Abstract
Arsenic is a toxic metalloid with differential biological effects, depending on speciation and concentration. Trivalent arsenic (arsenite, AsIII) is more toxic at lower concentrations than the pentavalent form (arsenate, AsV). In E. coli, the proteins encoded by the arsRBC operon are the major arsenic detoxification mechanism. Our previous transcriptional analyses indicate broad changes in metal uptake and regulation upon arsenic exposure. Currently, it is not known how arsenic exposure impacts the cellular distribution of other metals. This study examines the metalloproteome of E. coli strains with and without the arsRBC operon in response to sublethal doses of AsIII and AsV. Size exclusion chromatography coupled with inductively coupled plasma mass spectrometry (SEC-ICPMS) was used to investigate the distribution of five metals (56Fe, 24Mg, 66Zn, 75As, and 63Cu) in proteins and protein complexes under native conditions. Parallel analysis by SEC-UV-Vis spectroscopy monitored the presence of protein cofactors. Together, these data reveal global changes in the metalloproteome, proteome, protein cofactors, and soluble intracellular metal pools in response to arsenic stress in E. coli. This work brings to light one outcome of metal exposure and suggests that metal toxicity on the cellular level arises from direct and indirect effects.
1. Introduction
Arsenic is a well-known carcinogen and is one of the most prevalent environmental toxins [1,2,3]. In nature, the two main forms of inorganic arsenic are the trivalent form arsenite (AsIII) and the pentavalent form arsenate (AsV), the former being more toxic [4]. Both arsenic species enter the cell through adventitious mechanisms. AsV is structurally similar to phosphate and is uptaken by the phosphate transporters, Pst and Pit. AsIII enters the cell through the glycerol facilitator membrane protein GstF [5,6,7].
Because arsenic is readily assimilated, organisms have evolved to detoxify arsenic. In bacteria, this is primarily accomplished by proteins encoded by ars genes. There are many forms of the ars operon, but in its simplest form, only three proteins are encoded. This is known as the arsRBC locus. In a counterintuitive approach, AsV must first be reduced to the more toxic AsIII by ArsC, and is then removed through the efflux pump ArsB (or ArsAB). ArsR is a DNA-binding regulator protein that controls the expression of the other ars operon proteins [7,8,9,10]. Previous studies of the bacterium Agrobacterium tumefaciens 5A have shown that an intertwined genetic circuit exists between phosphorous uptake genes and arsenic detoxification genes [11,12,13]. A more recent study has shown that the ArsR protein also affects metabolism [9]. Using RNAseq, this study found that the ArsR proteins of Agrobacterium tumefaciens 5A also influence many biological processes, including sugar transport, chemotaxis, copper tolerance, and iron homeostasis.
Organisms use metalloproteins to achieve distinct and specific biologic functions. As a consequence of this, metal ions are crucial in biology, and organisms have evolved mechanisms to maintain metal homeostasis [14]. Imbalances in this homeostasis have effects on other metals, protein expression, and cofactor abundance. For example, high levels of zinc in E. coli lead to an increased demand for iron, along with decreased copper levels [15]. In Caenorhabditis elegans, exposure to Mn alters the Ca and Fe balances, as well as protein homeostasis [16]. To date, there are limited studies of how arsenic affects other metals. Unraveling the interplay between arsenic and other metals will provide a deeper understanding of arsenic toxicity.
In the present study, an E. coli strain containing the arsRBC operon K-12 and a strain lacking the operon AW3110 were cultured in sublethal doses of trivalent arsenic AsIII or pentavalent arsenic AsV. Using inductively coupled plasma mass spectrometry (ICP-MS), size-exclusion chromatography coupled to ICP-MS (SEC-ICPMS), and SEC coupled to a diode array UV detector (SEC-UV), changes were detected in the metalloproteome of five metals (75As, 56Fe, 24Mg, 66Zn, and 63Cu). The observed changes were different between the two strains of E. coli and depended on treatment with trivalent or pentavalent arsenic. Our findings extend understanding of the intricate biological coordination underlying arsenic regulation while opening a window for understanding the interplay between metal balances and cellular metabolism.
2. Materials and Methods
2.1. E. coli Cell Growths
E. coli K12 cultures were grown in 200 mL of LB broth as batch cultures at 37 °C. Cultures were inoculated from an overnight culture that was started from a single colony selected from an LB agar plate. A total of 2 h after inoculation (early log phase), cells were treated with 1 mM AsIII, 1.5 mM AsV, or left untreated. Batch cultures to establish arsenic concentrations and timing for the experiment were repeated over the span of months. For the cultures used for analysis, each condition had three biological replicates. Replicates for a given condition were grown on the same day. Cells were pelleted via centrifugation at 4000 x g for 10 min, 4 h post-arsenic administration. Cell pellets were carefully washed with 1 mM EDTA in PBS. A final centrifugation step was performed to harvest the cells. The cells were stored at −80 °C until further analysis. E. coli AW3110 cultures were grown under chloramphenicol selection in 200 mL of LB broth at 37 °C in the same fashion as for K-12. At 2 h post-inoculation (early log phase), 100 µM AsIII or 500 µM AsV was administered to the test cultures, while the control culture was left untreated. The AsV condition was grown in quadruplicate, while the control and AsIII conditions were grown in triplicate. At 4 h after arsenic administration, or 6 h after inoculation for control cultures, cells were pelleted, washed, harvested, and stored in the same manner as the K12 cultures. The K12 and AW3110 cell lines were a gift from Dr. Barry Rosen and are as described in the original publication [17].
2.2. Protein Extraction
Cell pellets were resuspended in 1.5 mL of 200 mM ammonium acetate pH 7 in the presence of the protease inhibitors (VWR) and immediately lysed using matrix E (MPBio Irvine, California, USA) and FastPrep-24 5G bead beater (MPBio Irvine, California, USA). Cells were then lysed with cycles at a speed of 6.0 m/s for 30 s on a FastPrep-24 5G bead beater (MPbio Irvine, California, USA). The cell debris and the matrix were pelleted at 18,000× g for 20 min and the soluble fraction was collected. Protein concentration was determined using a Bradford assay.
2.3. Metalloproteome Analysis
The metalloproteomes for each condition were analyzed via SEC-ICP-MS using an Agilent Infinity II LC coupled to an Agilent 7800 ICP-MS. The methods were inspired by work done by the Roberts lab [18,19]. A total of 160 µg of soluble protein (entire proteome) was injected onto an Agilent Bio-SEC 3 (3 µm, 300 Å, 4.6 × 300 mm) column and proteins were separated for 25 min using 200 mM ammonium acetate pH 8 as the mobile phase, with a flow rate of 0.4 mL/min. The five metal ion signals were monitored, and these included: 24Mg, 56Fe, 63Cu, 66Zn, and 75As, with an integration time/mass of 1.5 s per analyte. The monitoring of ion signals was performed using the Agilent MassHunter 4.6 (version C.01.06). Metal signals were manually integrated in MassHunter, and all other data workup was performed in Microsoft Excel. Statistical differences were determined using a Student’s t test at a 95% confidence interval.
