Comprehensive Genomics and Proteomics Analysis Reveals the Multiple Response Strategies of Endophytic Bacillus sp. WR13 to Iron Limitation
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacteria Growth and Treatment
2.2. Draft Genome Sequencing of WR13
2.3. Siderophore Determination
2.4. RNA Isolation and qPCR Assay
2.5. Total Protein Extraction, Digestion and iTRAQ Labeling
2.6. High pH Reverse Phase Separation and LC-MS/MS Analysis
2.7. Protein Identification and Differentially Abundant Protein (DAP) Screening
2.8. Statistical Analysis
3. Results
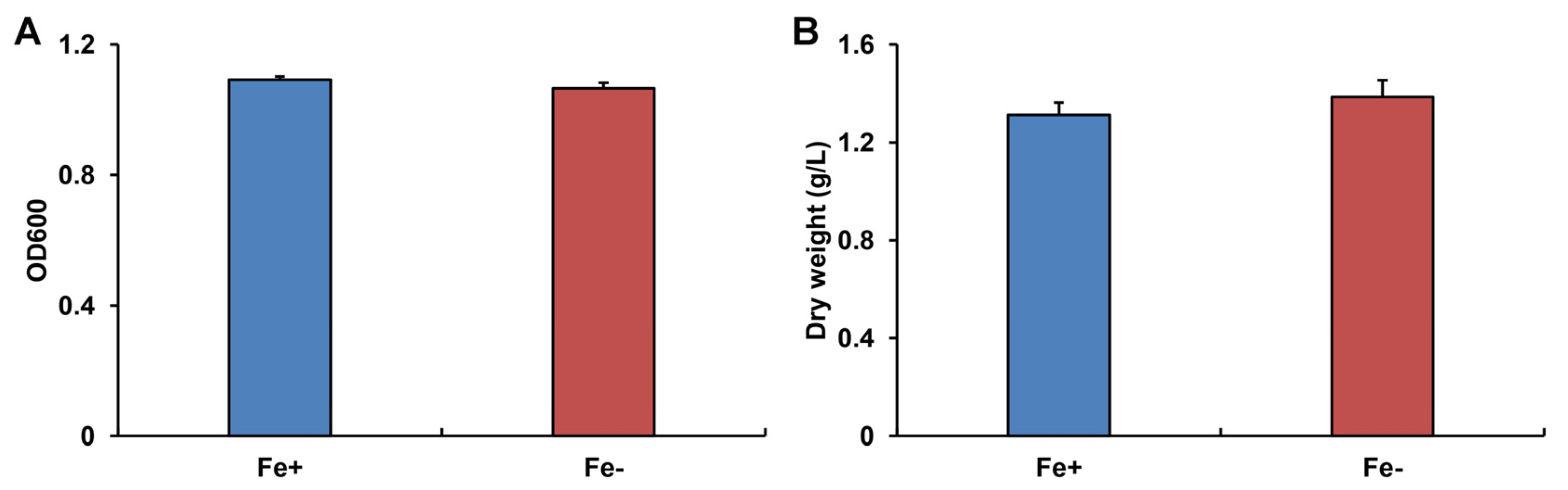
3.1. The Growth of WR13 under Different Fe Conditions
3.2. Genome Analysis of WR13
3.3. Siderophore Production and Siderophore-Related Genes Expression in WR13 under Fe Limitation
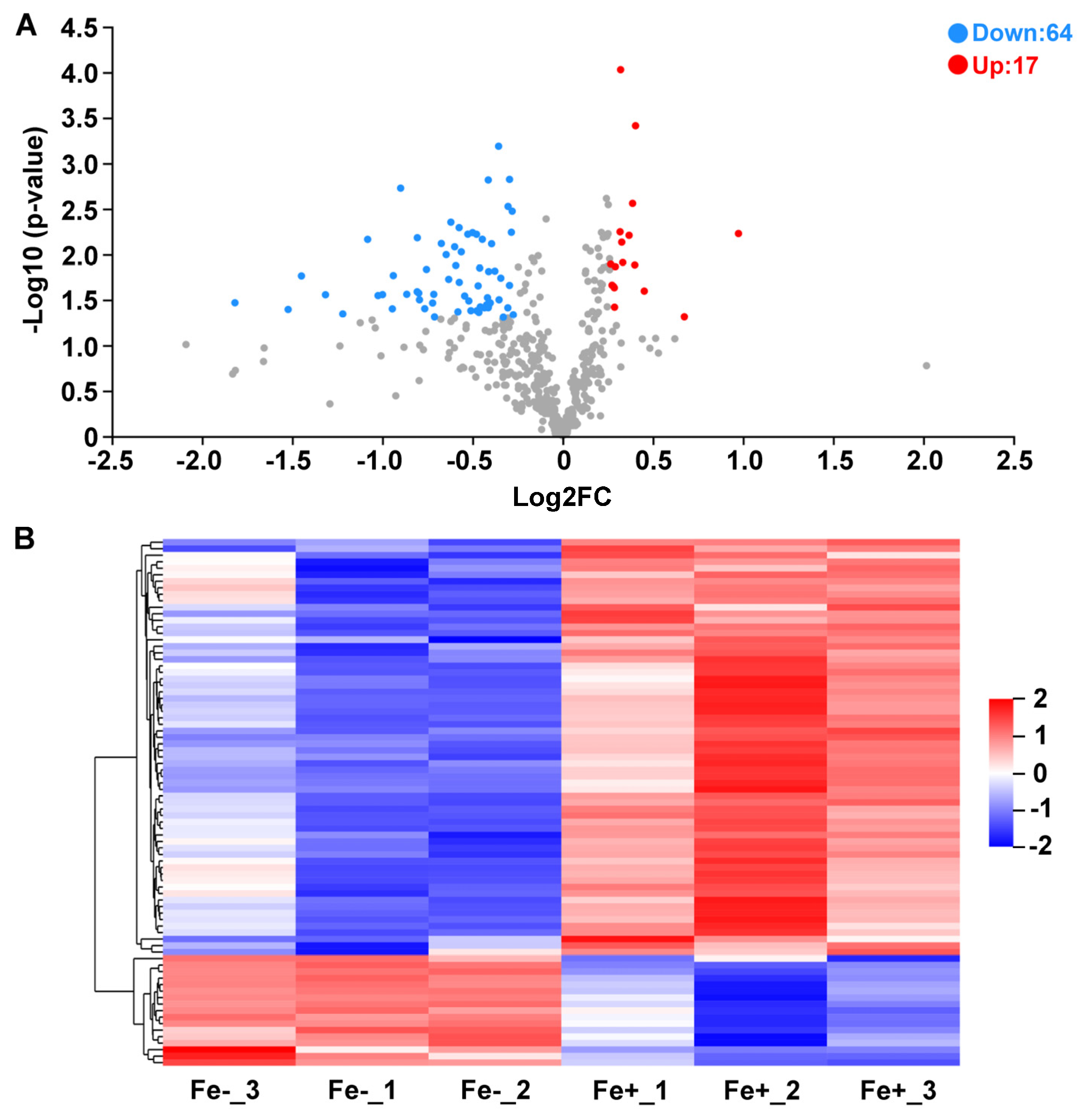
3.4. Quantification of Total Proteins and Protein Differential Analysis by iTRAQ-Labeled Proteomics
3.5. Functional Annotation of DAPs
3.6. The Key DAPs Involved in Carbohydrate Metabolism, Amino Acid Metabolism, Lipid Metabolism, and Fe-Containing Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aguado-Santacruz, G.A.A.; Moreno-Gómez, B.A.; Jiménez-Francisco, B.B.; García-Moya, E.B.; Preciado-Ortiz, R.E. Impact of the microbial siderophores and phytosiderophores on the iron assimilation by plants: A synthesis. Rev. Fitotec. Mex. 2012, 35, 9–21. [Google Scholar]
- Weinberg, E.D. Suppression of bacterial biofilm formation by iron limitation. Med. Hypotheses 2004, 63, 863–865. [Google Scholar] [CrossRef] [PubMed]
- Albelda-Berenguer, M.; Monachon, M.; Joseph, E. Siderophores: From Natural Roles to Potential Applications, 1st ed.; Elsevier: Amsterdam, The Netherlands, 2019; Volume 106, ISBN 9780128169759. [Google Scholar]
