Abstract
Luteibacter is a genus of the Rhodanobacteraceae family. The present study describes a novel species within the genus Luteibacter (EIF3T). The strain was analyzed genomically, morphologically and physiologically. Average nucleotide identity analysis revealed that it is a new species of Luteibacter. In silico analysis indicated two putative prophages (one incomplete, one intact). EIF3T cells form an elliptical morphotype with an average length of 2.0 µm and width of 0.7 µm and multiple flagella at one end. The bacterial strain is an aerobic Gram-negative with optimal growth at 30 °C. EIF3T is resistant towards erythromycin, tetracycline and vancomycin. We propose the name Luteibacter flocculans sp. nov. with EIF3T (=DSM 112537T = LMG 32416T) as type strain. Further, we describe the first known Luteibacter-associated bacteriophage called vB_LflM-Pluto.
1. Introduction
The genus Luteibacter is part of the family Rhodanobacteraceae, which belongs to the γ-subclass of the Proteobacteria. The family contains 17 genera, Aerosticca, Ahniella, Aquimonas, Chiayiivirga, Denitratimonas, Dokdonella, Dyella, Frateuria, Fulvimonas, Luteibacter, Oleiagrimonas, Pinirhizobacter, Pseudofulvimonas, Rehaibacterium, Rhodanobacter, Rudaea and Tahibacter, of which two are not validly published (Denitratimonas and Pinirhizobacter) [1]. The genus Luteibacter was established by Johansen et al. [2] based on the species Luteibacter rhizovicinus DSM 16549T. It currently comprises five species of which three are validly published: L. rhizovicinus DSM 16549T [2], L. yeojuensis DSM 17673T [3,4], L. anthropi CCUG 25036T [4], as well as L. jiangsuensis [5] and L. pinisoli [6]. Members of the Luteibacter genus were isolated from various environments such as rhizospheric soil [2,6], greenhouse soil [3] and human blood [4]. They are described as motile, aerobic Gram-negatives with a rod-like shape and yellow coloring. Further, they are catalase- and oxidase-positive and urease-negative.
To date, Luteibacter- or even Rhodanobacterceae-associated phages are unknown. Phages or bacteriophages are viruses that infect bacteria. While temperate phages can incorporate into the bacterial genome, lytic phages begin multiplying directly after infection. The temperate phages replicate their incorporated genome alongside the host genome, leading to a prophage and a lysogenic bacterium. Through the addition of its genetic material, a prophage can provide new abilities, defending the host from infection by related and unrelated viruses [7].
In a previous study, we were able to isolate an environmental Luteibacter sp. nov. strain from a eutrophic pond located in Göttingen, Germany. The Luteibacter strain was isolated as a prospective model strain to investigate the local viral diversity associated with it. Despite the fact that 16S rRNA gene analysis validated its species assignment, no additional characterization was performed [8].
Here, we describe a novel environmental Luteibacter isolate, which was characterized morphologically, physiologically and genomically. In addition, we investigated the potential of the host strain to access the environmental diversity of Luteibacter-associated phages.
2. Materials and Methods
2.1. Luteibacter flocculans EIF3 Strain Isolation, DNA Extraction and 16S rRNA Gene Sequencing
Luteibacter flocculans EIF3T (Figure S1) was isolated from the surface water of a eutrophic pond located at the North Campus of the Georg-August University in Göttingen, Germany (51°33′29″ N 9°56′41″ E 173 m, collected on 24 September 2018) [8]. This study was conducted at a public pond in Göttingen that required no specific permissions; 25 mL LB (1% peptone from casein, 0.5% yeast extract, 0.5% NaCl) was used as a culture medium. DNA was extracted and the 16S rRNA gene sequenced was as described by Friedrich et al., 2021 [8].
2.2. Sequencing, Assembly and Annotation of Bacterial and Phage Genome
Friedrich et al. 2021 describe the genome sequencing, assembly and annotation procedures. Briefly, Illumina paired-end sequencing libraries were generated using the Nextera XT DNA Sample Preparation kit. For sequencing, the MiSeq System and Reagent Kit version 3 (2 × 300 bp) were used according to the manufacturer’s instructions (Illumina, San Diego, CA, USA) [8]. For Nanopore sequencing, the Ligation Sequencing Kit (SQK-LSK109) and the Native Barcode Expansion Kit EXP-NBD114 (Oxford Nanopore Technologies, Oxford, UK) were utilized [8]. The same kit was used to prepare total and specific nucleic acids from the bacteriophage. To remove proteins, 5 µL of Proteinase K (20 mg/mL) were added to 300 µL of 2X T and C Lysis. This solution was applied to 300 µL of phage suspension. The pure viral genomic DNA was extracted from total nucleic acids using RNase A (DNase free).
CRISPRCasFinder [9] was used to identify potential CRISPR areas. Assembled bacterial and viral genomes as well PHASTER-predicted prophages were quality-checked with CheckM v1.1.2 [10] and CheckV v1.0.1, respectively [11]. Genome annotation was performed using the Prokaryotic Genome Annotation Pipeline v4.13 (PGAP) [12].
Raw bacteriophage reads were quality-processed using Trimmomatic v0.39 [13] and paired-end reads were merged using FLASH v1.2.11 [14]. The quality-processed reads served as input for the Unicycler v0.4.9 assembly pipeline in normal mode [15], which consisted of Spades v3.13.0 [16], makeblastdb v2.11.0+ and tblastn v2.11.0+ [17], bowtie2-build v2.4.4 and bowtie2 v2.4.4 [18], SAMtools v1.12 [19], java v.11.0.13 [20] and Pilon v1.23 [21]. The quality of assembly was evaluated using QualiMap v2.2.2 [22]. Annotations were performed using VIBRANT [23] and InterProScan v5.55-88.0 [24].