2.4. Total Metal Analysis of Soluble Fraction Digest and Media
Biologic replicates were diluted to the same protein concentration. Then equal parts were pooled to create one sample per condition. The metals were liberated from biologic molecules using a 5% nitric acid digestion. The precipitate was pelleted via centrifugation, and the supernatant was collected for analysis. Metal analysis was performed using an Agilent Infinity II autosampler coupled to an Agilent 7800 ICP-MS. The mobile phase was 2% HNO3, 0.5% HCl in water, with a flow rate of 1 mL/min. The signals of five metal ions were monitored for 30 s, and included: 24Mg, 56Fe, 63Cu, 66Zn, and 75As, with an integration time/mass of 0.5 s per analyte. The quantification of analytes was performed in Agilent MassHunter 4.6 (version C.01.06) against a standard curve. Media with and without arsenic was digested in the same fashion The metals of interest were analyzed using an Agilent SPS 4 autosampler coupled to an Agilent 7800 ICP-MS in helium mode. The quantification of analytes was performed against a standard curve in MassHunter.
2.5. ICP-MS Instrument Parameters
For small volume LC analysis (Agilent Infinity II coupled to Agilent 7800 ICP-MS), the following instrument parameters were used: RF power 1550 V, RF matching 1.00 V, nebulizer gas 0.99 L/min, option gas 0.0%, nebulizer pump 0.30 rps, S/C temp 2 °C, makeup gas 0.00 L/min, extract 1 lens 0.0 V, extract 2 lens −195.0 V, omega bias −95 V, omega lens 8.4 V, cell entrance −40 V, cell exit −60 V, deflect 0 V, plate bias −55 V, He flow rate 4.0 mL/min, octupole bias −18 V, octupole RF 200 V, and energy discrimination 3.0 V.
For large volume analysis (Agilent SPS 4 coupled to Agilent 7800 ICP-MS), the following instrument parameters were used: RF power 1550, RF matching 1 V, nebulizer gas 1.00 L/min, option gas, 0.0 %, nebulizer pump 0.10 rps, S/C temp 2 °C, makeup gas 0.00 L/min, extract 1 lens 0.0 V, extract 2 lens −200.0 V, omega bias −105 V, omega lens 7.5 V, cell entrance −40, cell exit −60 V, deflect −0.6 V, plate bias −55 V, He flow rate 4.5 mL/min, octupole bias −18.0 V, octupole RF 200 V, and energy discrimination 5.0 V.
2.6. Proteome Analysis
The native-state soluble proteome was fractionated by collecting samples from the Bio-SEC 3 column with the same method used for ICP-MS, except using a different LC system (Agilent 1290 Infinity) with an Agilent Infinity diode array detector. A total of 200 µg of total protein was manually injected, and tube lengths were matched to ensure profiles were comparable with the SEC-ICPMS profiles. Starting at 4 min post-injection, 1 min fractions were collected manually for a total of 10 min. Fractions were freeze-dried for subsequent SDS-PAGE analysis. The freeze-dried samples were resuspended in a 240 mM Tris HCl pH 6.8 buffer containing 277 mM SDS and 40% SDS, heated at 99 °C for 6 min, and loaded onto a 4–20% Mini-Protean ® TGX ™ Precast 15 µL-well (Bio-Rad Hercules, California, USA). Gels were stained with GelCode ™ Blue Stain Reagent (ThermoFisher Waltham, Massachusetts, USA).
3. Results
3.1. Arsenic Induced Changes in the Metalloproteome of E. coli K-12
E. coli strains K-12 and AW3110 were selected for this study because AW3110 lacks the arsRBC operon, while K-12 has an intact arsRBC operon [17]. A metalloproteomics workflow was developed for this study (Figure 1). Batch cultures were grown in sublethal concentrations of AsIII or AsV (Supplemental Figure S1). After 4 h of exposure to arsenic, the soluble fraction of the proteome was extracted under native conditions to keep protein complexes and protein-metal interactions intact. The metalloproteomes from different conditions were analyzed by size exclusion chromatography coupled with inductively coupled plasma mass spectrometry (SEC-ICPMS), while acid digestion following ICP-MS was used to quantify total cellular metals. SEC coupled to a UV-Vis diode array detector (SEC-UV) was used to monitor the wavelengths of the protein-bound cofactors and to fractionate the soluble proteome. Denaturing SDS-PAGE was used to visualize the soluble proteome and the proteins in each SEC fraction. Together, these techniques provide insight into the changes induced by AsIII and AsV treatments in E. coli K-12 and AW3110.
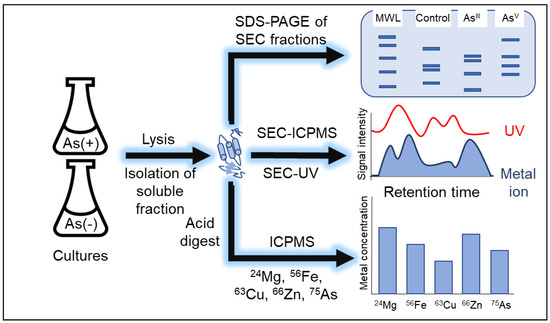
Figure 1.
Metalloproteomics workflow. Replicate cultures of K12 and AW3110 +/− As were cultured. Cells were lysed, and the soluble fraction was collected. Size-exclusion chromatography (SEC) was used for fractionating the native proteome prior to analysis by SDS-PAGE. SEC was used in-line with inductively coupled plasma mass spectrometry (SEC-ICPMS) for analysis of the native metalloproteome and in-line with a diode array detector for analysis of UV active cofactors (SEC-UV). The soluble fractions were digested with nitric acid for the analysis of cellular metal content via inductively coupled plasma mass spectrometry (ICP-MS).
The isolated soluble native protein fractions of E. coli K-12 untreated, AsIII exposed, and AsV exposed were analyzed using SEC-ICPMS to determine the effects of AsIII and AsV on the metalloproteome of E. coli. Specifically, 56Fe, 24Mg, 66Zn, and 63Cu ions were monitored, as proteins containing these metals are involved in DNA/RNA metabolism, oxidation/reduction, and other critical biological functions, while 75As was monitored to detect the integration of arsenic into the proteins. SEC-ICPMS was used under non-denaturing conditions to investigate the native proteome and maintain metal–protein interactions.
As expected, there was a negligible arsenic signal for the control sample, while both arsenic treatments showed arsenic inclusion into the proteome (Figure 2). The arsenic-containing protein peak observed in the AsV treatment peaked slightly before minute 9 on the SEC, while the AsIII treatment peak occurred slightly before minute 10. A Student’s t-test (n = 3) showed a significant increase (p-value = 0.011) in arsenic in the soluble fraction from the AsV treatment compared to the AsIII treatment group (Supplemental Figure S2). These results show that cells treated with arsenic incorporate the toxic metal into their cellular proteome.
