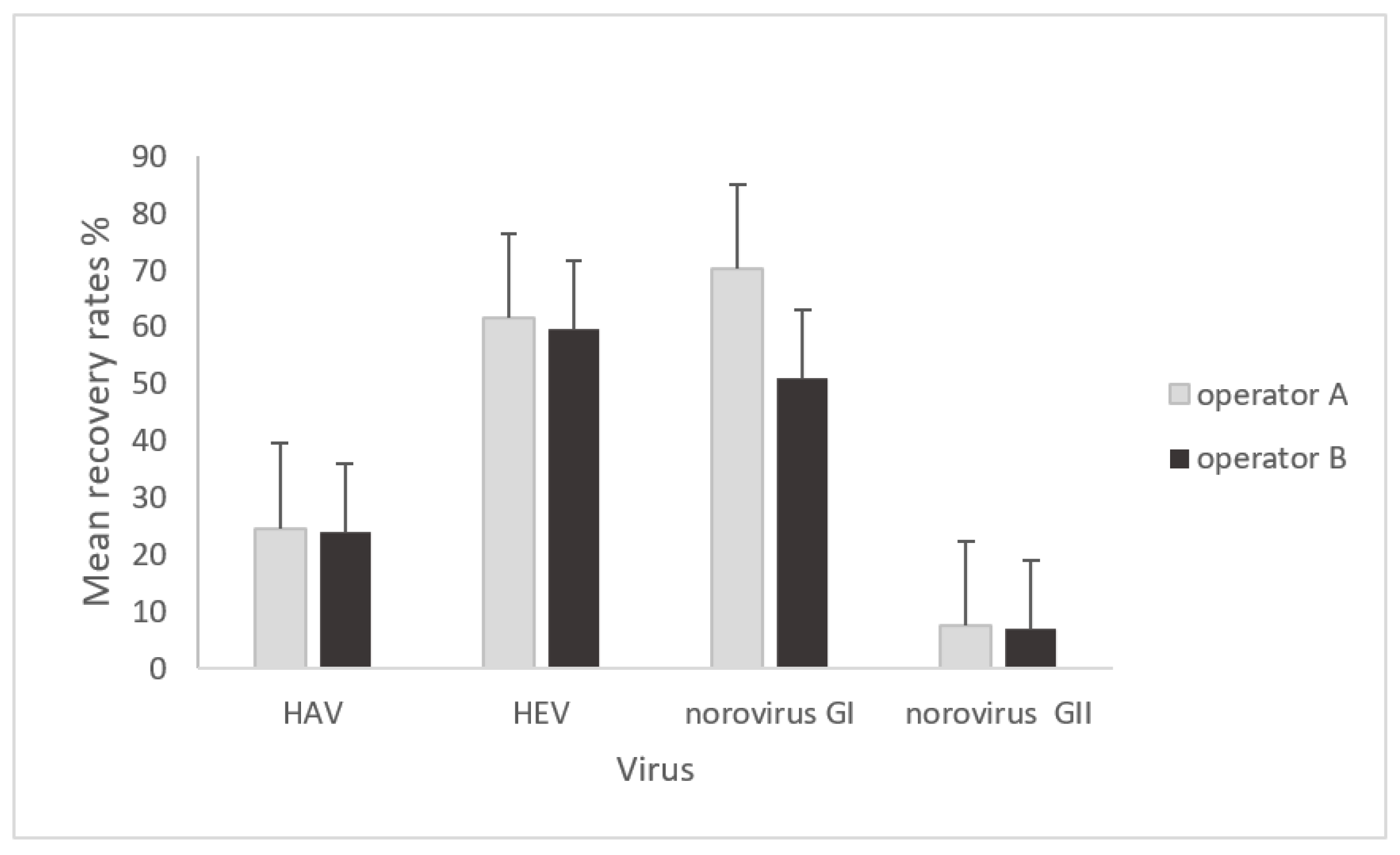
3.2.1. Mean Virus Recoveries of HAV, HEV, and Norovirus by Operator
3.2.2. MNV-1 Recovery Rates and Recovery Rates of the EAC
3.2.3. Viral Recovery Rates and Limits of Detection
3.3. Influence of Experimental Factors on Extraction Yield of Viruses
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barrett, K.A.; Nakao, J.H.; Taylor, E.V.; Eggers, C.; Gould, L.H. Fish-Associated Foodborne Disease Outbreak: United States, 1998–2015. Foodborne Pathog. Dis. 2017, 14, 537–543. [Google Scholar] [CrossRef]
- EFSA, European Food Safety Authority; European Centre for Disease Prevention and Control. The European union one health 2019 Zoonoses Report. EFSA J. 2021, 27, 19. [Google Scholar]
- Santé Publique France, Surveillance des Toxi-Infections Alimentaires Collectives. Données de la Déclaration Obligatoire. 2020. Available online: https://www.santepubliquefrance.fr/maladies-et-traumatismes/maladies-infectieuses-d-origine-alimentaire/toxi-infections-alimentaires-collectives/documents/bulletin-national/surveillance-des-toxi-infections-alimentaires-collectives.-donnees-de-la-declaration-obligatoire-2020 (accessed on 21 December 2021).