The whole-genome sequence of Luteibacter flocculans EIF3T has been submitted to GenBank under the accession number CP063231. The BioSample (SAMN16456042) is part of the BioProject with the accession number PRJNA669578. The raw reads have been submitted to the NCBI SRA database with the accession numbers SRR12951264 (Oxford Nanopore) and SRR12951265 (Illumina), as well as BioProject PRJNA669578. The strain was deposited at the DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany) with the collection number DSM 112537 and at the BCCM/LMG (Belgian Coordinated Collections of Microorganisms) with the collection number LMG 32416. The whole-genome sequence of Luteibacter-associated bacteriophage vB_LflM-Pluto is available at GenBank under the accession number ON529861 at GenBank.
2.3. Luteibacter flocculans sp. nov. EIF3T Phylogenetic Classification
The Genome Taxonomy Database Toolkit (GTDB-Tk) v1.0.2 [25] as well as whole-genome-based phylogeny with the Type (Strain) Genome Server (TYGS [26], accessed on 10 July 2022) were used to provide an initial taxonomic classification of the Luteibacter flocculans isolate. The ANIm method, which is provided by pyani v0.2.10 [27], was used with a species boundary of 95% ANI for in-depth phylogenetic analysis [25]. Based on the DSMZ and the NCBI, the genome of the isolate was compared to all available type strain and reference genomes (accessed on 10 July 2022) comprising Frateuria flava (GCF_017837635), F. defendens (GCF_001182895), Dyella solisilvae (GCA_003351225), D. kyungheensis (GCF_016905005), Luteibacter pinisoli (GCF_006385595), L. jiangsuensis (GCA_011742555), L. yeojuensis (GCA_011742875), L. anthropi (GCA_011759365), D. terrae (GCA_004322705), Fulvimonas soli (GCA_003148905), D. thiooxydans (GCA_001641285), as well as L. rhizovicinus (GCA_001010405).
2.4. Genomic Characterization
BlastKOALA v2.2 [28] was used to study the metabolic capacities of Luteibacter flocculans (Figure S2). AntiSMASH v6.0.0 [29] was employed to identify putative secondary metabolite biosynthetic gene clusters. PHASTER [30] was utilized to identify putative phage regions. Resfams v1.2.2 [31] was applied to examine presence of antibiotic resistance genes.
2.5. Cell Morphology and Gram Staining Techniques
Microscopy (Primo Star, Zeiss, Carl Zeiss Microscopy, Jena, Germany) was used to examine the morphology of single colonies after 72 hours of growth on LB solid medium (Fluka, Munich, Germany). Hucker’s crystal violet, an iodine and safranin solution and 1-propanol were used for Gram staining [32]. Microscopic images and stains were processed and analyzed using the software ZEISS Labscope (Carl Zeiss).
2.6. Bacterial and Phage Isolate Transmission Electron Microscopy
The morphology of Luteibacter flocculans and Luteibacter phage vB_LflM-Pluto was studied using transmission electron microscopy (TEM). The digital Micrograph software (Gatan GmbH, Munich, Germany) was used for imaging. Luteibacter flocculans was cultivated overnight in liquid LB medium at 30 °C. A negative staining was then conducted using a 5 µL cell or phage suspension. The suspension was mixed with an equal amount of diluted 0.5% (for bacterial isolate) or 1% (for viral isolate) phospho-tungstic acid (3% stock, pH 7). The mixture was transferred to a vaporized carbon mica for one minute. Before placing the mica on a thin copper-coated grid (PLANO GmbH, Marburg, Germany), it was gently cleaned with demineralized water. The coated grids were allowed to dry at room temperature and examined with a Jeol 1011 TEM (Georgia Electron Microscopy, Freising, Germany).
2.7. Determination of Salt Tolerance and Optimal Temperature
EIF3T was incubated at 30 °C in 4 mL LB medium adjusted with 0 and 5 g/L NaCl and 10 to 60 g/L NaCl in increments of 10 g to determine its salt tolerance. The optical density of the cell suspensions was measured at 600 nm (OD600) using the Ultraspec 3300 pro photometer (Amersham Pharmacia Biotec Europe GmbH, Munich, Germany). At the start of the experiment, the OD600 of the cell suspensions was set to 0.1 [33], followed by a 12-h incubation period at 30 °C and 180 rpm in an Infors HT shaker (Orbitron, Einsbach, Germany). To determine growth, the OD600 was measured after 12 h of incubation [33]. Every measurement was carried out in biological replicates. The temperature optimum was determined by culturing the isolate in 4 mL LB-0 medium under shaking (180 rpm) at 10, 20, 30, 37, 40 and 50 °C. The cultures’ starting OD600 was set to 0.1. After 12 h, the optical cell density of EIF3T was determined. R studio version 4.0.0 [34] and the ggplot2 package v3.3.6 [35] were used to visualize the data.
2.8. Growth Kinetics Determination
The cell growth quantifier (CGQuant 8.1; Aquila Biolabs GmbH, Baesweiler, Germany) was used to evaluate growth kinetics in liquid cultures under shaking (180 rpm) for 47 h at 30 °C. 250 mL shake flasks were filled with 25 mL of EIF3T culture in LB-0 medium (final OD600 of 0.1) and placed for measurement on the CGQuant sensor plate. Experiments were carried out using three biological replicates. The CGQuant uses a dynamic method of backscattered light measurement, allowing real-time monitoring of growth in liquid culture [36]. All data were plotted with R studio version 4.0.0 [34] and the ggplot2 package v3.3.6 [35].