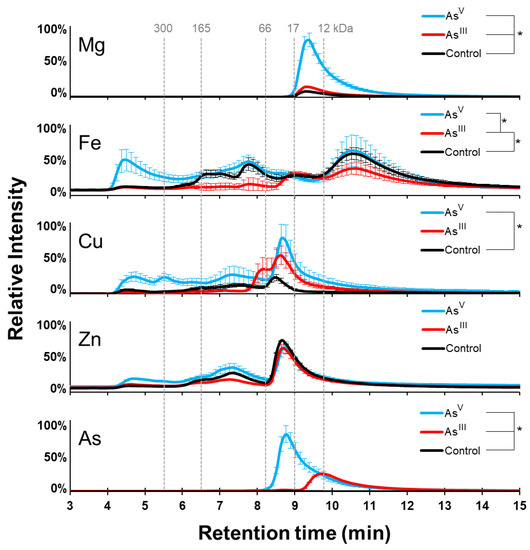
Figure 2.
K-12 metalloproteome response to arsenic treatment. SEC-ICP-MS analysis shows the size distribution of protein-bound copper, magnesium, arsenic, iron, and zinc. AsV treatment is shown in cyan, the AsIII treatment in red, and the control in black. Error bars are +/−1 standard deviation. * Indicates significant differences (p < 0.05) determined by a Student’s t-test (n = 3).
We also investigated the effect of arsenic treatment on the magnesium proteome. There was only one elution peak for magnesium-containing proteins, and it showed a maximum signal between minutes 9 and 10 in every condition tested (Figure 2). This peak was significantly increased (p-value = 0.012) in the AsIII treatment compared to the control group, and was further increased with AsV treatment (p-value = 0.00087). This result shows that arsenic treatments increase the abundance of magnesium or magnesium-containing proteins, specifically in the case of AsV treatment.
The integrated iron signal of the proteome was significantly higher (p-value = 0.040) in the control cell cultures compared to the AsIII treated groups, and the iron signal was significantly higher in the AsV treatment than the AsIII treatment group (p-value = 0.039) (Figure 2). Total iron was not significantly different between the AsV treatment and the control; however, the distribution of iron throughout the native proteome showed noticeable changes between all three groups. The maximum in the iron signal was observed in all three conditions between minutes 10 and 11. The maximum was near the same intensity in both the control and AsV treated cell cultures, with a decreased signal in the cells treated with AsIII. An increase in iron signal intensity was observed between minutes 4 and 5 in the AsV group, with only a minor signal observed in the control and AsIII treated groups. A decrease in the iron signal between minutes 6 and 8.5 was present in the AsIII treatment group as well. Together, the data suggest that arsenic treatment changes the iron proteome of E. coli K12 cells, with the greatest effects being noted in proteins and protein complexes greater than 66 kDa in size.
We next examined the effects of arsenic treatment on copper-containing proteins and zinc-containing proteins. AsV treatment significantly increased copper (p-value = 0.031) across the entire elution profile compared to the control group (Figure 2). In contrast, AsIII treatment caused an increase in the copper signal between minutes 8 and 10; however, the abundance of copper in the proteome was not significantly different. These results suggest that AsV treatment has a larger effect on the copper proteome than does the AsIII treatment, which increases the abundance of copper-containing proteins that are ~66 kDa in size. The zinc signal under all conditions was at a maximum near minute 9. This peak was indistinguishable between conditions, and no significant differences (p-value < 0.05) in the zinc-containing proteins were observed between conditions. This data indicates that in K-12, the zinc proteome is unchanged following arsenic treatments.
3.2. Arsenic Induced Changes in the Metalloproteome of E. coli AW3110
E. coli AW3110 lacks the genes primarily responsible for the cellular detoxification of arsenic. We used this strain to address how the metalloproteome changes in response to arsenic when the arsRBC operon is absent, in theory increasing the arsenic available to the proteome soluble protein fractions of E. coli AW3110. The concentrations of AsIII and AsV were chosen to elicit similar levels of arsenic stress as K-12. The AW3110 control, AsIII, and AsV exposed cultures were analyzed with SEC-ICPMS, as performed above for the E. coli K-12 strain.
Treatment with both species of arsenic resulted in arsenic incorporation into the soluble proteome. The AsIII condition showed a signal between minutes 9 and 13, with the highest intensity between minutes 10 and 11 and a shoulder between minutes 9 and 10 (Figure 3). AsV treatment, however, showed significantly fewer (p-value < 0.0016) arsenic-containing proteins, which eluted primarily between minutes 9 and 10, and a smaller maximum peak between minutes 8 and 9. As expected, there was a negligible arsenic signal in the proteins from the control group. These data are consistent with the arsenic incorporation into the proteome of E. coli K-12 cells.
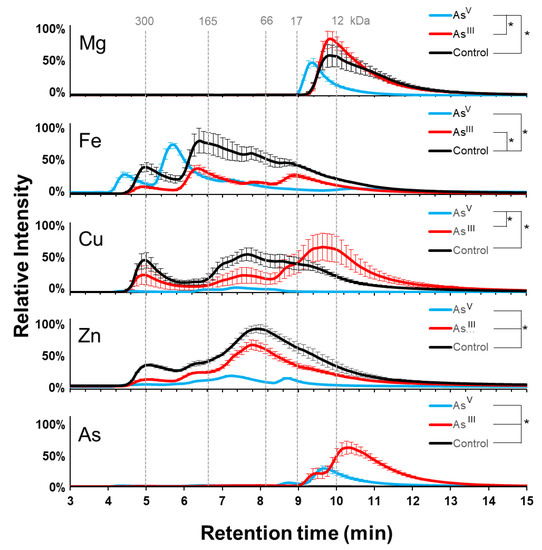
Figure 3.
AW3110 Metalloproteome response to arsenic treatment: Metal profiles for copper, magnesium, arsenic, iron, and zinc from SEC-ICP-MS analysis. AsV treatment is shown in cyan, AsIII treatment is in red, and control is in black. Error bars are +/−1 standard deviation. * Indicates significant differences (p < 0.05) determined by a Student’s t-test (n = 3 for AsIII and control; n = 4 for AV treatment).
The profiles of the magnesium-containing protein appeared to be very similar between the control and AsIII treated groups, while the AsV treatment caused a slight shift towards earlier eluting proteins (Figure 3). AsV treatment also caused a significant decrease (p-value = 0.0098) in the total amount of magnesium-bound protein and a decrease in total soluble magnesium (Supplemental Figure S3). The iron signals showed that the AsIII treatment and the control were very similar, except for a reduced signal intensity in the AsIII treatment (p-value < 0.0028). The AsV treatment group yielded two strong iron signals, with maxima between minutes 4 and 5 and between minutes 5 and 6. Proteins and protein complexes eluting after six minutes displayed a diminished signal for iron compared to that in the control group. Despite the different elution profiles, there was no significant difference in total iron signal between AsV and AsIII treatments (p-value = 0.51). These results indicate that arsenic imparts changes in the AW3110 magnesium metalloproteome that are distinct from those observed for E. coli K-12.
