Development of A Rapid, Low-Cost Portable Detection Assay for Enterococci in Wastewater and Environmental Waters
Abstract
1. Introduction
2. Material and Methods
2.1. Samples
2.2. Bacterial Isolates
2.3. Standard Plate Count for Enterococcus faecalis
2.4. Enterolert-E Test
2.5. Total DNA Extraction from Selected Bacteria
2.6. Designing and Screening of RPA Primers
2.7. Recombinase Polymerase Amplification-Agarose Gel Electrophoresis (RPA-AGE)
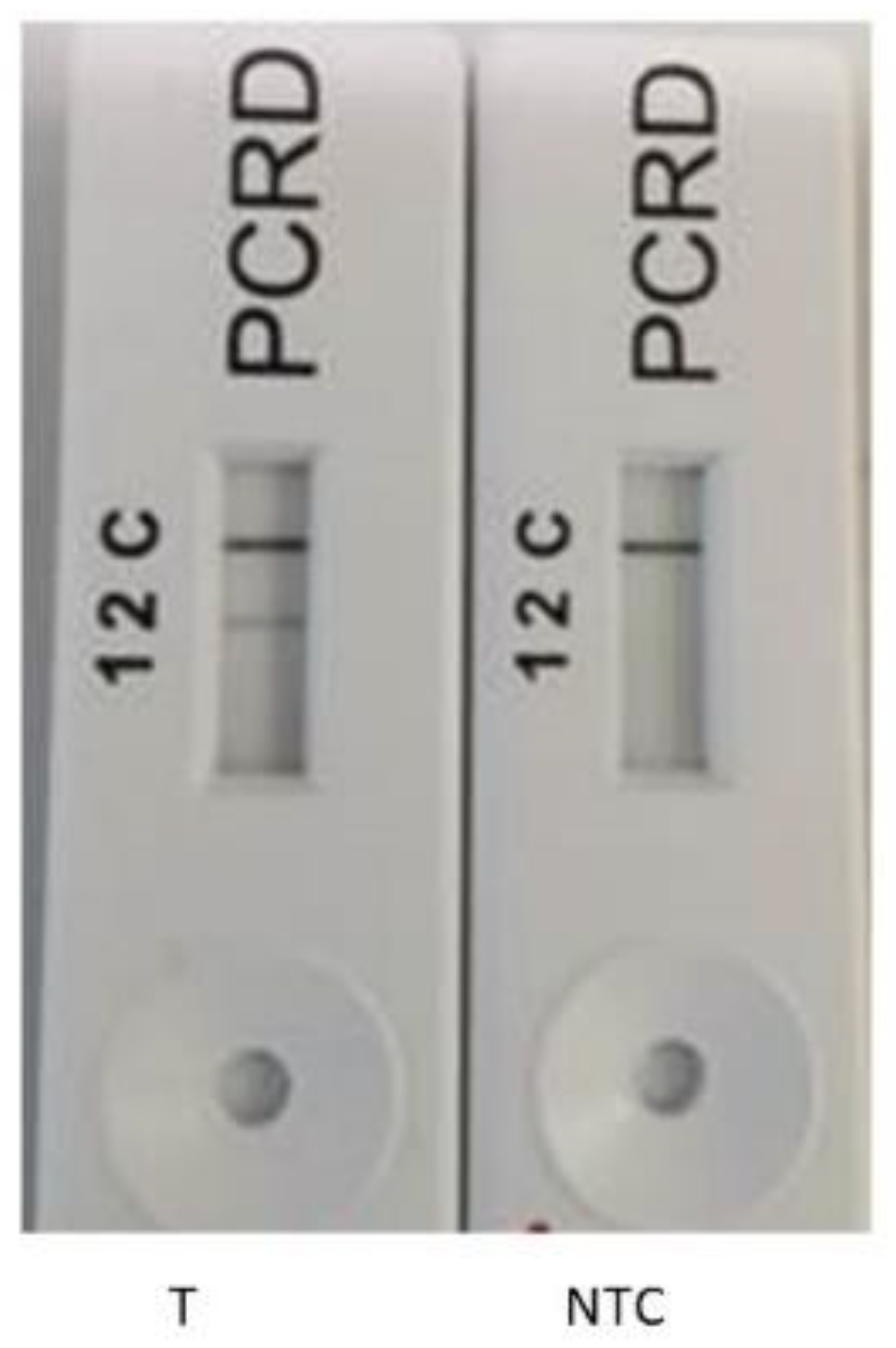
2.8. Recombinase Polymerase Amplification-Lateral Flow Assay (RPA-LFA)
2.9. Selectivity and Sensitivity of Recombinase Polymerase Amplification (RPA)
2.10. Quantification of the Recombinase Polymerase Amplification-Lateral