2.9. Antibiotic Resistances and Metabolic Activity
For assessment of metabolic activity, API ZYM and API 20 NE tests (BioMérieux, Nuertingen, Germany) were used. Both tests were carried out according to the manufacturer’s instructions. Catalase activity was measured with 3% H2O2 [37]. Antibiotic resistances with discs and strips (Oxoid, Thermo Fisher Scientific, Waltham, MA, USA) were determined using a soft-agar (0.4% (w/v) agarose in LB medium) overlay technique. Discs and strips contained ampicillin (25 µg), kanamycin (30 µg), oxytetracycline (30 µg), rifampicin (2 µg), streptomycin (10 µg), vancomycin (30 µg), tetracycline (0.015–256 µg) and erythromycin (0.015–256 µg). 2.5 mL of soft agar with a final OD600 of 0.1 was utilized. Discs or strips containing an antibiotic substance were then added on top of the soft agar. Antibiotic resistances were determined after overnight incubation at 30 °C.
2.10. Examination of Plaques
The approach described by Willms and Hertel, 2016 [38] and Willms et al., 2017 [38,39] was used for phage enrichment. Sewage samples were collected in February 2022. In order to identify plaque morphologies such as clear or turbid, plaque size and halo presence, plaque assays generally require the ability of the host to grow in bacterial lawns [40]. Phages were isolated via agar overlay plaque assay as described elsewhere [40] using host-specific culture media for the base agar (1.5% (w/v) agar) and overlay (0.4% (w/v) agarose). Infected overlay plates were incubated overnight at 30 °C. Individual phages appeared as morphologically distinct plaques, which were picked with a sterile toothpick and transferred into 500 µL sterile LB-0 medium. Reinfection was repeated three times to purify the phage strain.
2.11. Naming of Bacteriophage Isolate
The bacteriophage was named according to Adriaenssens and Brister’s informal guide [41]. As a result, vB stands for virus of bacteria, Lfl for the host organism L. flocculans, M for the virus family Myo-morphotype and Pluto is an individual name. As a result, the complete name of the virus is vB LflM-Pluto, abbreviated to Pluto in the following.
3. Results and Discussion
3.1. Morphological Characterization
EIF3T colonies were spherical and yellow with an average diameter of 1.93 mm on solid LB medium. The ability of Luteibacter flocculans sp. nov. to flocculate was apparent during growth in liquid LB or TSB media (Figure S1). Gram staining of EIF3T resulted in red/pink cells indicating a Gram-negative bacterial species. Cells were straight rods with rounded ends and ranged from 5.3 to 5.8 µm in size (Figure 1). The isolate matched typical morphological features of the family Rhodanobacteraceae such as motility via polar flagella, a cell size ranging from 1 to 4.5 µm and rod-shaped cells with rounded ends [42].
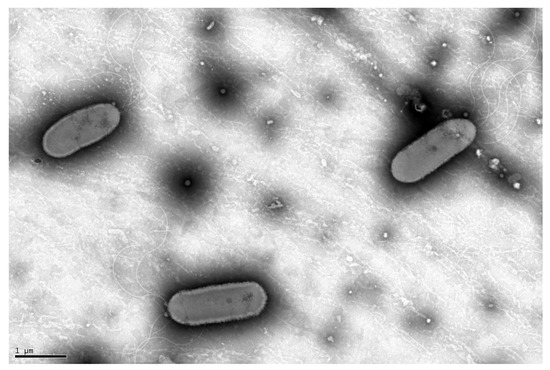
Figure 1.
Transmission electron micrograph of EIF3T. The micrograph depicts the rod-shaped, flagellated morphotype of the Luteibacter EIF3 isolate. The image was taken using TEM after 24 h of cell growth at 30 °C in LB medium followed by negative staining.
3.2. Physiological Characterization
EIF3T grew in LB medium with up to 4% (w/v) NaCl, with optimal growth achieved in the absence of added NaCl (Figure 2A). The strain is a mesophilic organism since it can grow at temperatures between 20 and 40 °C. The largest cell densities were observed at 30 °C with an OD600 of 3.403 (which is a ratio of 34.033, (Figure 2B). This observation is consistent with results derived from closely related strains [2].
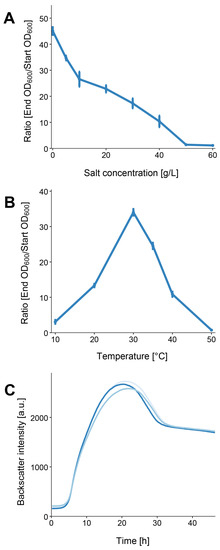
Figure 2.
L. flocculans sp. nov. growth characteristics. (A) Growth of EIF3T in 4 mL LB medium after 12 h of incubation at 180 rpm and 30 °C with different salt concentrations. (B) EIF3T growth at various temperatures in LB-0 medium after 12 h of incubation at 180 rpm. (C) Growth characterization of EIF3T in 25 mL LB-0 medium at the optimal temperature (30 °C). Triplicate measurements were conducted. Standard deviations in (A,B) are indicated by the error bars. In (C) various shades of blue represent each biological replicate.