The effect arsenic treatment had on copper-containing proteins showed that the AsV treatment significantly diminished most of the copper-containing proteins (p-value = 0.0042) (Figure 3). The copper signal observed in the control at minute 5 and between minutes 7 and 8 were decreased in the AsIII treated sample. AsIII treatment also yielded lower molecular weight copper-containing proteins. AsIII treatment significantly reduced the zinc-containing proteins compared to the control sample, but maintained a similar elution profile (p-value = 0.0062). AsV treatment resulted in a significantly decreased zinc signal compared to the AsIII (p-value = 0.000083) and control (p-value = 0.000022) conditions. It is clear that arsenic treatment has dramatic effects on the copper proteome, and that both arsenic treatments reduce the abundance of zinc or zinc-containing proteins across the entire elution profile, with AsV having the largest effect.
3.3. Changes in Total Cellular Metal
SEC-ICPMS experiments provide information about the metalloproteome, but provide no information about metal intake inside the cell. To establish this, we determined the total cellular pool of the metal in each group. To measure the total soluble metal pools of each treatment group, the metals were liberated from biomolecules using a nitric acid digestion. Biological replicates were analyzed by ICPMS for magnesium, zinc, iron, arsenic, and copper (Figure 4). The pooled K-12 control sample had a 5.7 ppm magnesium concentration. Arsenic treatment caused a 2.5-fold and a 10.5-fold increase in magnesium concentration relative to the control for the AsIII and AsV treatments, respectively. In AW3110, the pooled control sample had a magnesium concentration of 7.2 ppm. There was an increase in the AsIII treatment samples and a slight decrease for the AsV treatment samples (Figure 4a). Iron concentration increased slightly from 1.29 ppm in the AsIII and AsV conditions (Figure 4b). Cellular copper levels rose with both arsenic treatments, with the largest increase seen in the AsIII treatment for both cell strains (Figure 4c). In E. coli K-12, the AsIII treatment was associated with an increase in zinc concentration in both treatment groups. In the E. coli AW3110 strain, the control and AsIII treatment zinc concentrations were 304 and 317 ppb, respectively, while the AsV treatment reduced the zinc concentration to approximately one-third of this value (Figure 4d). Less than 5 ppb of arsenic was observed in the control samples of both cell lines. AsIII treatment resulted in 3.47 ppm and 3.77 ppm arsenic concentrations for the K-12 and AW3110 samples, respectively. AsV treatment led to an arsenic concentration of 6.5 ppm in the E. coli K-12 and 1.5 ppm in the AW3110 cells (Figure 4e). Media with and without AsIII or AsV was analyzed by ICP-MS to check for background arsenic in the stock solutions (Supplemental Figure S4). These results demonstrate that arsenic treatment not only changes the concentration and abundance of metal-bound proteins, but also alters the cellular metal concentration.
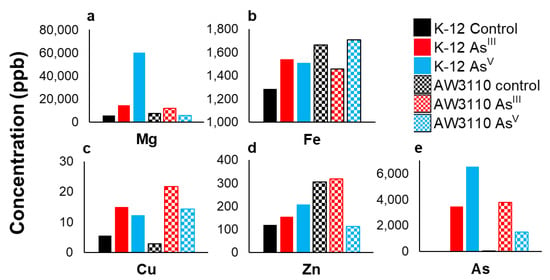
Figure 4.
Total intracellular soluble metal. Pooled soluble fractions for K-12 (solid fill) and AW3110 (checkerboard) growths analyzed by ICP-MS for magnesium (a), iron (b), copper (c), zinc (d), and arsenic (e). AsV treatment is shown in cyan, AsIII treatment is in red, and control is in black. The concentration is given above each sample in ppb.
3.4. Changes in the Soluble Proteome
In addition to changes in metal abundance, the SEC-ICPMS profile of the metalloproteome can be affected by differences in the abundance of proteins and the presence of protein complexes. To determine if and how the soluble proteome changed, an SDS-PAGE analysis was conducted on the total soluble proteome of the collected SEC fractions for the control, AsIII, and AsV treated cells for both K-12 and AW3110 E. coli strains. SDS-PAGE analysis is performed under denaturing conditions breaking about protein complexes. Differences in the soluble proteome were observed between all conditions for K-12 and AW3110 (Supplemental Figure S5). Gel lanes corresponding to the SEC fractions made those differences more apparent. In the soluble protein fractions of K-12, a protein band near 30 kDa was present in the AsV conditions, but absent in the control and AsIII conditions (Supplemental Figure S5a). Similarly, there was a protein band near 50 kDa that was present in fractions 4–7, which was absent or minimally present in the arsenic conditions. The banding patterns in the AsIII and AsV conditions between 37 and 50 kDa were also different in fraction 4. In the soluble protein fractions of AW3110, a protein band near 50 ka was present in fraction 2 of the control and AsV treated cells, but was minimally present in the cells treated with AsIII (Supplemental Figure S5b). In fraction 3, there was a protein band near 50 kDa that was present in the AsIII and AsV treated groups, but was more abundant in the control group. The protein bands between 10 and 15 kDa in fraction 3 were nearly absent in the AsV treated cells, but were present in the control and AsIII treated cells. The SDS-PAGE analysis indicated significant differences in the soluble proteome profiles across all conditions.
3.5. Protein Cofactor Changes
Organisms use enzymes to achieve specific cellular outcomes, and enzymes often require cofactors to function properly. Because of this, environmental conditions (such as the presence of heavy metals) will influence protein expression and cofactor abundance, as well as the redox status of cells. Many biological cofactors absorb in the UV and visible spectrum, and often, the cofactor will change its spectroscopic properties depending on its redox status. For example, oxidized flavin has two absorption peaks near 365 nm and 460 nm, while reduced flavin becomes spectroscopically inactive at these wavelengths [20]. Another important cofactor is heme, which absorbs at 405 nm [21]. Samples that were analyzed by SEC-ICPMS were pooled for each condition and analyzed by SEC-UV to determine whether there was a change in the detectable differences of protein-bound cofactors (Figure 5). In K-12 cells, AsV treatment decreases in absorbance at 365 nm between minutes 9 and 13, at 405 nm between minutes 6 and 12, and at 460 nm between minutes 6 and 11. AsIII treatment of K-12 resulted in very similar UV signal at 365 nm, but a decreased signal at 450 nm between minutes 6 and 10. The signal profiles are different for 405 nm and 460 nm (Figure 5a). The growth of AW3110 in the presence of AsIII caused increases in the 365 nm signal between minutes 8 and 15, and in the 405 nm and 460 nm signals between minutes 8.5 and 15, while decreases in the 405 nm and 460 nm signals were observed between minutes 6 and 8.5 (Figure 5b). AsV treatment caused decreases in the 365, 405, and the 460 nm signals between minutes 5.5 and 15. Our data suggest that the cofactor distribution and abundance in the proteome of AW3110 is more susceptible to changes induced by arsenic treatments, with AsIII having a more profound effect than AsV and coinciding with Cu and Mg metal distributions.