Flow Assay (RPA-LFA) Using the Benchtop Reader
3. Results and Discussion
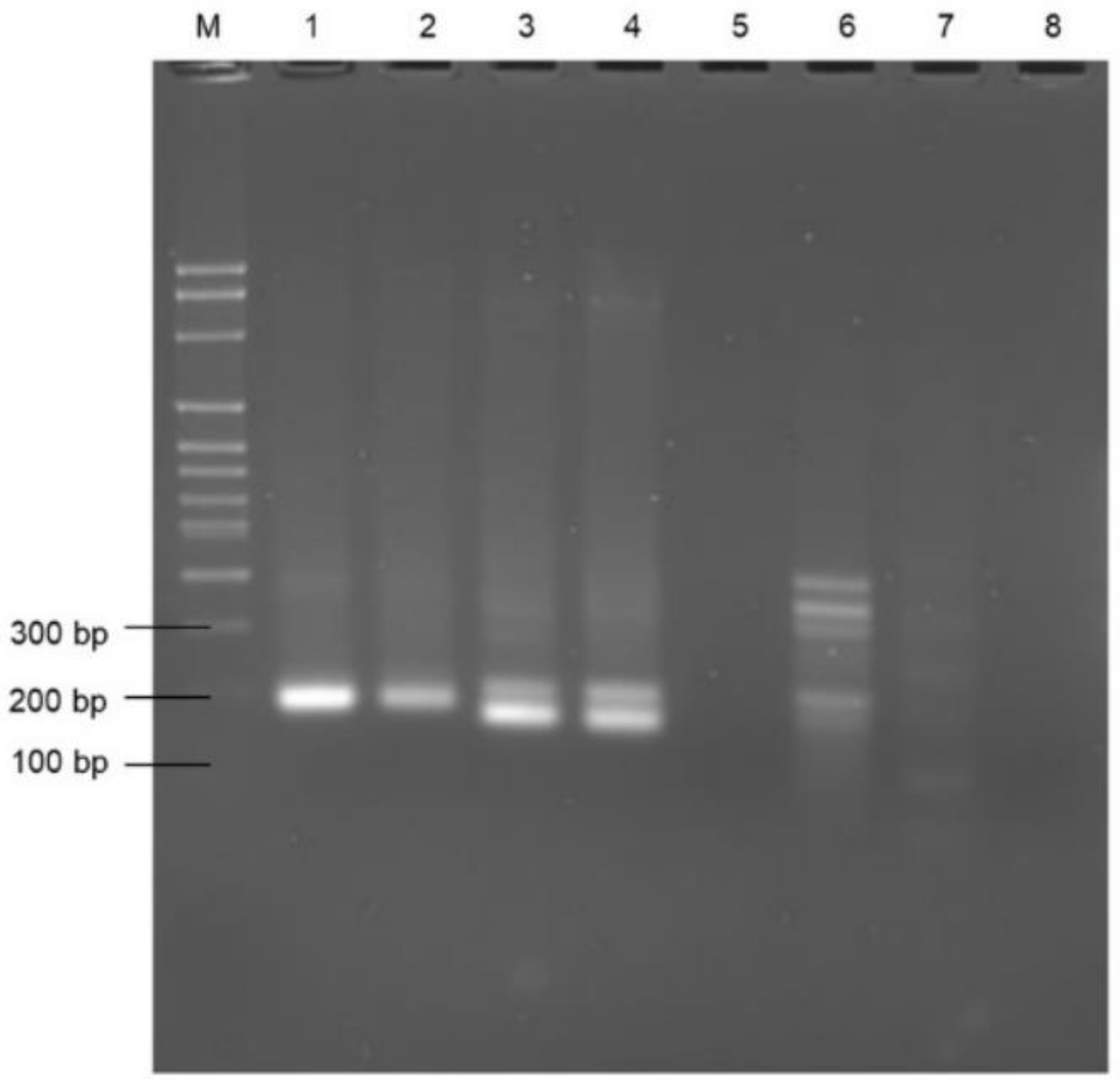
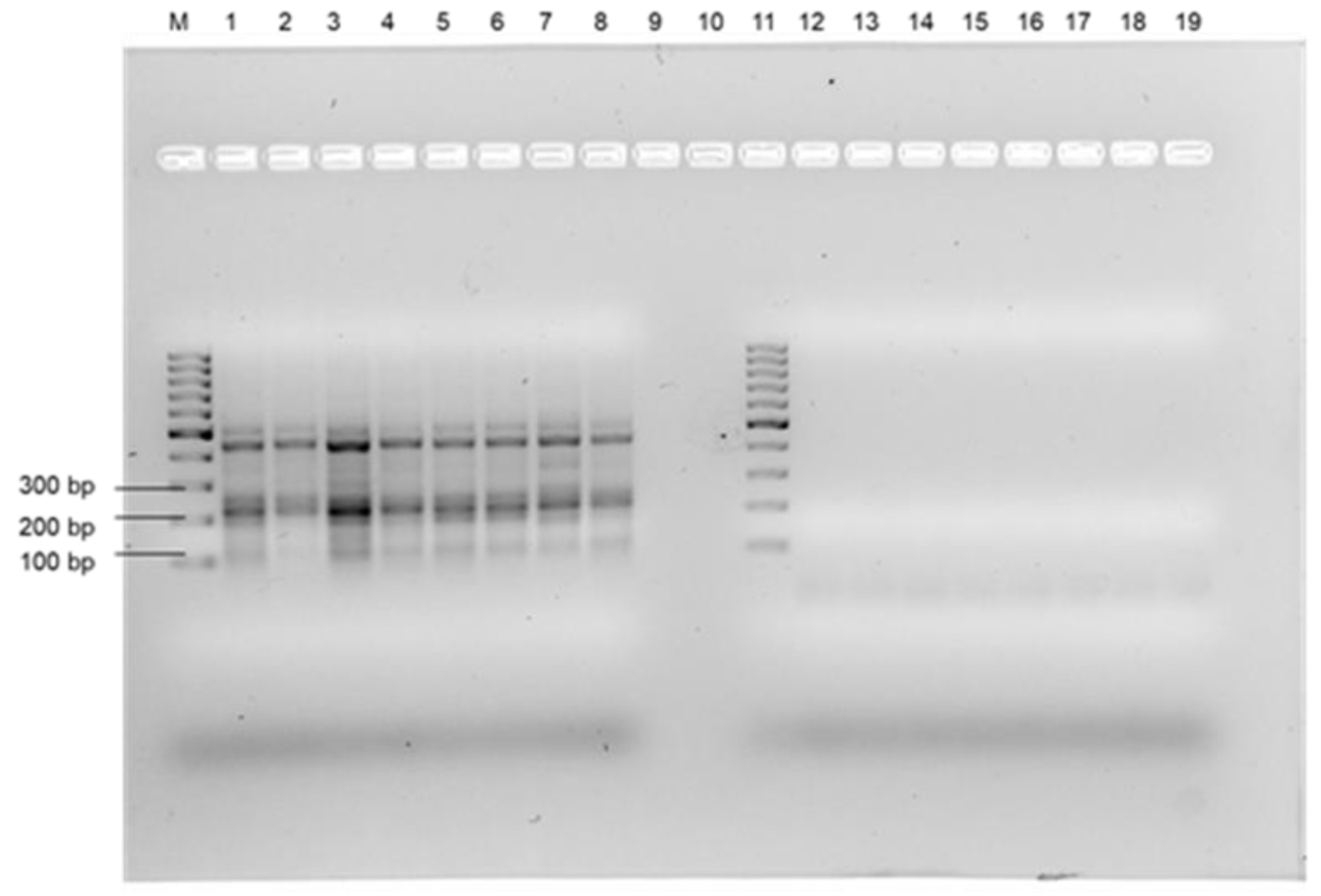
3.1. DNA Extraction and Selection of the Primers
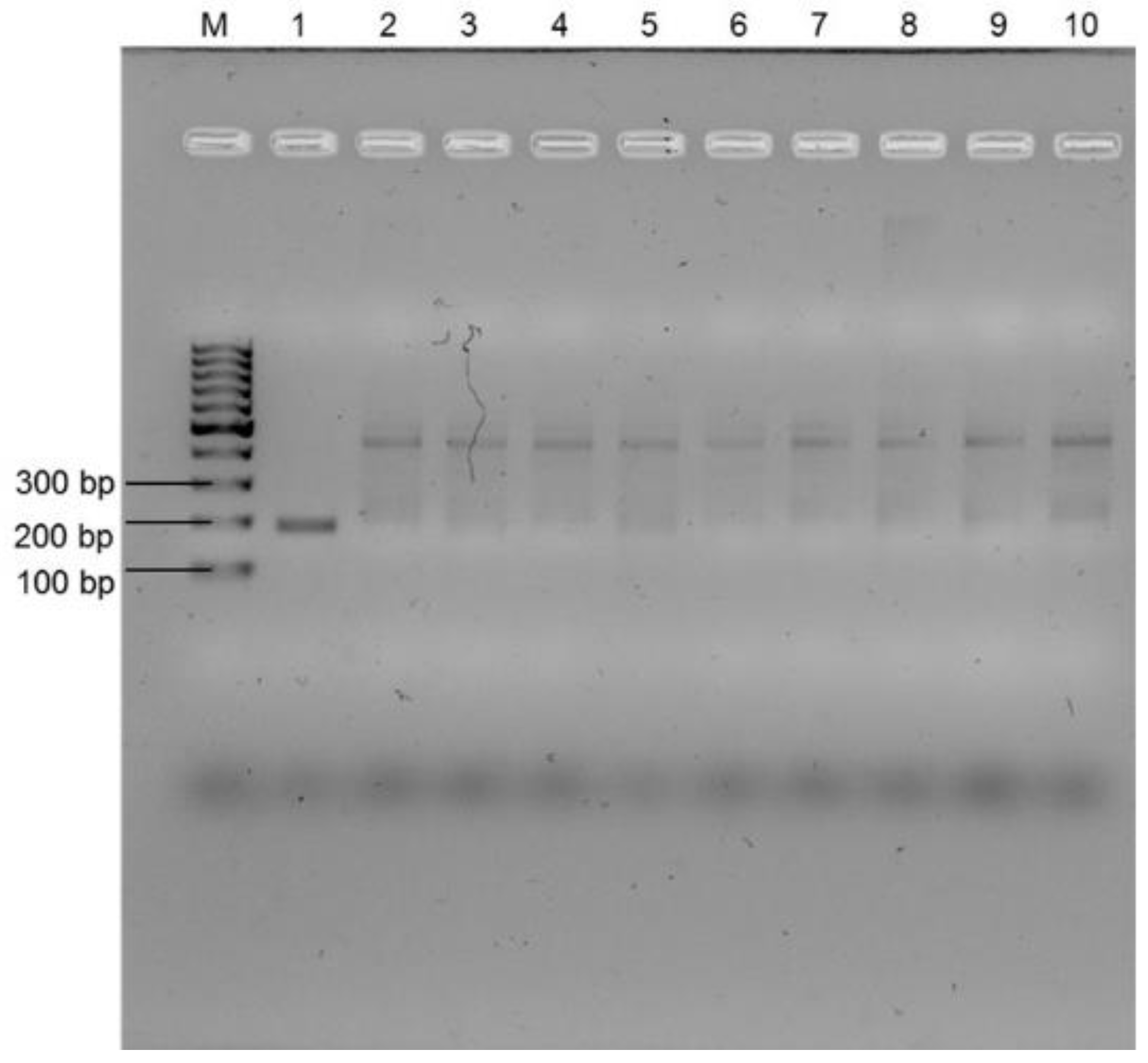
3.2. Selectivity Assessment of the RPA Using Selected Primer Set
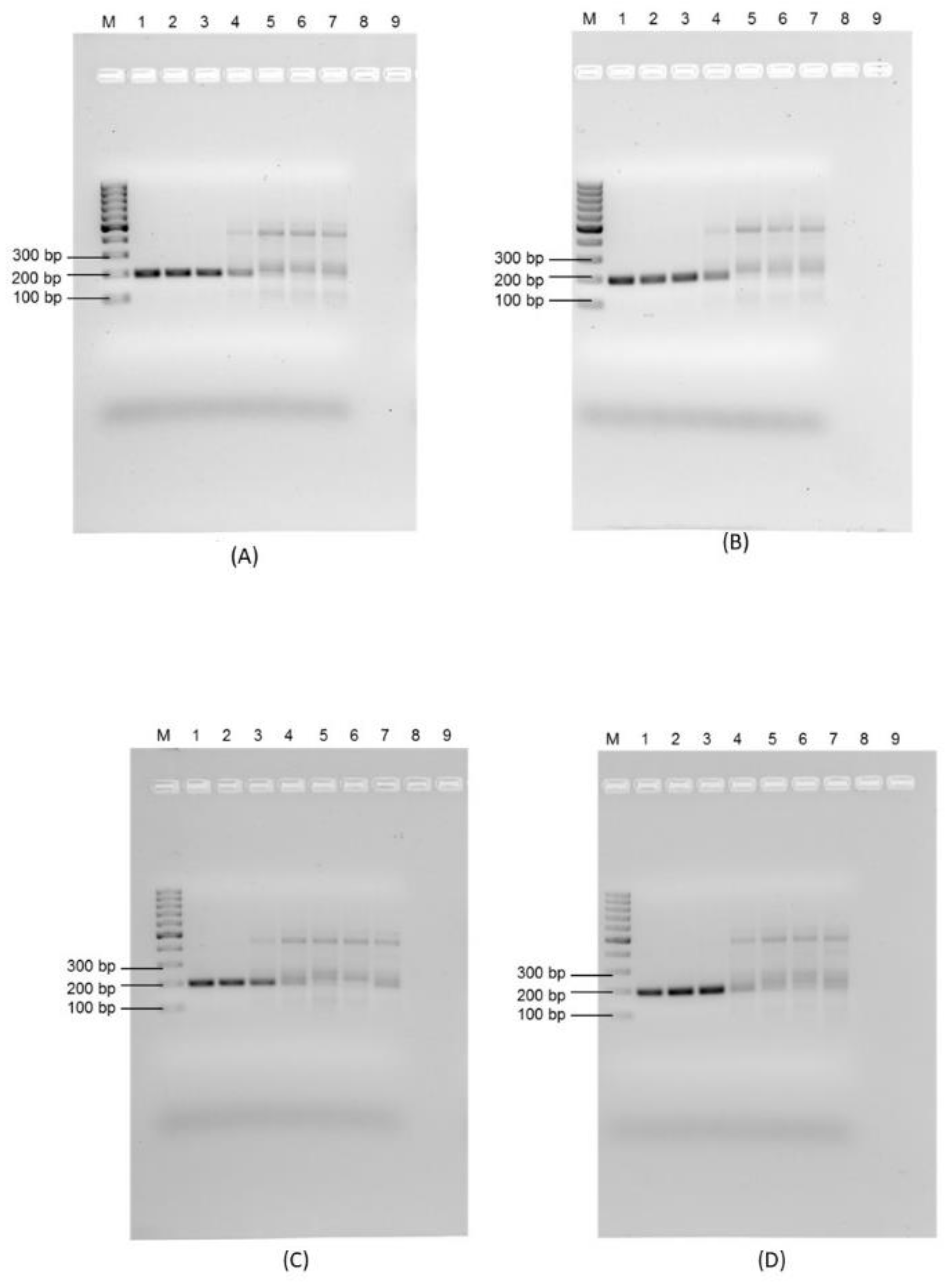