- Kotwal, G.; Cannon, J.L. Environmental persistence and transfer of enteric viruses. Curr. Opin. Virol. 2014, 4, 37–43. [Google Scholar] [CrossRef]
- Matthews, J.E.; Dickey, B.W.; Miller, R.D.; Felzer, J.R.; Dawson, B.P.; Lee, A.S.; Rocks, J.J.; Kiel, J.; Montes, J.S.; Moe, C.L. The epidemiology of published norovirus outbreaks: A review of risk factors associated with attack rate and genogroup. Epidemiol. Infect. 2012, 140, 1161–1172. [Google Scholar] [CrossRef]
- Dalton, H.R.; Hunter, J.G.; Bendall, R.P. Hepatitis E. Curr. Opin. Infect. Dis. 2013, 26, 471–478. [Google Scholar] [CrossRef]
- Meng, X.J. Hepatitis E virus: Animal reservoirs and zoonotic risk. Vet. Microbiol. 2010, 140, 256–265. [Google Scholar] [CrossRef] [PubMed]
- Pavio, N.; Meng, X.J.; Doceul, V. Zoonotic origin of hepatitis E. Curr. Opin. Virol. 2015, 10, 34–41. [Google Scholar] [CrossRef]
- Baert, L.; Uyttendaele, M.; Debevere, J. Evaluation of viral extraction methods on a broad range of ready-to-eat foods with conventional and real-time RT-PCR for Norovirus GII detection. Int. J. Food Microbiol. 2008, 123, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Barrabeig, I.; Rovira, A.; Buesa, J.; Bartolome, R.; Pinto, R.; Prellezo, H.; Dominguez, A. Foodborne norovirus outbreak: The role of an asymptomatic food handler. BMC Infect. Dis. 2010, 10, 269. [Google Scholar] [CrossRef] [PubMed]
- Franck, K.T.; Lisby, M.; Fonager, J.; Schultz, A.C.; Bottiger, B.; Villif, A.; Absalonsen, H.; Ethelberg, S. Sources of Calicivirus contamination in foodborne outbreaks in Denmark, 2005-2011-the role of the asymptomatic food handler. J. Infect. Dis. 2015, 211, 563–570. [Google Scholar] [CrossRef]
- Rönnqvist, M.; Aho, E.; Mikkela, A.; Ranta, J.; Tuominen, P.; Ratto, M.; Maunula, L. Norovirus transmission between hands, gloves, utensils, and fresh produce during simulated food handling. Appl. Environ. Microbiol. 2014, 80, 5403–5410. [Google Scholar] [CrossRef] [PubMed]
- Stals, A.; Uyttendaele, M.; Baert, L.; Van Coillie, E. Norovirus transfer between foods and food contact materials. J. Food Prot. 2013, 76, 1202–1209. [Google Scholar] [CrossRef] [PubMed]
- Pavoni, E.; Consoli, M.; Suffredini, E.; Arcangeli, G.; Serracca, L.; Battistini, R.; Rossini, I.; Croci, L.; Losio, M.N. Norovirus in seefood: A 9-year monitoring in Italy. Foodborne Pathog. Dis. 2013, 10, 533–539. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Stals, A.; Tang, Q.J.; Uyttendaele, M. Detection of noroviruses in shellfish and semiprocessed fishery products from a Belgian seafood company. J. Food Prot. 2014, 77, 1342–1347. [Google Scholar] [CrossRef] [PubMed]