We determined the bacterial growth of EIF3T at the optimal temperature and salt concentration. The lag phase lasted for approximately 3.5 h. It was followed by a 10 h log-phase and a transient phase with diminished growth. After approximately 21 h of incubation, the highest cell densities were recorded. The doubling time of our isolate was 221 min and the growth rate µ was 0.19 h−1.
Using the API ZYM and the API 20 NE assays, the metabolic capabilities of EIFT were examined. Twenty distinct enzyme activities were identified for the novel Luteibacter isolate using API ZYM. In six cases, no enzyme activity was detected. Activities of alkaline phosphatase, esterase, esterase lipase, leucine arylamidase, valine arylamidase, cysteine arylamidase, acid phosphatase, naphthol-AS-BI-phosphohydrolase, β-galactosidase, α-glucosidase, β-glucosidase and N-acetyl-β-glucosaminidase were recorded. Corresponding genes were identified in the genome (Table S1). In addition, Luteibacter flocculans shared features with closely related bacteria such as L. rhizovicinus, L. yeojuensis and L. jiangsuensis [2,3,5].
EIF3T was oxidase- and catalase-positive, which is characteristic of certain Rhodanobacteraceae family members [42]. Table 1 provides an overview of the enzymatic activities of the strain and closely related bacteria from TYGS [26]. According to the antibiogram, EIF3T was resistant to erythromycin (up to 4 µg/disc), tetracycline (up to 1 µg/disc) and vancomycin (30 µg/disc). Resfams in silico analysis [31] identified genes encoding an ABC transporter for erythromycin or vancomycin (PRJNAA669578|IM816_002307) and a tetracycline inactivation enzyme (IM816_003460; Table S2). The in silico analysis of EIF3T confirmed the measured antibiotic resistances (Table 1).

Table 1.
Phenotypic differences between strains EIF3T and phylogenetically related species L. yeojuensis DSM 17673T, L. jiangsuensis CGMCC 1.10133T, L. antrophi CCUG 25036T and L. rhizovicinus DSM 16549T. Taxa: 1, strain L. flocculans EIF3T; 2, strain L. yeojuensis DSM 17673T [3], 3, strain L. jiangsuensis CGMCC 1.10133T (data from [5,43]); 4, strain L. antrophi CCUG 25036T (data from [4] and BacDive [44] accessed on 31 July 2022); 5, strain L. rhizovicinus DSM 16549T (data from [2] and BacDive [44] accessed on 31 July 2022); +, Positive; –, negative; n/a, data not available.
Further, it suggested, that EIF3T can generate secondary metabolites such as aryl-polyene xanthomonadin (Table S3). Xanthomonadin is a yellow membrane-bound pigment, which is insoluble in water. Rajagopal et al. discovered that xanthomonadin may protect Xanthomonas oryzae against photodamage [45]. Moreover, this discovery is consistent with the characteristics of the family Rhodanobacteraceae [42].
Fatty acid analysis confirmed typical Luteibacter characteristics of our isolate and related strains. The most abundant fatty acids were branched fatty acids iso-C15:0 with 18.3%, iso-C17:1 ω9c with 29.4% and iso-C17:0 with 18.2% (Table 2). These corresponded to the main branched fatty acids of the Luteibacter described by Johansen et al. [2].

Table 2.
Composition of cellular fatty acids (%) in strain EIF3T and phylogenetically related species L. yeojuensis DSM 17673T, L. jiangsuensis CGMCC 1.10133T, L. antrophi CCUG 25036T and L. rhizovicinus DSM 16549T. Taxa: 1, strain L. flocculans EIF3T; 2, strain L. yeojuensis DSM 17673T [3,4], 3, strain L. jiangsuensis CGMCC 1.10133T (data from [5]); 4, strain L. antrophi CCUG 25036T (data from [4]); 5, strain L. rhizovicinus DSM 16549T (data from [4]); –, not detected/not reported.
3.3. Genome Characterization
The closed genome of EIF3T comprised one circular chromosome (4,299,254 bp) with a GC content of 64.82%. It encoded 3672 putative proteins, 59 rRNAs and 49 tRNAs. No CRISPR regions and plasmids were identified (Table 3).

Table 3.
Genome statistics of the Luteibacter flocculans sp. nov. EIF3T chromosome.
GTDB-Tk was used for the genome-based taxonomic classification of strain EIF3T (Data File S1) [25]. It demonstrated an average nucleotide identity (ANI) of approximately 96% to the most closely related species, Luteibacter sp. UNCMF366Tsu5.1 (ANI value of 96.48). The digital DNA-DNA hybridization value (dDDH) calculated by the Type Strain Genome Server (TYGS) is 39.9% in comparison to L. jiangsuensis. As the new species criterion for dDDH is less than 70% [46], this suggested that our strain is a potential new species (Figure S2; Table S4). Figure 3 shows an ANI-analysis of 12 most closely related type strain genome sequences in the TYGS database [26] (data in Table S5). No clustering with any other described Luteibacter strain was recorded. EIF3T shares 85.52% ANI with L. yeojuensis DSM 17673T, 85.41% with L. jiangsuensis CGMCC 1.10133T, 84.73% with L. anthropic CCUG 25036T and 84.60% with L. rhizovicinus DSM 16549T.