3.3. Sensitivity of the Recombinase Polymerase Assay and Impact of Environmental Water Characteristics
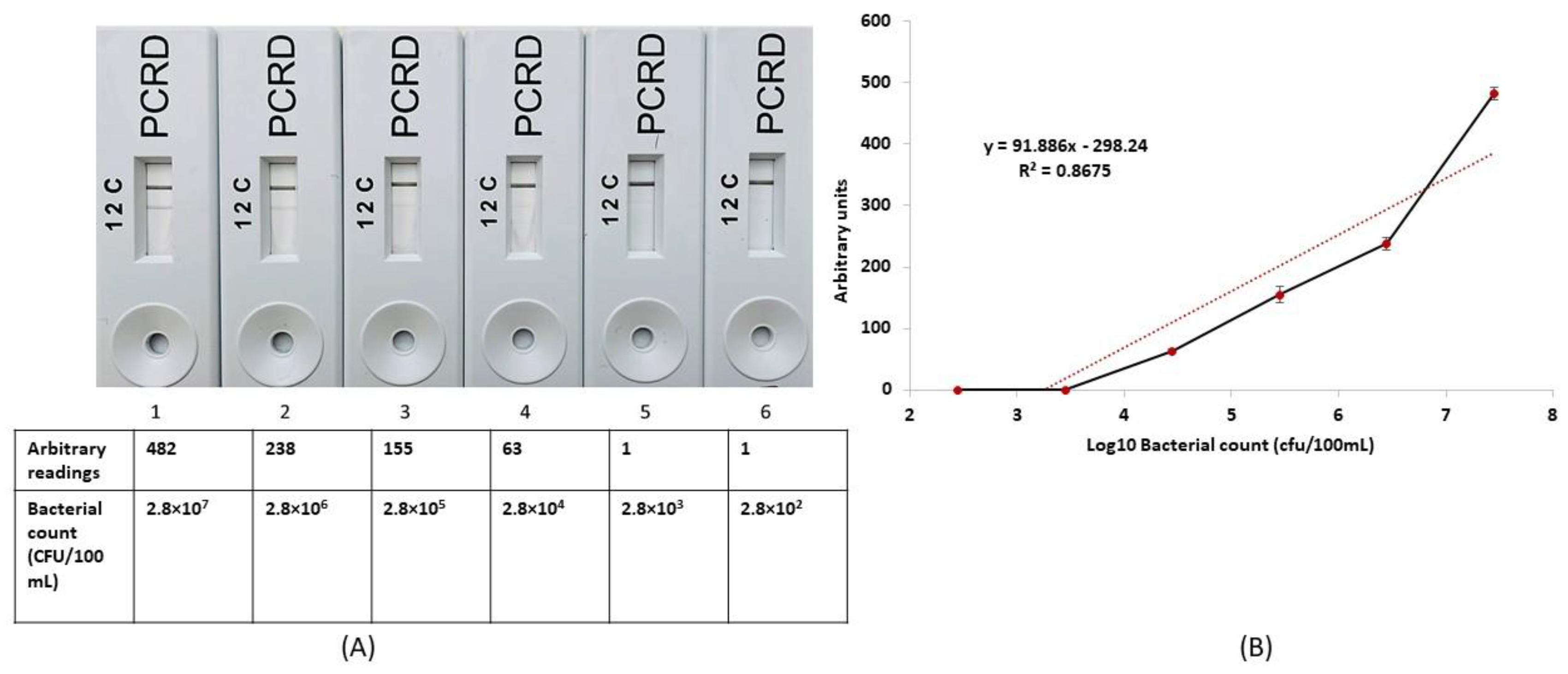
3.4. Potential for RPA-LFA as a Quantitative Measurement System for the Enumeration of E. faecalis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Drinking-Water; WHO: Geneva, Switzerland, 2022.