- Somura, Y.; Nagano, M.; Kimoto, K.; Oda, M.; Mori, K.; Shinkai, T.; Sadamasu, K. Detection of norovirus in food samples collected during suspected food-handler-involved foodborne outbreaks in Tokyo. Lett. Appl. Microbiol. 2019, 69, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Istre, G.R.; Hopkins, R.S. An outbreak of foodborne hepatitis A showing a relationship between dose and incubation period. Am. J. Public Health 1985, 75, 280–281. [Google Scholar] [CrossRef] [PubMed]
- EN ISO 15216-1:2017; Microbiology of the Food Chain–Horizontal Method for Determination of Hepatitis A Virus and Norovirus Using Real-Time RT-PCR—Part1: Method for Quantification. International Organization for Standardization: Geneva, Switzerland, 2017.
- EN ISO 15216-2:2019; Microbiology of the Food Chain-Horizontal Method for Determination of Hepatitis A Virus and Norovirus Using Real-Time RT-PCR—Part2: Method for Detection. International Organization for Standardization: Geneva, Switzerland, 2019.
- Hennechart-Collette, C.; Martin-Latil, S.; Guillier, L.; Perelle, S. Determination of which virus to use as a process control when testing for the presence of hepatitis A virus and norovirus in food and water. Int. J. Food Microbiol. 2015, 202, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Hennechart-Collette, C.; Martin-Latil, S.; Guillier, L.; Perelle, S. Multiplex real-time RT-qPCR for the detection of Norovirus in bottled and tap water using murine norovirus as a process control. J. Appl. Microbiol. 2014, 116, 179–190. [Google Scholar] [CrossRef] [PubMed]
- Kingsley, D.H. An RNA extraction protocol for shellfish-borne viruses. J. Virol. Methods 2007, 141, 58–62. [Google Scholar] [CrossRef]
- Martin-Latil, S.; Hennechart-Collette, C.; Guillier, L.; Perelle, S. Duplex RT-qPCR for the detection of hepatitis E virus in water, using a process control. Int. J. Food Microbiol. 2012, 157, 167–173. [Google Scholar] [CrossRef]
- Wobus, C.E.; Thackray, L.B.; Virgin, H.W. Murine norovirus: A model system to study norovirus biology and pathogenesis. J. Virol. 2006, 80, 5104–5112. [Google Scholar] [CrossRef]
- PR ISO/DIS 16140-4: 2018-01; Microbiology of the Food Chain-Method Validation-Part 4: Protocol for Single-Laboratory (In-House) Method Validation. International Organization for Standardization: Geneva, Switzerland, 2018.