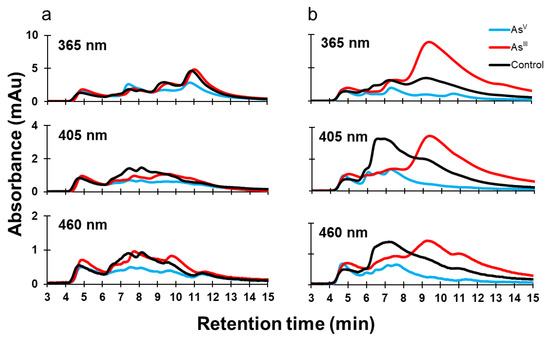
Figure 5.
Protein-bound cofactor distribution changes caused by arsenic treatment. Soluble proteomes were analyzed on an SEC coupled with a DAD detector. The wavelengths 365, 405, and 460 nm were monitored for K-12 (a) and AW3110 (b) under three conditions: control (black), AsV (cyan), and AsIII (red).
4. Discussion
E. coli strains K-12 and AW3110 were given sub-lethal doses of the most prevalent arsenic toxins, arsenite (AsIII) and arsenate (AsV). K-12 has an intact arsenic detoxification mechanism that is encoded by the arsRBC genes. AW3110 lacks these genes and is more susceptible to arsenic toxicity. Little is known about how arsenic affects the assimilation of other metals, or how the arsenic detoxification machinery is involved with metal perturbations inside the cells. Our experiments shed new light on the perturbations of cellular metal homeostasis caused by arsenic.
The foundation for our work came from the transcriptomic study of Rawle et al. on Agrobacterium tumefaciens 5A. This bacterium has been used as a model organism for studying arsenic detoxification [9,12,13,22,23,24]. The genome encodes two ars loci, and each locus contains the basic components of arsenic detoxification: a reductase, an efflux pump, and repressor proteins, along with other proteins not discussed here. In total, A. tumefaciens has four arsR protein-encoding genes; arsR1, arsR2, arsR3, and arsR4. Rawle’s study knocked out each arsR gene individually prior to transcriptomic analysis. Their work showed that the arsR genes were involved in nearly every aspect of gene regulation and cell physiology when A. tumefaciens 5A was exposed to AsIII [13]. This involvement includes both the repression and activation of metal-containing proteins. Their data revealed that genes coding for arsenic, copper, zinc, and iron-dependent proteins were both repressed and/or activated when AsIII was introduced. Copper metabolism and iron transport/regulation genes were primarily downregulated with the highest AsIII levels tested. While our current study lacks transcriptomic analyses, we see clear changes in the metalloproteomes of both E. coli strains upon arsenic treatments, except for the zinc metalloproteome of E. coli K-12 cells.
4.1. Arsenic
It has been demonstrated previously that cytosolic proteins isolated from rabbit liver incubated with AsIII bound 13 times more arsenic than proteins incubated with AsV [25]. In K-12 cells, we see a noticeably higher arsenic protein signal for AsV treatment compared to AsIII treatment, whereas with E. coli AW3110, the converse is true. K-12 encodes for the AsV reductase ArsC, which converts AsV into AsIII. K-12 cells were given 50% more arsenic in the AsV treatment than was provided during the AsIII treatment. This may explain why there is more arsenic incorporation in K-12 than in AW3110 E. coli cells. Interestingly, there is a trend in both K-12 and AW3110, where AsV treatment incorporates arsenic into proteins with higher molecular weights (or complexes). While the expression of ArsC and a higher concentration of arsenic in the AsV treatment compared to the AsIII treatment may explain increased arsenic incorporation in proteins, the observed trend of arsenic incorporation into higher molecular weight proteins in AsV treatment compared to AsIII treatment in both cell strains remains enigmatic. Our data clearly show that arsenic incorporates into the proteome differently depending on the arsenic species provided to the cells, regardless of whether an arsenic detoxification mechanism is present or not.
4.2. Copper
Copper is coordinated by proteins, primarily through histidine, methionine, and cysteine side chains [26]. AsIII may bind to the sulfhydryl groups of cysteines and displace liganded copper. A 2004 study looked at trivalent arsenicals binding to rat hemoglobin [27]. The research found that trivalent arsenicals bind only at cysteine residues, and that less than 10% of inorganic AsIII is bound to rat hemoglobin, while monomethylarsonous acid and dimethylarsonous acid were bound to nearly 50% and 80%, respectively. This may explain why AsIII treatment of AW3110 does not drastically reduce the proteome profile of copper-containing proteins. Rawle et al. saw both upregulation and downregulation by the arsR genes of transcripts encoding copper-containing proteins involved in copper transport, electron-transport, and copper detoxification in AsIII treated cells [13]. It is plausible that this transcriptome pattern is present in AsV treated A. tumefaciens as well. In AsV treated K-12 cells, an increase in higher molecular weight copper-containing proteins was observed. In AW3110, however, AsV treatment drastically reduced the amount of copper-containing proteins. Interestingly, AsIII treatment in AW3110 resulted in similar concentrations of copper-containing proteins, but with a different elution profile.
4.3. Magnesium
One of the most surprising results included the large increase in the magnesium bound proteins when K-12 E. coli cells were treated with AsV (Figure 2), as well a large increase the total intracellular soluble magnesium intake (Figure 4a). Magnesium influx has been reported as a response to ribosomal stress in Bacillus subtilitis, which prevents the hyperpolarization of the membrane potential [28]. Furthermore, AsV has been shown to degrade ribosomes in E. coli, releasing phosphate into the cell [29]. This was demonstrated by growing E. coli in the presence of [3H]-uridine. The authors analyzed acid-soluble material released following ribosome degradation using thin-layer chromatography (TLC) and observed that AsV-induced ribosomal degradation led to increased radioactivity in the TLC chromatogram, which corresponded to uracil, but not to UTP, UDP, or UMP. A different study using untargeted proteomic analysis revealed a downregulation of ribosomal protein-encoding genes in AsIII treated yeast [30]. This was proposed to reflect a protection mechanism, rather than a toxic effect from AsIII. In the transcriptomic analysis by Rawle et al., only three magnesium related proteins were shown to be affected by AsIII treatment. The three proteins were a putative adenylate cyclase, GroES, and a nudix hydrolase. Adenylate cyclases convert ATP into cAMP [31,32]. GroES is part of the GroEL-GroES chaperonin machine which provides a nano-cage for the correct folding of proteins [33]. Nudix hydrolases are a large class of enzymes which catalyze the hydrolysis of nucleoside diphosphates [34,35]. Our data, in conjunction with current literature, suggest that the presence of the arsRBC operon is likely involved in the regulation of nucleotide pools and magnesium transporters as a mechanism to cope with ribosomal stress caused by arsenic, which is exacerbated when pentavalent arsenic, which is a strong phosphate imitator, is present.