- Salgot, M.; Huertas, E.; Weber, S.; Dott, W.; Hollender, J. Wastewater reuse and risk: Definition of key objectives. Desalination 2006, 187, 29–40. [Google Scholar] [CrossRef]
- Fernandes, L.S.; Galvão, A.; Santos, R.; Monteiro, S. Impact of water reuse on agricultural practices and human health. Environ. Res. 2023, 216, 114762. [Google Scholar] [PubMed]
- Alegbeleye, O.; Sant’Ana, A.S. Microbiological quality of irrigation water for cultivation of fruits and vegetables: An overview of available guidelines, water testing strategies and some factors that influence compliance. Environ. Res. 2022, 220, 114771. [Google Scholar] [CrossRef] [PubMed]
- Uyttendaele, M.; Jaykus, L.A.; Amoah, P.; Chiodini, A.; Cunliffe, D.; Jacxsens, L.; Holvoet, K.; Korsten, L.; Lau, M.; McClure, P. Microbial hazards in irrigation water: Standards, norms, and testing to manage use of water in fresh produce primary production. Compr. Rev. Food Sci. Food Saf. 2015, 14, 336–356. [Google Scholar] [CrossRef]
- Foladori, P.; Cutrupi, F.; Cadonna, M.; Manara, S. Coronaviruses and SARS-CoV-2 in sewerage and their removal: Step by step in wastewater treatment plants. Environ. Res. 2022, 207, 112204. [Google Scholar] [CrossRef] [PubMed]
- An, X.-L.; Wang, J.-Y.; Pu, Q.; Li, H.; Pan, T.; Li, H.-Q.; Pan, F.-X.; Su, J.-Q. High-throughput diagnosis of human pathogens and fecal contamination in marine recreational water. Environ. Res. 2020, 190, 109982. [Google Scholar] [CrossRef]
- Tallon, P.; Magajna, B.; Lofranco, C.; Leung, K.T. Microbial indicators of faecal contamination in water: A current perspective. Water Air Soil Pollut. 2005, 166, 139–166. [Google Scholar] [CrossRef]
- Jokinen, C.C.; Cook, S.R.; Reuter, T.; Tymensen, L. Assessing enterococci as an alternative fecal indicator for irrigation water quality. Agric. Water Manag. 2020, 233, 106098. [Google Scholar] [CrossRef]
- Maheux, A.F.; Bissonnette, L.; Boissinot, M.; Bernier, J.-L.T.; Huppe, V.; Berube, E.; Boudreau, D.K.; Picard, F.J.; Huletsky, A.; Bergeron, M.G. Method for rapid and sensitive detection of Enterococcus sp. and Enterococcus faecalis/faecium cells in potable water samples. Water Res. 2011, 45, 2342–2354. [Google Scholar]
- Boehm, A.B.; Sassoubre, L.M. Enterococci as Indicators of Environmental Fecal Contamination; National Center fo Biotechnology Information: Bethesda, MD, USA, 2014.