- Hennechart-Collette, C.; Dehan, O.; Laurentie, M.; Fraisse, A.; Martin-Latil, S.; Perelle, S. Detection of norovirus, hepatitis A and hepatitis E viruses in multicomponent foodstuffs. Int. J. Food Microbiol. 2021, 337, 108931. [Google Scholar] [CrossRef] [PubMed]
- Martin-Latil, S.; Hennechart-Collette, C.; Guillier, L.; Perelle, S. Method for HEV detection in raw pig liver products and its implementation for naturally contaminated food. Int. J. Food Microbiol. 2014, 176, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Hennechart-Collette, C.; Martin-Latil, S.; Fraisse, A.; Perelle, S. Comparison of three extraction methods to detect noroviruses in dairy products. Food Microbiol. 2017, 61, 113. [Google Scholar] [CrossRef] [PubMed]
- Hennechart-Collette, C.; Niveau, F.; Martin-Latil, S.; Fraisse, A.; Perelle, S. Development of an extraction method to detect enteric viruses in dressed vegetables. Int. J. Food Microbiol. 2019, 311, 108349. [Google Scholar] [CrossRef]
- Hennechart-Collette, C.; Dehan, O.; Laurentie, M.; Fraisse, A.; Martin-Latil, S.; Perelle, S. Method for detecting norovirus, hepatitis A and hepatitis E viruses in tap and bottled drinking water. Int. J. Food Microbiol. 2022, 377, 109757. [Google Scholar] [CrossRef]
- Wilrich, C.; Wilrich, P.T. Estimation of the POD function and the LOD of a qualitative microbiological measurement method. J. AOAC Int. 2009, 92, 1763–1772. [Google Scholar] [CrossRef]
- Murchie, L.W.; Cruz-Romero, M.; Kerry, J.P.; Linton, M.; Patterson, M.F.; Smiddy, M.; Kelly, A.L. High pressure processing of shellfish: A review of microbiological and other quality aspects. Food Sci. Emerg. Technol. 2005, 6, 257–270. [Google Scholar] [CrossRef]
- Buras, N.; Duek, L.; Niv, S. Reactions of fish to microorganisms in wastewater. Appl. Environ. Microbiol. 1985, 50, 989–995. [Google Scholar] [CrossRef]
- Montalvo Villalba, M.C.; Cruz Martínez, D.; Ahmad, I.; Rodriguez Lay, L.A.; Bello Corredor, M.; Guevara March, C.; Martínez, L.S.; Martínez-Campo, L.S.; Jameel, S. Hepatitis E virus in bottlenose dolphins Tursiops truncatus. Dis. Aquat. Organ. 2017, 123, 13–18. [Google Scholar] [CrossRef]
- Anbazhagi, S.; Kamatchiammal, S. Contamination of seefood by norovirus in India. Int. J. Virol. 2010, 6, 138–149. [Google Scholar] [CrossRef]
- Lekshmi, M.; Das, O.; Kumar, S.; Nayak, B.B. Occurrence of human enterovirus in tropical fish and shellfish and their relationship with fecal indicator bacteria. Vet. World 2018, 11, 1285–1290. [Google Scholar] [CrossRef] [PubMed]