- Neilands, J.B. Siderophores: Structure and Function of Microbial Iron Transport Compounds. J. Biol. Chem. 1995, 270, 26723–26726. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Singh, P.; Srivastava, A. Synthesis, nature and utility of universal iron chelator—Siderophore: A review. Microbiol. Res. 2018, 212–213, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Khasheii, B.; Mahmoodi, P.; Mohammadzadeh, A. Siderophores: Importance in bacterial pathogenesis and applications in medicine and industry. Microbiol. Res. 2021, 250, 126790. [Google Scholar] [CrossRef] [PubMed]
- Liermann, L.J.; Kalinowski, B.E.; Brantley, S.L.; Ferry, J.G. Role of bacterial siderophores in dissolution of hornblende. Geochim. Cosmochim. Acta 2000, 64, 587–602. [Google Scholar] [CrossRef]
- Kraemer, S.M. Iron oxide dissolution and solubility in the presence of siderophores. Aquat. Sci. 2004, 66, 3–18. [Google Scholar] [CrossRef]
- May, J.J.; Wendrich, T.M.; Marahiel, M.A. The dhb operon of Bacillus subtilis encodes the biosynthetic template for the catecholic siderophore 2,3-dihydroxybenzoate-glycine-threonine trimeric ester bacillibactin. J. Biol. Chem. 2001, 276, 7209–7217. [Google Scholar] [CrossRef]
- Pi, H.; Helmann, J.D. Sequential induction of Fur-regulated genes in response to iron limitation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2017, 114, 12785–12790. [Google Scholar] [CrossRef]
- Massé, E.; Salvail, H.; Desnoyers, G.; Arguin, M. Small RNAs controlling iron metabolism. Curr. Opin. Microbiol. 2007, 10, 140–145. [Google Scholar] [CrossRef]
- Coronel-Tellez, R.H.; Pospiech, M.; Barrault, M.; Liu, W.; Bordeau, V.; Vasnier, C.; Felden, B.; Sargueil, B.; Bouloc, P. sRNA-controlled iron sparing response in Staphylococci. Nucleic Acids Res. 2022, 50, 8529–8546. [Google Scholar] [CrossRef] [PubMed]
- da Silva Neto, J.F.; Lourenço, R.F.; Marques, M.V. Global transcriptional response of Caulobacter crescentus to iron availability. BMC Genom. 2013, 14, 549. [Google Scholar] [CrossRef] [PubMed]
- Gaballa, A.; Antelmann, H.; Aguilar, C.; Khakh, S.K.; Song, K.B.; Smaldone, G.T.; Helmann, J.D. The Bacillus subtilis iron-sparing response is mediated by a Fur-regulated small RNA and three small, basic proteins. Proc. Natl. Acad. Sci. USA 2008, 105, 11927–11932. [Google Scholar] [CrossRef] [PubMed]