- Collender, P.A.; Kirby, A.E.; Addiss, D.G.; Freeman, M.C.; Remais, J.V. Methods for quantification of soil-transmitted helminths in environmental media: Current techniques and recent advances. Trends Parasitol. 2015, 31, 625–639. [Google Scholar] [CrossRef]
- Young, S.; Nayak, B.; Sun, S.; Badgley, B.D.; Rohr, J.R.; Harwood, V.J. Vancomycin-resistant enterococci and bacterial community structure following a sewage spill into an aquatic environment. Appl. Environ. Microbiol. 2016, 82, 5653–5660. [Google Scholar] [CrossRef] [PubMed]
- Teoh, B.-T.; Sam, S.-S.; Tan, K.-K.; Danlami, M.B.; Shu, M.-H.; Johari, J.; Hooi, P.-S.; Brooks, D.; Piepenburg, O.; Nentwich, O. Early detection of dengue virus by use of reverse transcription-recombinase polymerase amplification. J. Clin. Microbiol. 2015, 53, 830–837. [Google Scholar] [CrossRef] [PubMed]
- Piepenburg, O.; Williams, C.H.; Stemple, D.L.; Armes, N.A. DNA detection using recombination proteins. PLoS Biol. 2006, 4, e204. [Google Scholar] [CrossRef] [PubMed]
- Lobato, I.M.; O’Sullivan, C.K. Recombinase polymerase amplification: Basics, applications and recent advances. TrAC Trends Anal. Chem. 2018, 98, 19–35. [Google Scholar] [CrossRef]
- Li, J.; Macdonald, J.; von Stetten, F. A comprehensive summary of a decade development of the recombinase polymerase amplification. Analyst 2019, 144, 31–67, Correction in Analyst 2020, 145, 1950–1960. [Google Scholar] [CrossRef]
- Daher, R.K.; Stewart, G.; Boissinot, M.; Bergeron, M.G. Recombinase polymerase amplification for diagnostic applications. Clin. Chem. 2016, 62, 947–958. [Google Scholar] [CrossRef] [PubMed]
- Kersting, S.; Rausch, V.; Bier, F.F.; von Nickisch-Rosenegk, M. Multiplex isothermal solid-phase recombinase polymerase amplification for the specific and fast DNA-based detection of three bacterial pathogens. Microchim. Acta 2014, 181, 1715–1723. [Google Scholar] [CrossRef] [PubMed]
- Euler, M.; Wang, Y.; Heidenreich, D.; Patel, P.; Strohmeier, O.; Hakenberg, S.; Niedrig, M.; Hufert, F.T.; Weidmann, M. Development of a panel of recombinase polymerase amplification assays for detection of biothreat agents. J. Clin. Microbiol. 2013, 51, 1110–1117. [Google Scholar] [CrossRef]
- Wang, J.; Liu, L.; Wang, J.; Sun, X.; Yuan, W. Recombinase polymerase amplification assay—A simple, fast and cost-effective alternative to real time PCR for specific detection of feline herpesvirus-1. PLoS ONE 2017, 12, e0166903. [Google Scholar] [CrossRef]
- James, A.; Macdonald, J. Recombinase polymerase amplification: Emergence as a critical molecular technology for rapid, low-resource diagnostics. Expert Rev. Mol. Diagn. 2015, 15, 1475–1489. [Google Scholar] [CrossRef]
- Rani, A.; Ravindran, V.B.; Surapaneni, A.; Shahsavari, E.; Haleyur, N.; Mantri, N.; Ball, A.S. Evaluation and comparison of recombinase polymerase amplification coupled with lateral-flow bioassay for Escherichia coli O157: H7 detection using different genes. Sci. Rep. 2021, 11, 1881. [Google Scholar] [CrossRef]
- Rani, A.; Dike, C.C.; Mantri, N.; Ball, A. Point-of-Care Lateral Flow Detection of Viable Escherichia coli O157: H7 Using an Improved Propidium Monoazide-Recombinase Polymerase Amplification Method. Foods 2022, 11, 3207. [Google Scholar] [CrossRef]
- Rani, A.; Ravindran, V.B.; Surapaneni, A.; Mantri, N.; Ball, A.S. Trends in point-of-care diagnosis for Escherichia coli O157: H7 in food and water. Int. J. Food Microbiol. 2021, 349, 109233. [Google Scholar] [CrossRef]
- Ravindran, V.B.; Khallaf, B.; Surapaneni, A.; Crosbie, N.D.; Soni, S.K.; Ball, A.S. Detection of helminth ova in wastewater using recombinase polymerase amplification coupled to lateral flow strips. Water 2020, 12, 691. [Google Scholar] [CrossRef]
- Lau, H.Y.; Botella, J.R. Advanced DNA-based point-of-care diagnostic methods for plant diseases detection. Front. Plant Sci. 2017, 8, 2016. [Google Scholar] [CrossRef]
- Rani, A.; Donovan, N.; Mantri, N. The future of plant pathogen diagnostics in a nursery production system. Biosens. Bioelectron. 2019, 145, 111631. [Google Scholar] [CrossRef] [PubMed]
- Tu, P.-A.; Shiu, J.-S.; Lee, S.-H.; Pang, V.F.; Wang, D.-C.; Wang, P.-H. Development of a recombinase polymerase amplification lateral flow dipstick (RPA-LFD) for the field diagnosis of caprine arthritis-encephalitis virus (CAEV) infection. J. Virol. Methods 2017, 243, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Xing, W.; Yu, X.; Fu, W.; Wang, Y.; Zou, M.; Luo, Z.; Xu, D. Recombinase polymerase amplification combined with a lateral flow dipstick for rapid and visual detection of Schistosoma japonicum. Parasites Vectors 2016, 9, 476. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Song, L. Recombinase polymerase amplification combined with a lateral flow dipstick for rapid and visual detection of adenovirus. Mil. Med. Sci. 2017, 547–551. [Google Scholar]
- Rohrman, B.A.; Leautaud, V.; Molyneux, E.; Richards-Kortum, R.R. A lateral flow assay for quantitative detection of amplified HIV-1 RNA. PLoS ONE 2012, 7, e45611. [Google Scholar] [CrossRef]