- Lowther, J.A.; Bosch, A.; Butot, S.; Ollivier, J.; Mäde, D.; Rutjes, S.A.; Hardouin, G.; Lombard, B.; in’t Veld, P.; Leclercq, A. Validation of ISO 15216 part 1—Quantification of hepatitis A virus and norovirus in food martrices. Int. J. Food Microbiol. 2017, 288, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Fraisse, A.; Niveau, F.; Hennechart-Collette, C.; Coudray-Meunier, C.; Martin-Latil, S.; Perelle, S. Discrimination of infectious and heat-treated norovirus by combining platinum compounds and real-time RT-PCR. Int. J. Food Microbiol. 2018, 269, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Razafimahefa, R.M.; Ludwig-Begall, L.F.; Le Guyader, F.S.; Farnir, F.; Mauroy, A.; Thiry, E. Optimisation of a PMAxx™-RT-qPCR assay and the preceding extraction method to selectively detect infectious murine norovirus particles in mussels. Food Environ. Virol. 2021, 13, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Comelli, H.L.; Rimstad, E.; Larsen, S.; Myrmel, M. Detection of norovirus genotype I.3b and II.4 in bioaccumulated blue mussels using different virus recovery methods. Int. J. Food Microbiol. 2008, 127, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Langlet, J.; Kaas, L.; Croucher, D.; Hewitt, J. Effect of the Shellfish Proteinase K Digestion Method on Norovirus Capsid Integrity. Food Environ. Virol. 2018, 10, 151–158. [Google Scholar] [CrossRef]
- Nuanualsuwan, S.; Cliver, D.O. Pretreatment to avoid positive RT-PCR results with inactivated viruses. J. Virol. Methods 2002, 104, 217–225. [Google Scholar] [CrossRef]
- Hennechart-Collette, C.; Gonzalez, G.; Fourniol, L.; Fraisse, A.; Beck, C.; Moutailler, S.; Bournez, L.; Dheilly, N.M.; Lacour, S.A.; Lecollinet, S.; et al. Method for tick-borne encephalitis virus detection in raw milk products. Food Microbiol. 2022, 104, 104003. [Google Scholar] [CrossRef]
- Uhrbrand, K.; Myrmel, M.; Maunula, L.; Vainio, K.; Trebbien, R.; Nørrung, B.; Schultz, A.C. Evaluation of a rapid method for recovery of norovirus and hepatitis A virus from oysters and blue mussels. J. Virol. Methods 2010, 169, 70–78. [Google Scholar] [CrossRef]
- Le Guyader, F.S.; Parnaudeau, S.; Schaeffer, J.; Bosch, A.; Loisy, F.; Pommepuy, M.; Atmar, R.L. Detection and quantification of noroviruses in shellfish. Appl. Environ. Microbiol. 2009, 75, 618–624. [Google Scholar] [CrossRef]
- Blaise-Boisseau, S.; Hennechart-Collette, C.; Guillier, L.; Perelle, S. Duplex real-time qRT-PCR for the detection of hepatitis A virus in water and raspberries using the MS2 bacteriophage as a process control. J. Virol. Methods 2010, 166, 48–53. [Google Scholar] [CrossRef]
- Butot, S.; Putallaz, T.; Sanchez, G. Procedure for rapid concentration and detection of enteric viruses from berries and vegetables. Appl. Environ. Microbiol. 2007, 73, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Summa, M.; von Bonsdorff, C.H.; Maunula, L. Evaluation of four virus recovery methods for detecting noroviruses on fresh lettuce, sliced ham, and frozen raspberries. J. Virol. Methods 2012, 183, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Yavarmanesh, M.; Abbaszadegan, M.; Alum, A.; Mortazavi, A.; Habibi Najafi, M.B.; Bassami, M.R.; Nassiri, M.R. Impact of milk components on recovery of viral RNA from MS2 bacteriophage. Food Environ. Virol. 