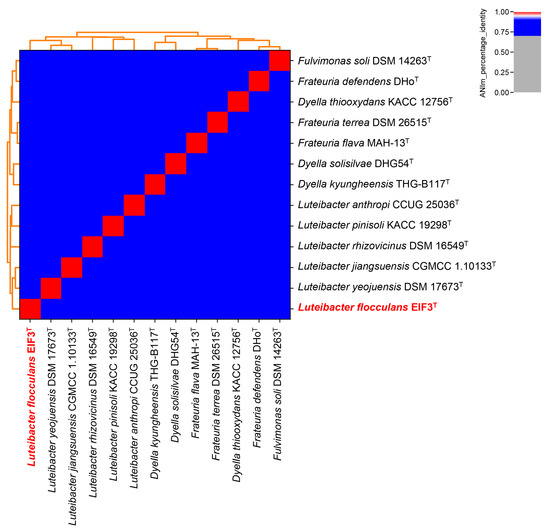
Figure 3.
Genome-based classification of Luteibacter flocculans EIF3T. All genome sequences from available type strains (T) listed in the TYGS database [26] were examined. Pyani [27,47] was used to calculate relatedness employing the ANIm technique and default settings. EIF3T is highlighted in bold red letters.
Luteibacter flocculans sp. nov. EIF3T was therefore considered as a novel type strain within the Luteibacter genus.
3.4. Prophages Analysis
EIF3T represented a prospective host system for the investigation of the environmental diversity of phages. Thus, the capabilities to host prophages was of particular interest in this study. Initial analysis of prophage regions with PHASTER [30] revealed two putative prophage regions (region 1, 1,300,438–1,322,193; region 2, 1,306,588–1,352,491). The regions were 21.7 and 45.9 kb in size. It was estimated that region 1 was incomplete (20.97% completeness estimation by CheckV [11]) and region 2 intact (62.83% completeness estimation by CheckV [11]). In addition, region 1 was part of region 2 (Table S6). Since prophages can provide phage-resistance to the host, the genome of a potential host strain for phage isolation must have a low number of prophages or none at all [7], which is applicable for EIF3T. A repressor protein cI was predicted (locus tag: IM816_005715) in both prophages. Bacterial cells which harbor a lysogenic lambda phage are immune to further lambda phage infections [48]. In addition, a gene encoding RecA-dependent nuclease (locus tag: IM816_005740), which is also encoded by the temperate bacteriophage P1 [49]. was predicted. The terminase (large subunit) gene (locus tag: IM816_005805) involved in the initiation of phage DNA packaging into the prohead indicated that prophage 2 is an active prophage. The phage portal protein (locus tag: IM816_005810) binding to the terminase subunit to form the packaging machinery was also identified [50]. A phage major capsid protein (locus tag: IM816_005820) indicated an icosahedral head structure, placing the prophage withing the Claudoviricetes. The tail assembly chaperone protein (locus tag: IM816_005855) suggested a Sipho-morphotype. Lastly, a gene coding for a tail tape protein (locus tag: IM816_005865) was also present. During infection, the protein determines tail length and facilitates DNA transit [51].
3.5. Phage Isolation and Characterization
Luteibacter flocculans has proven to be an organism with minimal requirements for cultivation and proliferation. This provided a solid foundation for its prospective use in molecular biology. To evaluate its viability as a host strain for environmental phage isolations. EIF3T was infected with a viral suspension obtained from raw sewage. To determine cell infection, an overlay plaque test was performed. To avoid redundancies during phage isolation, picked plaques were assessed by identifying the unique genetic restriction patterns of each phage isolate. This approach led to the isolation of a novel Luteibacter-associated phage.
Transmission electron microscopy shows a Myo-morphotype head-tail morphology (Figure 4). The phage consisted of an icosahedral head, contractile tail and tail spikes. The diameter of the capsid was 75 nm and the length of the tail 110 nm, resulting in a total length of 185 nm. We sequenced the viral DNA and assembled the genome with a high coverage of 690.1-fold. The assembled viral genome exhibited a size of 67,528 bp, a completeness estimate of 100% (according to the CheckV report [11]) and a G + C content of 57.7% (host G + C content 64.8%). A total of 99 CDS of which 79 encode hypothetical proteins were detected. We detected genes similar to phage-related genes. These included a spike protein, showing the highest sequence identity to the corresponding protein of the Xanthomonas phage FoX6, which also has a Myo-morphotype [52]. Further, the terminase (large subunit), baseplate protein J, major capsid protein and DNA polymerase shared an average sequence identity of 60% to the corresponding proteins of FoX6. The host Xanthomonas (family: Lysobacteraceae) belongs as Luteibacter to the order Lysobacterales. The terminase (large subunit) suggested an icosahedral head morphotype while the baseplate protein J indicated a Myo-morphotype. An overview of all genes is shown in Table S7. Resulting from the morphological and genomic investigations, our phage was named vB_LflM-Pluto (vB = virus of bacter, Lfl = L. flocculans, M = Myo-morphotype, Pluto = specific phage name). Our results represented the first description of a phage from the bacterial Rhodanobacteraceae family. In addition, we showed that Luteibacter flocculans sp. nov. is a suitable host strain for phage isolation.
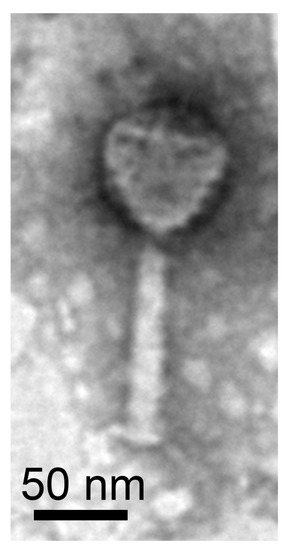
Figure 4.
Luteibacter-associated bacteriophage vB_LflM-Pluto.