- Bahadır, E.B.; Sezgintürk, M.K. Lateral flow assays: Principles, designs and labels. TrAC Trends Anal. Chem. 2016, 82, 286–306. [Google Scholar]
- Urusov, A.E.; Zherdev, A.V.; Dzantiev, B.B. Towards lateral flow quantitative assays: Detection approaches. Biosensors 2019, 9, 89. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Du, J.; Peng, Z. Correlation between Enterococcus faecalis and persistent intraradicular infection compared with primary intraradicular infection: A systematic review. J. Endod. 2015, 41, 1207–1213. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Hu, J.; Li, X.; Li, X.; Wang, L.; Fan, S.; Jin, X.; Wang, K.; Zhao, W.; Zhu, W. Rapid and specific detection of Enterococcus faecalis with a visualized isothermal amplification method. Front. Cell. Infect. Microbiol. 2022, 12, 1339. [Google Scholar] [CrossRef] [PubMed]
- Panpru, P.; Srisrattakarn, A.; Panthasri, N.; Tippayawat, P.; Chanawong, A.; Tavichakorntrakool, R.; Daduang, J.; Wonglakorn, L.; Lulitanond, A. Rapid detection of Enterococcus and vancomycin resistance using recombinase polymerase amplification. PeerJ 2021, 9, e12561. [Google Scholar] [CrossRef]
- Raja, B.; Goux, H.J.; Marapadaga, A.; Rajagopalan, S.; Kourentzi, K.; Willson, R.C. Development of a panel of recombinase polymerase amplification assays for detection of common bacterial urinary tract infection pathogens. J. Appl. Microbiol. 2017, 123, 544–555. [Google Scholar] [CrossRef] [PubMed]
- Yin, K.; Ding, X.; Xu, Z.; Li, Z.; Wang, X.; Zhao, H.; Otis, C.; Li, B.; Liu, C. Multiplexed colorimetric detection of SARS-CoV-2 and other pathogens in wastewater on a 3D printed integrated microfluidic chip. Sens. Actuators B Chem. 2021, 344, 130242. [Google Scholar] [CrossRef]
- Ke, D.; Picard, F.J.; Martineau, F.; Ménard, C.; Roy, P.H.; Ouellette, M.; Bergeron, M.G. Development of a PCR assay for rapid detection of enterococci. J. Clin. Microbiol. 1999, 37, 3497–3503. [Google Scholar] [CrossRef]
- Naser, S.M.; Thompson, F.L.; Hoste, B.; Gevers, D.; Dawyndt, P.; Vancanneyt, M.; Swings, J. Application of multilocus sequence analysis (MLSA) for rapid identification of Enterococcus species based on rpoA and pheS genes. Microbiology 2005, 151, 2141–2150. [Google Scholar] [CrossRef]
- Urusova, D.V.; Merriman, J.A.; Gupta, A.; Chen, L.; Mathema, B.; Caparon, M.G.; Khader, S.A. Rifampin resistance mutations in the rpoB gene of Enterococcus faecalis impact host macrophage cytokine production. Cytokine 2022, 151, 155788. [Google Scholar] [CrossRef]
- Kosecka-Strojek, M.; Wolska, M.; Żabicka, D.; Sadowy, E.; Międzobrodzki, J. Identification of clinically relevant Streptococcus and Enterococcus species based on biochemical methods and 16S rRNA, sodA, tuf, rpoB, and recA gene sequencing. Pathogens 2020, 9, 939. [Google Scholar] [CrossRef] [PubMed]
- Edge, T.A.; Boehm, A.B. Classical and molecular methods to measure fecal bacteria. In The Fecal Bacteria; Wiley Online Library: New York City, NY, USA, 2010; pp. 241–273. [Google Scholar] [CrossRef]
- Munawar, M.A. Critical insight into recombinase polymerase amplification technology. Expert Rev. Mol. Diagn. 2022, 22, 725–737. [Google Scholar] [CrossRef]
- Carey, S.A.; Goldstein, R.E.R.; Gibbs, S.G.; Claye, E.; He, X.; Sapkota, A.R. Occurrence of vancomycin-resistant and-susceptible Enterococcus spp. in reclaimed water used for spray irrigation. Environ. Res. 2016, 147, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Domingo, J.W.S.; Siefring, S.C.; Haugland, R.A. Real-time PCR method to detect Enterococcus faecalis in water. Biotechnol. Lett. 2003, 25, 261–265. [Google Scholar] [CrossRef]
- He, J.-W.; Jiang, S. Quantification of enterococci and human adenoviruses in environmental samples by real-time PCR. Appl. Environ. Microbiol. 2005, 71, 2250–2255. [Google Scholar] [CrossRef] [PubMed]
- Mason, M.G.; Botella, J.R. A simple, robust and equipment-free DNA amplification readout in less than 30 seconds. RSC Adv. 2019, 9, 24440–24450. [Google Scholar] [CrossRef]
| Primer Name | Targeted Gene | Oligonucleotide Sequence (5′ to 3′) | Direction |
|---|---|---|---|
| Enttuf F1 | tuf | TGACGATAAAGAAGAGAGCGGAGACACGAATC | Forward |
| Enttuf R1 | tuf | CCCCATCTTTTTCATTTGGAGCGATAGTTTTT | Reverse |
| Enttuf F2 | tuf | GACGATAAAGAAGAGAGCGGAGACACGAATCC | Forward |
| Enttuf R2 | tuf | GCCCCATCTTTTTCATTTGGAGCGATAGTTTT | Reverse |
| Ent rpoA F1 | rpoA | GCAGTGAAAACCGAAGCAAGCGCCATTCAAAT | Forward |
| Ent rpoA R1 | rpoA | TAAGGCATCAAATTCTTCCTCCAATAAAATAT | Reverse |
| Ent rpoB F1 F1 | rpoB | GGACCCGCTACCGTGACTGCCGGCGATATTATCG | Forward |
| Ent rpoB R1 R1 | rpoB | Biotin-GAATCAACTGGAAGTACACCGATTGGCATATC | Reverse |
| Ent rpoB P1 | 5′-FAM -GATGTTGAGATCCTAAATAAAGATTTAGTTAT/dSpacer/TGTAGTGTTGCTGAAGGAG-C3 spacer-3′ | Probe |
| Sample Dilution (from A Culture Containing 2.8 × 108 Organisms/100 mL) Calculated Using Selective Isolation Plating. Actual Numbers are Shown in Brackets | Estimated Numbers in Spiked Tap Water | Estimated Numbers in Spiked Saline | Estimated Numbers in Spiked Koo Wee Rup Wastewater | Estimated Numbers in Non-Spiked Lang Lang Wastewater |