2013, 5, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Yavarmanesh, M.; Abbaszadegan, M.; Mortazavi, A.; Najafi, M.B.; Bassami, M.R.; Nassiri, M.R. Impact of milk components in recovery of the MS2 bacteriophage as an indicator of enteric viruses. J. Virol. Methods 2010, 168, 103–107. [Google Scholar] [CrossRef] [PubMed]

| Processing Factor | Matrix Types |
|---|---|
| Tin can | Fish dumpling Tuna Tuna rillette Salmon |
| Ready to eat meal | Salmon sorrel sauce |
| Raw and smoked (+4 °C) | Tuna albacore Cod Halibut Kipper Haddock |
| Fresh (+4 °C) | Shelled pink shrimp Surimi |
| Repeat Experiments | |||||
|---|---|---|---|---|---|
| Virus contamination levels | R1 | R2 | R3 | R4 | |
| High | Fish dumpling | Tuna | Tuna rillette | Kipper | |
| Medium | Tuna albacore | Shelled pink shrimp | Surimi | Haddock | |
| Low | Cod | Halibut | Salmon sorrel sauce | Salmon | |
| Very low | Shelled pink shrimp | Surimi | Haddock | Tuna albacore | |
| Salmon | Herring | ||||
|---|---|---|---|---|---|
| Method | RNA Extracts | MNV-1 Extraction Yield (% ± SD) | Factor (F) (Diluted/Undiluted) | MNV-1 Extraction Yield (% ± SD) | Factor (F) (Diluted/Undiluted) |
| Method 1 | Undiluted | 55.85 ± 19.09 | 1.04 | 8.37 ± 7.18 | 3.11 |
| 10-fold diluted | 58.17 ± 24.93 | 26.10-± 13.51 | |||
| Method 2 | Undiluted | 3.03 ± 2.09 | 0.88 | 36.77 ± 28.16 | 1.47 |
| 10-fold diluted | 2.68 ± 1.29 | 54.28 ± 28.49 | |||
| Method 3 | Undiluted | 26.48 ± 13.46 | 1.34 | 82.22 ± 27.71 | 0.97 |
| 10-fold diluted | 35.57 ± 10.29 | 79.45 ± 27.93 | |||
| Method 4 | Undiluted | 97.02 ± 7.30 | 0.94 | 86.17 ± 21.43 | 1.05 |
| 10-fold diluted | 91.62 ± 13.00 | 93.50 ± 16.88 | |||
| Virus | Number of Genome Copies/Sample | RNA Extracts | Repeated Experiment R1 (% ± SD) | (F) | Repeated Experiment R2 (% ± SD) | (F) | Repeated Experiment R3 (% ± SD) | (F) | Repeated Experiment R4 (% ± SD) | (F) |
|---|---|---|---|---|---|---|---|---|---|---|
| HAV | 2.9 × 106 | pure | 32.06 ± 11.10 (4/4) | 1.1 | 45.39 ± 10.06 (4/4) | 1.1 | 0.18 ± 0.01 (2/4) | 4.2 | 36.85 ± 5.50 (4/4) | 1.34 |
| 10-fold diluted | 36.56 ± 10.86 (4/4) | 49.48 ± 6.65 (4/4) | 0.75 ± 0.62 (4/4) | 49.36 ± 4.82 (4/4) | ||||||
| 2.9 × 105 | pure | 3.93 ± 0.63 (4/4) | 1.7 | 23.89 ± 6.16 (4/4) | 1.2 | 66.19 ± 6.38 (4/4) | >1.5 | 15.17 ± 3.14 (4/4) | 3.08 | |
| 10-fold diluted | 6.52 ± 1.49 (4/4) | 28.73 ± 10.78 (4/4) | 100.00 (4/4) | 46.86 ± 36.07 (4/4) | ||||||
| 2.9 × 104 | pure | 12.13 ± 9.04 (4/4) | 0.7 | 2.25 ± 2.55 (2/4) | 5.4 | 40.52 ± 9.17 (4/4) | 2.2 | 6.34 ± 3.34 (4/4) | 1.1 | |
| 10-fold diluted | 8.16 ± 6.75 (3/4) | 12.09 ± 12.52 (2/4) | 88.06 ± 61.91 (4/4) | 7.03 ± 1.2 (2/4) | ||||||