While Luteibacter phage LflM-Pluto is of the Myo-morphotype, the prophage of Luteibacter flocculans likely has a Sipho-morphotype. Both showed no gene or protein similarities and therefore were not genetically related. No genes encoding for antibiotic resistances were detected in either phage genome.
4. Conclusions
The results demonstrated the suitability of the novel Luteibacter species L. flocculans for the isolation of environmental phages. The isolation and characterization of a novel Luteibacter-associated phage vB_LflM-Pluto, the first documented Luteibacter-associated phage, further supported this.
Description of Luteibacter flocculans sp. nov.
Luteibacter flocculans (floc.cuĭlans N.L. part. adj. flocculans, flocculating, pertaining to the organism’s ability to flocculate in liquid cultures). L. flocculans cells were Gram-negative rod-shaped, 2.0 µm long and 0.7 µm wide. They did not form spores and were motile by means of lophotrichous bacteria. After 72 h of growth on LB medium, colonies were 1.93 mm in diameter and showed yellow pigmentation. Cells grew at 10–45 °C (optimum 30 °C) and at 0–4% NaCl (optimum without addition of NaCl). The strain was catalase and oxidase positive. Cell growth occurred on R2A agar, TSA and LB agar. The strain was susceptible to erythromycin, tetracycline and vancomycin, but not to ampicillin, kanamycin, oxytetracycline, rifampicin and streptomycin. It utilized esculin/ferric citrate, D-glucose (assimilation), D-mannose, N-acetyl-D-glucosamine and malic acid employing the API 20NE test system. Alkaline phosphatase, esterase, esterase lipase, lipase, leucine arylamidase, valine arylamidase, cysteine arylamidase, acid phosphatase, naphthol-AS-BI-phosphohydrolase, β-galactosidase, α-glucosidase, β-glucosidase and N-acetyl-β-glucosaminidase activities were detected with API ZYM test system. In Table 1 and Table 2, additional phenotypic characteristics are depicted.
The type strain EIF3T (=DSM 112537T = LMG 32416T), was isolated from a eutrophic pond located on the North Campus of the Georg-August University in Göttingen, Germany. The major fatty acids were C15:0 iso, C17:0 iso and summed feature 9 (C17:1 iso ω9c). The genome of the type strain showed a DNA G + C content of 64.8 mol%.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms11020307/s1, Figure S1: Flocculation of EIF3T in LB medium; Figure S2: Phylogenetic classification of Luteibacter flocculans EIF3T. Table S1: KEGG Mapper reconstruction result of Luteibacter flocculans EIF3T chromosome [26,53,54]; Table S2: Resfams prediction of Luteibacter flocculans EIF3T; Table S3: List of putative biosynthetic gene clusters in Luteibacter flocculans EIF3T; Table S4: List of putative biosynthetic gene clusters in Luteibacter flocculans EIF3T [26]; Table S5: Phylogenetic analysis for Luteibacter flocculans EIF3T; Table S6: PHASTER analysis of Luteibacter flocculans EIF3T; Table S7: Genes and putative their functions of Luteibacter-associated phage vB_LflM-Pluto and its closest relatives; Data File S1: GTDB-Tk of Luteibacter flocculans sp. nov. isolate.
Author Contributions
Conceptualization: I.F., R.H. and R.D.; Investigation: I.F. and A.K.; Data curation: I.F. and A.K.; Validation: R.H., R.D. and I.F.; Visualization: I.F.; Formal analysis: I.F., R.H. and R.D.; Writing original draft: I.F.; Writing—review & editing: R.H., I.F., A.K. and R.D.; Project administration: R.D.; Supervision: R.H. and R.D. All authors have read and agreed to the published version of the manuscript.
Funding
The research received no special funding. We acknowledge support by the Open Access Publication Funds of the University of Göttingen. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Data Availability Statement
The whole-genome shotgun project of Luteibacter flocculans sp. nov. EIF3T has been deposited at GenBank under the accession numbers CP063231 and the BioProject accession number PRJNA669578. BioSample accession number is SAMN16456042. The raw reads have been submitted to the NCBI SRA database with the accession numbers SRR12951264 (Oxford Nanopore) and SRR12951265 (Illumina), as well as BioProject PRJNA669578. The genome of Luteibacter phage vB_LflM-Pluto has been deposited at GenBank under the accession number ON529861.