|---|---|---|---|---|
| Control (0/100 mL) | 0 | 0 | 0 | 0 |
| 10−6 (2.8 × 100) | 0 | 0 | 1222.0–1986.3 | 0–1 |
| 10−5 (2.8 × 101) | 120–175 | 0 | 1630.4–2419.6 | 0.7–3.1 |
| 10−4 (2.8 × 102) | 1203–1750 | 20–30.4 | >2419.6 to infinite | 55.0–77.1 |
| 10−3 (2.8 × 103) | >2419.6 to infinite | 143.7–181.9 | >2419.6 to infinite | 126.1–187.2 |
| 10−2 (2.8 × 104) | >2419.6 to infinite | >2419.6 to infinite | >2419.6 to infinite | 120.8–179.3 |
| 10−1 (2.8 × 105) | >2419.6 to infinite | >2419.6 to infinite | >2419.6 to infinite | 126.1–187.2 |
| 100 (2.8 × 106) | >2419.6 to infinite | >2419.6 to infinite | >2419.6 to infinite | 206.6–325.5 |
| Technique | Description | Advantages | Disadvantages | Website |
|---|---|---|---|---|
| Selective Plate Isolation | Use of a dilution series of sample plated onto selective isolation media enabling the counting of colonies present (colony-forming units). | Simple detection system. Can be accurate. | Time consuming—24–48 h. Requires experienced laboratory technician and laboratory. Expensive in terms of consumables. May underestimate the number of enterococci in samples with high turbidity. | https://www.merckmillipore.com/AU/en/products/industrial-microbiology/culture-media/, (accessed on 20 November 2019) |
| Membrane Filtration | Use of membranes (pore size 0.22 −0.45 µm) to filter water after following by selective isolation plating (colony-forming units). | Efficient system. Can be accurate. | Time consuming—24–48 h. Requires experienced laboratory technician and laboratory. Expensive in terms of consumables. May underestimate the number of enterococci in samples with high turbidity. | www.oxoid.com/UK/blue/prod_detail/, (accessed on 20 November 2019) |
| IDEXX Enterolert-E | Based on the use of a defined substrate technology nutrient indicator to detect enterococci. This nutrient indicator fluoresces when metabolised by enterococci. (most probable number of cells). | No media preparation. Sensitive to 1 enterococcus per 100 mL. Less subjective interpretation. 50% fewer false positives and 95% fewer false negatives than the standard membrane filtration (MF) method. | Time consuming—24–48 h. Requires experienced laboratory technician and laboratory. Expensive in terms of consumables. Overestimation/underestimation of Enterococci is possible. | https://www.idexx.com.au/en-au/water/water-products-services/enterolert/, (accessed on 24 November 2019) |
| Polymerase Chain Reaction | Based on the use of 16S rDNA real-time PCR (gene copies). | Relatively fast (3–5 h), specific and sensitive. Multiplex PCR is possible. Quantitative detection (qPCR) of target pathogen is rapid (number of gene copies). | Well-equipped laboratory required. Requires primer optimisation and DNA extraction. Requirement of skilled personnel. | https://www.genesig.com/, (accessed on 4 December 2019) |
| Recombinase Polymerase Amplification | Versatile isothermal DNA/RNA amplification by TwistDx DNA polymerase. Based on the use of rpoB (RNA polymerase subunit B) specific gene target. | Rapid, less than 1 h.Carried out at 37–42 °C Requires very limited equipment and training. | Requires primer optimisation and DNA extraction. | http://www.twistdx.co.uk, (accessed on 21 September 2020) |
| Parameters | Sensitivity (CFU/100 mL) | Time Required | PON Applicability | Approximate Cost USD/Reaction | Temperature Required | References | |
|---|---|---|---|---|---|---|---|
| Assay | |||||||
| PCR | 104–105 | 120–150 min | NP | 5 | Thermal cycling (95 °C, 55 °C, 72 °C) | [39,40] | |
| RPA-EF | 2.8 × 103–2.8 × 103 | 80 min | NP | 5.5 | 38 °C | This study | |
| RPA-LFA | 2.8 × 103 | 25–30 min | P | 6 | 38 °C | This study | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Batra, A.R.; Cottam, D.; Lepesteur, M.; Dexter, C.; Zuccala, K.; Martino, C.; Khudur, L.; Daniel, V.; Ball, A.S.; Soni, S.K. Development of A Rapid, Low-Cost Portable Detection Assay for Enterococci in Wastewater and Environmental Waters. Microorganisms 2023, 11, 381. https://doi.org/10.3390/microorganisms11020381
Batra AR, Cottam D, Lepesteur M, Dexter C, Zuccala K, Martino C, Khudur L, Daniel V, Ball AS, Soni SK. Development of A Rapid, Low-Cost Portable Detection Assay for Enterococci in Wastewater and Environmental Waters. Microorganisms. 2023; 11(2):381. https://doi.org/10.3390/microorganisms11020381
Chicago/Turabian StyleBatra, Alka Rani, Darren Cottam, Muriel Lepesteur, Carina Dexter, Kelly Zuccala, Caroline Martino, Leadin Khudur, Vivek Daniel, Andrew S. Ball, and Sarvesh Kumar Soni. 2023. "Development of A Rapid, Low-Cost Portable Detection Assay for Enterococci in Wastewater and Environmental Waters" Microorganisms 11, no. 2: 381. https://doi.org/10.3390/microorganisms11020381
APA StyleBatra, A. R., Cottam, D., Lepesteur, M., Dexter, C., Zuccala, K., Martino, C., Khudur, L., Daniel, V., Ball, A. S., & Soni, S. K. (2023). Development of A Rapid, Low-Cost Portable Detection Assay for Enterococci in Wastewater and Environmental Waters. Microorganisms, 11(2), 381. https://doi.org/10.3390/microorganisms11020381