| 2.9 × 103 | pure | 6.06 ± 5.21 (3/4) | 0.1 | 21.13 ± 27.82 (4/4) | - | 24.73 ± 25.49 (4/4) | 4.0 | 24.51 ± 22.96 (4/4) | 4.1 | |
| 10-fold diluted | 0.40 (1/4) | nd (0/4) | 100.00 (1/4) | 100.00 (1/4) | ||||||
| MNV-1 | MNV | 77.58 ± 31.29 | 71.21 ± 32.47 | 68.07 ± 52.74 | 43.25 ± 22.23 | |||||
| Total samples with recovery rates > 1% | 30/32 | |||||||||
| HEV | 1.0 × 106 | pure | 100 (4/4) | 1.0 | 55.48 ± 13.42 (4/4) | 0.97 | 19.69 ± 4.83 (4/4) | 0.5 | 94.59 ± 10.82 (4/4) | 0.73 |
| 10-fold diluted | 100 (4/4) | 54.09 ± 18.35 (4/4) | 9.93 ± 7.05 (4/4) | 69.40 ± 33.75 (4/4) | ||||||
| 1.0 × 105 | pure | nd (0/4) | - | 34.46 ± 24.41 (4/4) | - | 100 (4/4) | - | 100 (4/4) | 0.48 | |
| 10-fold diluted | nd (0/4) | nd (0/4) | 0.99 (1/4) | 48.06 ± 41.98 (3/4) | ||||||
| 1.0 × 104 | pure | 100 (1/4) | - | nd (0/4) | - | 4.04 ± 6.21 (4/4) | - | 87.15 (1/4) | - | |
| 10-fold diluted | nd (0/4) | 10.08 ± 0.55 (2/4) | nd (0/4) | nd (0/4) | ||||||
| 1.0 × 103 | pure | nd (0/4) | - | nd (0/4) | - | 4.39 (1/4) | - | nd (0/4) | - | |
| 10-fold diluted | nd (0/4) | nd (0/4) | nd (0/4) | nd (0/4) | ||||||
| MNV-1 | MNV | 61.18 ± 36.35 | 58.04 ± 24.79 | 45.40 ± 38.41 | 48.29 ± 27.67 | |||||
| Total samples with recovery rates > 1% | 31/32 | |||||||||
| norovirus GI | 1.6 × 106 | pure | 82.48 ± 13.79 (2/4) | 1.21 | 97.30 ± 3.83 (4/4) | 1.0 | 4.79 ± 3.07 (4/4) | 8.9 | 63.01 ± 21.77 (4/4) | 1.4 |
| 10-fold diluted | 100.00 (2/4) | 100.00 (4/4) | 42.51 ± 31.69 (4/4) | 88.58 ± 13.24 (4/4) | ||||||
| 1.6 × 105 | pure | 49.27 ± 1.17 (2/4) | 1.06 | 71.87± 32.51 (4/4) | 0.93 | 68.60 ± 61.29 (4/4) | 1.1 | 100.00 (4/4) | 0.8 | |
| 10-fold diluted | 52.47 ± 62.22 (2/4) | 66.92 ± 54.90 (3/4) | 77.11 ± 14.90 (4/4) | 83.50 ± 32.93 (4/4) | ||||||
| 1.6 × 104 | pure | 2.18 (1/4) | - | nd (0/4) | - | 97.32 (1/4) | - | 34.41 ± 27.98 (3/4) | - | |
| 10-fold diluted | nd (0/4) | nd (0/4) | nd (0/4) | nd (0/4) | ||||||
| 1.6 × 103 | pure | nd (0/4) | - | nd (0/4) | - | nd (0/4) | - | nd (0/4) | - | |
| 10-fold diluted | nd (0/4) | nd (0/4) | nd (0/4) | nd (0/4) | ||||||
| MNV-1 | MNV | 100 | 100 | 100 | 80.29 ± 56.88 | |||||
| Total samples with recovery rates > 1% | 32/32 | |||||||||
| norovirus GII | 1.3 × 106 | pure | 11.34 ± 2.43 (4/4) | 2.3 | 5.77 ± 1.30 (4/4) | 1.0 | 0.17 ± 0.06 (4/4) | 1.2 | 12.49 ± 1.21 (4/4) | 1.1 |
| 10-fold diluted | 26.14 ± 33.00 (4/4) | 5.67 ± 1.36 (4/4) | 1.96 ± 0.37 (4/4) | 14.09 ± 1.60 (4/4) | ||||||
| 1.3 × 105 | pure | 8.00 ± 1.23 (4/4) | 1.0 | 6.51 ± 1.88 (4/4) | 1.0 | 4.65 ± 0.51 (4/4) | 0.7 | 7.66 ± 2.76 (4/4) | 1.2 | |
| 10-fold diluted | 8.45 ± 2.78 (4/4) | 6.55 ± 3.68 (4/4) | 3.31 ± 1.23 (3/4) | 8.98 ± 5.29 (4/4) | ||||||
| 1.3 × 104 | pure | 9.09 ± 5.35 (4/4) | 1.9 | 5.11 ± 5.22 (3/4) | 2.1 | 10.43 ± 4.05 (4/4) | 0.4 | 8.32 ± 2.04 (2/4) | - | |
| 10-fold diluted | 17.38 ± 20.49 (3/4) | 10.77 ± 8.29 (2/4) | 4.21 ± 5.91 (2/4) | nd (0/4) | ||||||