4.4. Zinc
There was a stark difference observed between the two strains of E. coli tested regarding the changes in the zinc metalloproteomes due to arsenic treatment. The zinc metalloproteins were significantly decreased with AsIII treatment and further decreased with AsV treatment in AW3110, but little to no difference in the zinc metalloproteome was observed in K-12 E. coli cells. Interestingly, arsenic treatment increased the total concentration of soluble zinc for K-12, and only the AsV treatment of AW3110 reduced the soluble zinc concentration (Figure 4d). An explanation for this may come from zinc studies in other organisms. Zinc has been shown to regulate the antioxidant defense system in rats in response to AsIII treatment [36]. Zinc was also reported to have antagonist effects against arsenic in preterm births of pregnant women in Bangladesh [37]. Transcriptomic analysis by Rawle et al. showed the increased transcription of an ABC transporter, zinc-containing alcohol dehydrogenases, and a TraR/DksA C4-type zinc finger protein [13]. Interestingly, AsIII has been shown to displace zinc in zinc-finger proteins containing three or more cysteines [38]. High levels of AsIII cause a scarcity of zinc in an arsenic-resistant Paenibacillus taichungensis isolated from a gold–copper mine [39]. P. taichungensis upregulates a zinc uptake system and a zinc metallochaperone, which have been hypothesized to be a coping mechanism against oxidative stress. Our data support an emerging notion in the literature that zinc plays an important role in the detoxification of arsenic. Our data further suggest that the arsRBC operon may regulate the zinc metalloproteome.
4.5. Iron
In the same arsenic-resistant P. taichungensis study, the downregulation of genes involved in iron uptake was observed following AsIII exposure, which was attributed to be part of an oxidative stress response mechanism [39]. The authors supported this interpretation of the data by pointing to iron as a major source of biological reactive oxygen species and the cells’ needs to limit the labile iron pool to avoid oxidative damage from both iron and arsenic reactive oxygen species generation. Our data on E. coli do not support this hypothesis because the total soluble iron increased in K-12 following AsIII and AsV treatment. K-12 cell treatment with AsIII, however, caused a significant decrease in iron-containing proteins, while AsV treatment led to an increase in high molecular weight proteins or protein complexes, with no significant difference in iron-protein abundance (Figure 2). A similar shift is observed in the absence of the arsRBC operon upon AsV treatment of AW3110 E. coli cells. Unlike in K-12, both AsIII and AsV treatment caused a significant decrease in iron-bound proteins in AW3110 (Figure 3). The presence of the arsR genes in the gene knockout study by Rawle et al. showed increased and decreased expression of gene transcripts coding for iron transporters, depending on which arsR gene was present. Our data demonstrate that there are detectable changes in the iron proteome following arsenic treatments. Additionally, the data support involvement of the arsRBC operon in iron homeostasis, as put forth by Rawle et al.
4.6. Protein Cofactor and Redox Status
As a final analysis of intracellular metalloproteome status, we monitored three wavelengths to probe protein cofactor changes in K-12 and AW3110 E. coli as a function of arsenic treatment. The wavelengths monitored were chosen because absorption at 360 nm and 460 nm will detect oxidized flavin [20], and monitoring at 405 nm can detect heme [21]; however, this method is non-specific, and other molecules can influence the monitored wavelengths [40]. Despite this non-specificity, there are clear differences between the two strains of E. coli. In K-12, the biggest changes appear to be from AsV treatment. These changes were mostly reflected in spectral intensity changes, while shifts in the elution time associated with these signals were minimal (Figure 5a). In AW3110, however, AsIII treatment induced a shift in absorbance to lower molecular weight protein/protein complexes for all wavelengths, while AsV treatment mainly reduced the signal intensity over the entire elution window (Figure 5b). It is difficult to definitively say whether the observed changes are due to changing redox status, but the current literature shows that arsenic does influence the redox status of organisms. A study examining the long-term effect of AsIII treatment in adult rats showed that AsIII inhibits several intermediary enzymes of heme metabolism, as well as alters the intra- and extracellular porphyrin content [41]. Beyond heme, other changes of cellular redox status due to arsenic are expected. The ArsC protein in the E. coli K-12 strain, for example, uses reduced glutathione and glutaredoxin as electron donors [42]. Glutathione is important because it is involved in ZnII and CuII homeostasis and toxicity resistance in E. coli [43]. AsIII exposure has been shown to lower the ratio of reduced to oxidized glutathione, enhance glutathione transferase activity, as well as increase the oxidation of protein thiols in Rhodotorula mucilaginous [44]. This, together with the conserved zinc metalloproteome of K-12 E. coli cells following arsenic treatment, suggests that there may be an interplay between the arsRBC operon, metal homeostasis, and protein-bound cofactor abundance.
5. Concluding Remarks
To the best of our knowledge, this is the first SEC-ICPMS analysis of E. coli bacterial metalloproteome in response to arsenic stress. The data clearly demonstrate that arsenic treatment causes global changes in the metalloproteome of E. coli, and that these changes differ between the two strains of E. coli (K-12 and AW3110) when the cells are under similar levels of metal-induced stress. Our work further supports the hypothesis postulated by Rawle et al. that the ars operon is involved in the widespread regulation in bacteria, and that arsenic detoxification is a complex process involving more than just the three proteins encoded by the arsRBC gene locus. This study sets the stage for future experiments investigating the role of individual arsR, arsB, and arsC genes regarding protein expression and the metalloproteome.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms11020382/s1.
Author Contributions
Conceptualization, J.L., H.F., and M.T.-L.; methodology, J.L., H.F., and M.T.-L.; validation, J.L., H.F., and M.T.-L.; formal analysis, J.L.; investigation, J.L., H.F., and M.T.-L.; resources, B.B.; data curation, S.S., S.C., and G.C.; writing—original draft preparation, J.L.; writing—review and editing, J.L., M.T.-L., H.F., S.S., S.C., G.C., B.B., and V.C.; visualization, J.L. and S.C.; supervision, J.L., H.F., and B.B.; project administration, J.L. and H.F.; funding acquisition, B.B. and V.C. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the U.S. Department of Energy, Office of Science, Basic Energy Sciences, under Award DESC0020246 to B.B. Additional support was provided in part by NSF grant award MCB-1714556 to V.C. Funding for the Proteomics, Metabolomics, and Mass Spectrometry Facility used in this publication was made possible in part by the MJ Murdock Charitable Trust and the National Institute of General Medical Sciences of the National Institutes of Health, under Award Numbers P20GM103474 and S10OD28650, and the MSU Office of Research, Economic Development, and Graduate Education. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The raw data generated during the current study are available from the corresponding author upon request.