Acknowledgments
We thank Anja Poehlein for sequencing, Mechthild Bömeke for technical assistance, Dominik Schneider for his help with bioinformatics and Michael Hoppert for support with TEM-imaging. We thank Avril von Hoyningen-Huene for her meticulous proofreading of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Parte, A.C.; Carbasse, J.S.; Meier-Kolthoff, J.P.; Reimer, L.C.; Göker, M. List of Prokaryotic Names with Standing in Nomenclature (LPSN) Moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 2020, 70, 5607–5612. [Google Scholar] [CrossRef] [PubMed]
- Johansen, J.E.; Binnerup, S.J.; Kroer, N.; Mølbak, L. Luteibacter rhizovicinus gen. nov., sp. nov., a Yellow-Pigmented Gammaproteobacterium Isolated from the Rhizosphere of Barley (Hordeum vulgare L.). Int. J. Syst. Evol. Microbiol. 2005, 55, 2285–2291. [Google Scholar] [CrossRef]
- Kim, B.-Y.; Weon, H.-Y.; Lee, K.-H.; Seok, S.-J.; Kwon, S.-W.; Go, S.-J.; Stackebrandt, E. Dyella yeojuensis sp. nov., Isolated from Greenhouse Soil in Korea. Int. J. Syst. Evol. Microbiol. 2006, 56, 2079–2082. [Google Scholar] [CrossRef] [PubMed]
- Kämpfer, P.; Lodders, N.; Falsen, E. Luteibacter Anthropi Sp. Nov., Isolated from Human Blood and Reclassification of Dyella yeojuensis Kim et al. 2006 as Luteibacter yeojuensis comb. nov. Int. J. Syst. Evol. Microbiol. 2009, 59, 2884–2887. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, G.-L.; Li, S.-P.; Jiang, J.-D. Luteibacter jiangsuensis sp. nov.: A Methamidophos-Degrading Bacterium Isolated from a Methamidophos-Manufacturing Factory. Curr. Microbiol. 2011, 62, 289–295. [Google Scholar] [CrossRef]
- Akter, S.; Huq, A. Luteibacter pinisoli sp. nov., a Casein Degrading Bacterium Isolated from Rhizospheric Soil of Pinus koraiensis. Arch. Microbiol. 2018, 200, 1017–1023. [Google Scholar] [CrossRef]
- Kohm, K.; Hertel, R. The Life Cycle of SPβ and Related Phages. Arch. Virol. 2021, 166, 2119–2130. [Google Scholar] [CrossRef]
- Friedrich, I.; Klassen, A.; Neubauer, H.; Schneider, D.; Hertel, R.; Daniel, R. Living in a Puddle of Mud: Isolation and Characterization of Two Novel Caulobacteraceae Strains Brevundimonas pondensis sp. nov. and Brevundimonas goettingensis sp. nov. Appl. Microbiol. 2021, 1, 38–59. [Google Scholar] [CrossRef]
- Couvin, D.; Bernheim, A.; Toffano-Nioche, C.; Touchon, M.; Michalik, J.; Néron, B.; Rocha, E.P.C.; Vergnaud, G.; Gautheret, D.; Pourcel, C. CRISPRCasFinder, an Update of CRISRFinder, Includes a Portable Version, Enhanced Performance and Integrates Search for Cas Proteins. Nucleic Acids Res. 2018, 46, W246–W251. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the Quality of Microbial Genomes Recovered from Isolates, Single Cells and Metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Nayfach, S.; Camargo, A.P.; Schulz, F.; Eloe-Fadrosh, E.; Roux, S.; Kyrpides, N.C. CheckV Assesses the Quality and Completeness of Metagenome-Assembled Viral Genomes. Nat. Biotechnol. 2021, 39, 578–585. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI Prokaryotic Genome Annotation Pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast Length Adjustment of Short Reads to Improve Genome Assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving Bacterial Genome Assemblies from Short and Long Sequencing Reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Arnold, K.; Gosling, J.; Holmes, D. The Java Programming Language, 4th ed.; Addison-Wesley Professional: Lebanon, IN, USA, 2005; ISBN 978-0-321-34980-4. [Google Scholar]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Conesa, A.; García-Alcalde, F. Qualimap 2: Advanced Multi-Sample Quality Control for High-Throughput Sequencing Data. Bioinformatics 2016, 32, 292–294. [Google Scholar] [CrossRef] [PubMed]
- Kieft, K.; Zhou, Z.; Anantharaman, K. VIBRANT: Automated Recovery, Annotation and Curation of Microbial Viruses and Evaluation of Viral Community Function from Genomic Sequences. Microbiome 2020, 8, 90. [Google Scholar] [CrossRef] [PubMed]
- Zdobnov, E.M.; Apweiler, R. InterProScan—An Integration Platform for the Signature-Recognition Methods in InterPro. Bioinformatics 2001, 17, 847–848. [Google Scholar] [CrossRef] [PubMed]
- Chaumeil, P.-A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: A Toolkit to Classify Genomes with the Genome Taxonomy Database. Bioinformatics 2019, 36, 1925–1927. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS Is an Automated High-Throughput Platform for State-of-the-Art Genome-Based Taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Pritchard, L.; Glover, R.H.; Humphris, S.; Elphinstone, J.G.; Toth, I.K. Genomics and Taxonomy in Diagnostics for Food Security: Soft-Rotting Enterobacterial Plant Pathogens. Anal. Methods 2016, 8, 12–24. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. AntiSMASH 6.0: Improving Cluster Detection and Comparison Capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A Better, Faster Version of the PHAST Phage Search Tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef]
- Gibson, M.K.; Forsberg, K.J.; Dantas, G. Improved Annotation of Antibiotic Resistance Determinants Reveals Microbial Resistomes Cluster by Ecology. ISME J. 2015, 9, 207–216. [Google Scholar] [CrossRef]