| 1.3 × 103 | pure | 6.92 ± 4.12 (2/4) | 5.0 | 3.96 ± 0.33 (2/4) | - | nd (0/4) | - | nd (0/4) | - | |
| 10-fold diluted | 34.41 ± 19.47 (2/4) | nd (0/4) | nd (0/4) | nd (0/4) | ||||||
| MNV-1 | MNV | 55.73 ± 8.56 | 51.00 ± 14.69 | 14.76 ± 11.79 | 46.42 ± 30.50 | |||||
| Total samples with recovery rates > 1% | 32/32 | |||||||||
| Sample Analyzed | Mean Inhibition Recoveries (% ± SD) |
|---|---|
| Fish dumpling | 26.46 ± 33.77 (N = 8) |
| Albacore tuna | 39.03 ± 32.27(N = 16) |
| Cod | 26.60 ± 33.84 (N = 8) |
| Tuna | 30.10 ± 30.26 (N = 8) |
| Shelled pink shrimp | 36.83 ± 32.58 (N = 16) |
| Halibut | 12.93 ± 16.12 (N = 8) |
| Tuna rillette | 84.50 ± 34.35 (N = 8) |
| Surimi | 25.82 ± 29.19 (N = 16) |
| Salmon sorrel sauce | 20.56 ± 33.94 (N = 8) |
| Kipper | 15.40 ± 34.47 (N = 8) |
| Haddock | 15.73 ± 25.58 (N = 16) |
| Salmon | 20.12 ± 19.57 (N = 8) |
| Target Virus | Genome Copies/g | R1 | R2 | R3 | R4 | All Settings |
|---|---|---|---|---|---|---|
| HAV | LOD50 | 600 | 3.8 × 103 | <1.2 × 103 * | <1.2 × 103 * | 144 |
| LOD95 | 2.5 × 103 | 1.7 × 104 | <1.2 × 103 * | <1.2 × 103 * | 6.4 × 103 | |
| HEV | LOD50 | 8.4 × 104 | 1.2 × 102 | 7.2 × 102 | 7.6 × 103 | Between 7.2 × 102 and 8.4 × 104 ** |
| LOD95 | 3.6 × 105 | 5.2 × 104 | 3.2 × 103 | 3.3 × 104 | Between 3.2 × 103 and 3.6 × 105 ** | |
| norovirus GI | LOD50 | 7.2 × 103 | 1.9 × 104 | 1.2 × 104 | 3.5 × 103 | 1 × 104 |
| LOD95 | 3.1 × 104 | 8.4 × 104 | 5.2 × 104 | 1.2 × 104 | 4.4 × 104 | |
| norovirus GII | LOD50 | 520 | 2.2 × 103 | 1.5 × 103 | 5.6 × 103 | 2.0 × 103 |
| LOD95 | 2.2 × 103 | 9.6 × 103 | 6.4 × 103 | 2.5 × 104 | 8.8 × 103 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hennechart-Collette, C.; Dehan, O.; Fraisse, A.; Martin-Latil, S.; Perelle, S. Development of an Extraction Method to Detect Hepatitis A Virus, Hepatitis E Virus, and Noroviruses in Fish Products. Microorganisms 2023, 11, 624. https://doi.org/10.3390/microorganisms11030624
Hennechart-Collette C, Dehan O, Fraisse A, Martin-Latil S, Perelle S. Development of an Extraction Method to Detect Hepatitis A Virus, Hepatitis E Virus, and Noroviruses in Fish Products. Microorganisms. 2023; 11(3):624. https://doi.org/10.3390/microorganisms11030624
Chicago/Turabian StyleHennechart-Collette, Catherine, Océane Dehan, Audrey Fraisse, Sandra Martin-Latil, and Sylvie Perelle. 2023. "Development of an Extraction Method to Detect Hepatitis A Virus, Hepatitis E Virus, and Noroviruses in Fish Products" Microorganisms 11, no. 3: 624. https://doi.org/10.3390/microorganisms11030624
APA StyleHennechart-Collette, C., Dehan, O., Fraisse, A., Martin-Latil, S., & Perelle, S. (2023). Development of an Extraction Method to Detect Hepatitis A Virus, Hepatitis E Virus, and Noroviruses in Fish Products. Microorganisms, 11(3), 624. https://doi.org/10.3390/microorganisms11030624