Acknowledgments
The authors would like to thank Jordan Pauley and Kristen Greg for help in running the SDS-PAGE gels, as well as Jesse Thomas, Donald Smith, and Laura Dobeck of the Montana State University Mass Spectrometry Core Facility for technical advice regarding the ICP-MS. We also acknowledge the previous gene knockout and transcriptomic work by R. Rawle and T. McDermott, upon which this study is built.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hughes, M.F.; Beck, B.D.; Chen, Y.; Lewis, A.S.; Thomas, D.J. Arsenic Exposure and Toxicology: A Historical Perspective. Toxicol. Sci. 2011, 123, 305–332. [Google Scholar] [CrossRef]
- Sambu, S.; Wilson, R. Arsenic in Food and Water-a Brief History. Toxicol. Ind. Health 2008, 24, 217–226. [Google Scholar] [CrossRef]
- Chung, J.Y.; Do Yu, S.; Hong, Y.S. Environmental Source of Arsenic Exposure. J. Prev. Med. Public Health 2014, 47, 253. [Google Scholar] [CrossRef]
- Páez-Espino, D.; Tamames, J.; De Lorenzo, V.; Cánovas, D. Microbial Responses to Environmental Arsenic. BioMetals 2009, 22, 117–130. [Google Scholar] [CrossRef]
- Yang, H.C.; Fu, H.L.; Lin, Y.F.; Rosen, B.P. Chapter Twelve—Pathways of Arsenic Uptake and Efflux. Curr. Top. Membr. 2012, 69, 325. [Google Scholar] [CrossRef]
- Garbinski, L.D.; Rosen, B.P.; Chen, J. Pathways of Arsenic Uptake and Efflux. Environ. Int. 2019, 126, 585–597. [Google Scholar] [CrossRef]
- Rosen, B.P. Biochemistry of Arsenic Detoxification. FEBS Lett. 2002, 529, 86–92. [Google Scholar] [CrossRef]
- Yan, G.; Chen, X.; Du, S.; Deng, Z.; Wang, L.; Chen, S. Genetic Mechanisms of Arsenic Detoxification and Metabolism in Bacteria. Curr. Genet. 2019, 65, 329–338. [Google Scholar] [CrossRef]
- Rawle, R.; Saley, T.C.; Kang, Y.S.; Wang, Q.; Walk, S.; Bothner, B.; McDermott, T.R. Introducing the ArsR-Regulated Arsenic Stimulon. Front. Microbiol. 2021, 12, 162. [Google Scholar] [CrossRef]
- Murphy, J.N.; Saltikov, C.W. The ArsR Repressor Mediates Arsenite-Dependent Regulation of Arsenate Respiration and Detoxification Operons of Shewanella Sp. Strain ANA-3. J. Bacteriol. 2009, 191, 6722. [Google Scholar] [CrossRef]
- Hao, X.; Lin, Y.; Johnstone, L.; Liu, G.; Wang, G.; Wei, G.; McDermott, T.; Rensing, C. Genome Sequence of the Arsenite-Oxidizing Strain Agrobacterium Tumefaciens 5A. J. Bacteriol. 2012, 194, 903. [Google Scholar] [CrossRef]
- Kang, Y.S.; Heinemann, J.; Bothner, B.; Rensing, C.; McDermott, T.R. Integrated Co-Regulation of Bacterial Arsenic and Phosphorus Metabolisms. Environ. Microbiol. 2012, 14, 3097–3109. [Google Scholar] [CrossRef]
- Rawle, R.A.; Kang, Y.S.; Bothner, B.; Wang, G.; McDermott, T.R. Transcriptomics Analysis Defines Global Cellular Response of Agrobacterium Tumefaciens 5A to Arsenite Exposure Regulated through the Histidine Kinases PhoR and AioS. Environ. Microbiol. 2019, 21, 2659–2676. [Google Scholar] [CrossRef]
- Chandrangsu, P.; Rensing, C.; Helmann, J.D. Metal Homeostasis and Resistance in Bacteria. Nat. Rev. Microbiol. 2017, 15, 338. [Google Scholar] [CrossRef]
- Xu, Z.; Wang, P.; Wang, H.; HangYu, Z.; Au-Yeung, H.Y.; Hirayama, T.; Sun, H.; Yan, A. Zinc Excess Increases Cellular Demand for Iron and Decreases Tolerance to Copper in Escherichia Coli. J. Biol. Chem. 2019, 294, 16978–16991. [Google Scholar] [CrossRef]
- Angeli, S.; Barhydt, T.; Jacobs, R.; Killilea, D.W.; Lithgow, G.J.; Andersen, J.K. Manganese Disturbs Metal and Protein Homeostasis in Caenorhabditis Elegans. Metallomics 2014, 6, 1816–1823. [Google Scholar] [CrossRef] [PubMed]
- Carlin, A.; Shi, W.; Dey, S.; Rosen, B.P. The Ars Operon of Escherichia Coli Confers Arsenical and Antimonial Resistance. J. Bacteriol. 1995, 177, 981–986. [Google Scholar] [CrossRef]
- Lago, L.; Thomas, O.R.B.; Roberts, B.R. Choice of Mobile Phase: Implications for Size Exclusion Chromatography-Inductively Coupled Plasma-Mass Spectrometry Analyses of Copper, Zinc and Iron Metalloproteins. J. Chromatogr. A 2020, 1616, 460806. [Google Scholar] [CrossRef]
- Hare, D.J.; Grubman, A.; Ryan, T.M.; Lothian, A.; Liddell, J.R.; Grimm, R.; Matsuda, T.; Doble, P.A.; Cherny, R.A.; Bush, A.I.; et al. Profiling the Iron, Copper and Zinc Content in Primary Neuron and Astrocyte Cultures by Rapid Online Quantitative Size Exclusion Chromatography-Inductively Coupled Plasma-Mass Spectrometry. Metallomics 2013, 5, 1656–1662. [Google Scholar] [CrossRef]
- Chapman, S.K.; Reid, G.A.; Macheroux, P. UV-Visible Spectroscopy as a Tool to Study Flavoproteins. Methods Mol. Biol. 1999, 131, 1–7. [Google Scholar] [CrossRef]
- Hopp, M.T.; Schmalohr, B.F.; Kühl, T.; Detzel, M.S.; Wißbrock, A.; Imhof, D. Heme Determination and Quantification Methods and Their Suitability for Practical Applications and Everyday Use. Anal. Chem. 2020, 92, 9429–9440. [Google Scholar] [CrossRef]
- Shi, K.; Wang, Q.; Fan, X.; Wang, G. Proteomics and Genetic Analyses Reveal the Effects of Arsenite Oxidation on Metabolic Pathways and the Roles of AioR in Agrobacterium Tumefaciens GW4. Environ. Pollut. 2018, 235, 700–709. [Google Scholar] [CrossRef]
- Liu, G.; Liu, M.; Kim, E.H.; Maaty, W.S.; Bothner, B.; Lei, B.; Rensing, C.; Wang, G.; McDermott, T.R. A Periplasmic Arsenite-Binding Protein Involved in Regulating Arsenite Oxidation. Environ. Microbiol. 2012, 14, 1624–1634. [Google Scholar] [CrossRef] [PubMed]