- Claus, D. A Standardized Gram Staining Procedure. World J. Microbiol. Biotechnol. 1992, 8, 451–452. [Google Scholar] [CrossRef]
- Abraham, W.-R.; Strompl, C.; Meyer, H.; Lindholst, S.; Moore, E.R.B.; Christ, R.; Vancanneyt, M.; Tindall, B.J.; Bennasar, A.; Smit, J.; et al. Phylogeny and Polyphasic Taxonomy of Caulobacter Species. Proposal of Maricaulis gen. nov. with Maricaulis maris (Poindexter) comb. nov. as the Type Species and Emended Description of the Genera Brevundimonas and Caulobacter. Int. J. Syst. Evol. Microbiol. 1999, 49, 1053–1073. [Google Scholar] [CrossRef]
- R Core Team. R Core Team R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2020. [Google Scholar]
- Wickham, H. Ggplot2—Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009; Volume 77, p. 3. ISBN 978-0-387-98140-6. [Google Scholar]
- Bruder, S.; Reifenrath, M.; Thomik, T.; Boles, E.; Herzog, K. Parallelized Online Biomass Monitoring in Shake Flasks Enables Efficient Strain and Carbon Source Dependent Growth Characterization of Saccharomyces cerevisiae. Microb. Cell Factories 2016, 15, 127. [Google Scholar] [CrossRef]
- Clarke, P.H.; Cowan, S.T. Biochemical Methods for Bacteriology. Microbiol. Soc. 1952, 6, 187–197. [Google Scholar] [CrossRef]
- Willms, I.M.; Hertel, R. Phage VB_BsuP-Goe1: The Smallest Identified Lytic Phage of Bacillus subtilis. FEMS Microbiol. Lett. 2016, 363, fnw208. [Google Scholar] [CrossRef]
- Willms, I.; Hoppert, M.; Hertel, R. Characterization of Bacillus subtilis Viruses VB_BsuM-Goe2 and VB_BsuM-Goe3. Viruses 2017, 9, 146. [Google Scholar] [CrossRef]
- Kropinski, A.M.; Mazzocco, A.; Waddell, T.E.; Lingohr, E.; Johnson, R.P. Enumeration of Bacteriophages by Double Agar Overlay Plaque Assay. Methods Mol. Biol. 2009, 501, 69–76. [Google Scholar] [CrossRef]
- Adriaenssens, E.; Brister, J.R. How to Name and Classify Your Phage: An Informal Guide. Viruses 2017, 9, 70. [Google Scholar] [CrossRef]
- Naushad, S.; Adeolu, M.; Wong, S.; Sohail, M.; Schellhorn, H.E.; Gupta, R.S. A Phylogenomic and Molecular Marker Based Taxonomic Framework for the Order Xanthomonadales: Proposal to Transfer the Families Algiphilaceae and Solimonadaceae to the Order Nevskiales ord. nov. and to Create a New Family within the Order Xanthomonadales, the Family Rhodanobacteraceae fam. nov., Containing the Genus Rhodanobacter and Its Closest Relatives. Antonie Leeuwenhoek 2015, 107, 467–485. [Google Scholar] [CrossRef]
- Zhao, F.; Guo, X.; Wang, P.; He, L.; Huang, Z.; Sheng, X. Dyella jiangningensis sp. nov., a γ-Proteobacterium Isolated from the Surface of Potassium-Bearing Rock. Int. J. Syst. Evol. Microbiol. 2013, 63, 3154–3157. [Google Scholar] [CrossRef]
- Reimer, L.C.; Vetcininova, A.; Carbasse, J.S.; Söhngen, C.; Gleim, D.; Ebeling, C.; Overmann, J. BacDive in 2019: Bacterial Phenotypic Data for High-Throughput Biodiversity Analysis. Nucleic Acids Res. 2019, 47, D631–D636. [Google Scholar] [CrossRef] [PubMed]
- Rajagopal, L.; Sundari, C.S.; Balasubramanian, D.; Sonti, R.V. The Bacterial Pigment Xanthomonadin Offers Protection against Photodamage. FEBS Lett. 1997, 415, 125–128. [Google Scholar] [CrossRef] [PubMed]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; da Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.-W.; De Meyer, S.; et al. Proposed Minimal Standards for the Use of Genome Data for the Taxonomy of Prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rosselló-Móra, R. Shifting the Genomic Gold Standard for the Prokaryotic Species Definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef]
- Lewis, D.; Le, P.; Zurla, C.; Finzi, L.; Adhya, S. Multilevel Autoregulation of λ Repressor Protein CI by DNA Looping in Vitro. Proc. Natl. Acad. Sci. USA 2011, 108, 14807–14812. [Google Scholar] [CrossRef]
- Gruenig, M.C.; Lu, D.; Won, S.J.; Dulberger, C.L.; Manlick, A.J.; Keck, J.L.; Cox, M.M. Creating Directed Double-Strand Breaks with the Ref Protein. J. Biol. Chem. 2011, 286, 8240–8251. [Google Scholar] [CrossRef]
- Yeo, A.; Feiss, M. Specific Interaction of Terminase, the DNA Packaging Enzyme of Bacteriophage γ, with the Portal Protein of Theeh Prohead. J. Mol. Biol. 1995, 245, 141–150. [Google Scholar] [CrossRef]
- Mahony, J.; Alqarni, M.; Stockdale, S.; Spinelli, S.; Feyereisen, M.; Cambillau, C.; van Sinderen, D. Functional and Structural Dissection of the Tape Measure Protein of Lactococcal Phage TP901-1. Sci. Rep. 2016, 6, 36667. [Google Scholar] [CrossRef]
- Holtappels, D.; Fortuna, K.J.; Moons, L.; Broeckaert, N.; Bäcker, L.E.; Venneman, S.; Rombouts, S.; Lippens, L.; Baeyen, S.; Pollet, S.; et al. The Potential of Bacteriophages to Control Xanthomonas campestris pv. campestris at Different Stages of Disease Development. Microb. Biotechnol. 2022, 15, 1762–1782. [Google Scholar] [CrossRef]
- Lefort, V.; Desper, R.; Gascuel, O. FastME 2.0: A Comprehensive, Accurate, and Fast Distance-Based Phylogeny Inference Program. Mol. Biol. Evol. 2015, 32, 2798–2800. [Google Scholar] [CrossRef]
- Farris, J.S. Estimating phylogenetic trees from distance matrices. Am Nat. 1972, 106, 645–668. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).