- Kashyap, D.R.; Botero, L.M.; Franck, W.L.; Hassett, D.J.; McDermott, T.R. Complex Regulation of Arsenite Oxidation in Agrobacterium Tumefaciens. J. Bacteriol. 2006, 188, 1081–1088. [Google Scholar] [CrossRef]
- Bogdan, G.M.; Sampayo-Reyes, A.; Vasken Aposhian, H. Arsenic Binding Proteins of Mammalian Systems: I. Isolation of Three Arsenite-Binding Proteins of Rabbit Liver. Toxicology 1994, 93, 175–193. [Google Scholar] [CrossRef]
- Rubino, J.T.; Franz, K.J. Coordination Chemistry of Copper Proteins: How Nature Handles a Toxic Cargo for Essential Function. J. Inorg. Biochem. 2012, 107, 129–143. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Wang, H.; Li, X.-F.; Lu, X.; Cullen, W.R.; Arnold, L.L.; Cohen, S.M.; Le, X.C. Evidence of Hemoglobin Binding to Arsenic as a Basis for the Accumulation of Arsenic in Rat Blood. Chem. Res. Toxicol. 2004, 17, 1733–1742. [Google Scholar] [CrossRef]
- Lee, D.-Y.D.; Galera-Laporta, L.; Bialecka-Fornal, M.; Briggs, S.P.; Garcia-Ojalvo, J.; Moon, E.C.; Shen, Z.; Rol, G.; Sü El, M. Magnesium Flux Modulates Ribosomes to Increase Bacterial Survival In Brief Bacteria Cope against Ribosome-Targeting Antibiotics by Controlling the Influx of Intracellular Magnesium and Controlling Their Membrane Potential. Article Magnesium Flux Modulates Ribosomes to Increase Bacterial Survival. Cell 2019, 177, 352–360.e13. [Google Scholar] [CrossRef]
- Basturea, G.N.; Harris, T.K.; Deutscher, M.P. Growth of a Bacterium That Apparently Uses Arsenic Instead of Phosphorus Is a Consequence of Massive Ribosome Breakdown. J. Biol. Chem. 2012, 287, 28816–28819. [Google Scholar] [CrossRef] [PubMed]
- Guerra-Moreno, A.; Isasa, M.; Bhanu, M.K.; Waterman, D.P.; Eapen, V.V.; Gygi, S.P.; Hanna, J. Proteomic Analysis Identifies Ribosome Reduction as an Effective Proteotoxic Stress Response. J. Biol. Chem. 2015, 290, 29695. [Google Scholar] [CrossRef]
- Khannpnavar, B.; Mehta, V.; Qi, C.; Korkhov, V. Structure and Function of Adenylyl Cyclases, Key Enzymes in Cellular Signaling. Curr. Opin. Struct. Biol. 2020, 63, 34–41. [Google Scholar] [CrossRef]
- Steer, M.L. Adenyl Cyclase. Ann. Surg. 1975, 182, 603. [Google Scholar] [CrossRef]
- Hayer-Hartl, M.; Bracher, A.; Hartl, F.U. The GroEL-GroES Chaperonin Machine: A Nano-Cage for Protein Folding. Trends Biochem. Sci. 2016, 41, 62–76. [Google Scholar] [CrossRef]
- Carreras-Puigvert, J.; Zitnik, M.; Jemth, A.S.; Carter, M.; Unterlass, J.E.; Hallström, B.; Loseva, O.; Karem, Z.; Calderón-Montanõ, J.M.; Lindskog, C.; et al. A Comprehensive Structural, Biochemical and Biological Profiling of the Human NUDIX Hydrolase Family. Nat. Commun. 2017, 8, 1541. [Google Scholar] [CrossRef]
- Mildvan, A.S.; Xia, Z.; Azurmendi, H.F.; Saraswat, V.; Legler, P.M.; Massiah, M.A.; Gabelli, S.B.; Bianchet, M.A.; Kang, L.W.; Amzel, L.M. Structures and Mechanisms of Nudix Hydrolases. Arch. Biochem. Biophys. 2005, 433, 129–143. [Google Scholar] [CrossRef]
- Ganger, R.; Garla, R.; Mohanty, B.P.; Bansal, M.P.; Garg, M.L. Protective Effects of Zinc Against Acute Arsenic Toxicity by Regulating Antioxidant Defense System and Cumulative Metallothionein Expression. Biol. Trace Elem. Res. 2016, 169, 218–229. [Google Scholar] [CrossRef]
- Wei, Y.Y.; Huang, H.; Xia, Y.K.; Wei, L.M.; Chen, X.; Zhang, R.Y.; Duan, W.W.; Su, L.; Rahman, M.L.; Rahman, M.; et al. Antagonistic Effect of Early Stage Zinc on Arsenic Toxicity Induced Preterm Birth during Pregnancy: Evidence from a Rural Bangladesh Birth Cohort. Chin. Med. J. (Engl.) 2021, 134, 619–621. [Google Scholar] [CrossRef]
- Zhou, X.; Sun, X.; Cooper, K.L.; Wang, F.; Liu, K.J.; Hudson, L.G. Arsenite Interacts Selectively with Zinc Finger Proteins Containing C3H1 or C4 Motifs. J. Biol. Chem. 2011, 286, 22855. [Google Scholar] [CrossRef]
- Yu, Y.; Su, J.; Xu, J.; Li, Y.P.; Alwathnani, H.A.; Wu, Z.; Ji, C.; Feng, R.; Rensing, C.; Herzberg, M. As(III) Exposure Induces a Zinc Scarcity Response and Restricts Iron Uptake in High-Level Arsenic-Resistant Paenibacillus Taichungensis Strain NC1. Appl. Environ. Microbiol. 2022, 88, e00312-22. [Google Scholar] [CrossRef]
- Soltani, S.; Ojaghi, A.; Robles, F.E. Deep UV Dispersion and Absorption Spectroscopy of Biomolecules. Biomed. Opt. Express 2019, 10, 487. [Google Scholar] [CrossRef]
- Aguilar-González, M.G.; Hernández, A.; López, M.L.; Mendoza-Figueroa, T.; Albores, A. Arsenite Alters Heme Synthesis in Long-Term Cultures of Adult Rat Hepatocytes. Toxicol. Sci. 1999, 49, 281–289. [Google Scholar] [CrossRef]
- Shi, J.; Vlamis-Gardikas, A.; Åslund, F.; Holmgren, A.; Rosen, B.P. Reactivity of Glutaredoxins 1, 2, and 3 FromEscherichia Coli Shows That Glutaredoxin 2 Is the Primary Hydrogen Donor to ArsC-Catalyzed Arsenate Reduction. J. Biol. Chem. 1999, 274, 36039–36042. [Google Scholar] [CrossRef] [PubMed]
- Helbig, K.; Bleuel, C.; Krauss, G.J.; Nies, D.H. Glutathione and Transition-Metal Homeostasis in Escherichia Coli. J. Bacteriol. 2008, 190, 5431–5438. [Google Scholar] [CrossRef]
- Ilyas, S.; Rehman, A.; Coelho, A.V.; Sheehan, D. Proteomic Analysis of an Environmental Isolate of Rhodotorula Mucilaginosa after Arsenic and Cadmium Challenge: Identification of a Protein Expression Signature for Heavy Metal Exposure. J. Proteom. 2016, 141, 47–56. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).