- Massé, E.; Gottesman, S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 2002, 99, 4620–4625. [Google Scholar] [CrossRef] [PubMed]
- Wilderman, P.J.; Sowa, N.A.; FitzGerald, D.J.; FitzGerald, P.C.; Gottesman, S.; Ochsner, U.A.; Vasil, M.L. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc. Natl. Acad. Sci. USA 2004, 101, 9792–9797. [Google Scholar] [CrossRef]
- Sun, Z.; Liu, K.; Zhang, J.; Zhang, Y.; Xu, K.; Yu, D.; Wang, J.; Hu, L.; Chen, L.; Li, C. IAA producing Bacillus altitudinis alleviates iron stress in Triticum aestivum L. seedling by both bioleaching of iron and up-regulation of genes encoding ferritins. Plant Soil 2017, 419, 1–11. [Google Scholar] [CrossRef]
- Hertlein, G.; Müller, S.; Garcia-Gonzalez, E.; Poppinga, L.; Süssmuth, R.D.; Genersch, E. Production of the Catechol Type Siderophore Bacillibactin by the Honey Bee Pathogen Paenibacillus larvae. PLoS ONE 2014, 9, e108272. [Google Scholar] [CrossRef]
- Segond, D.; Khalil, E.A.; Buisson, C.; Daou, N.; Kallassy, M.; Lereclus, D.; Arosio, P.; Bou-Abdallah, F.; Le Roux, C.N. Iron Acquisition in Bacillus cereus: The Roles of IlsA and Bacillibactin in Exogenous Ferritin Iron Mobilization. PLoS Pathog. 2014, 10, e1003935. [Google Scholar] [CrossRef]
- Yue, Z.; Chen, Y.; Hao, Y.; Wang, C.; Zhang, Z.; Chen, C.; Liu, H.; Liu, Y.; Li, L.; Sun, Z. Bacillus sp. WR12 alleviates iron deficiency in wheat via enhancing siderophore- and phenol-mediated iron acquisition in roots. Plant Soil 2021, 471, 247–260. [Google Scholar] [CrossRef]
- Chen, L.; James, L.P.; Helmann, J.D. Metalloregulation in Bacillus subtilis: Isolation and characterization of two genes differentially repressed by metal ions. J. Bacteriol. 1993, 175, 5428–5437. [Google Scholar] [CrossRef]
- Miethke, M. Molecular strategies of microbial iron assimilation: From high-affinity complexes to cofactor assembly systems. Metallomics 2013, 5, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Koppisch, A.T.; Browder, C.C.; Moe, A.L.; Shelley, J.T.; Kinkel, B.A.; Hersman, L.E.; Iyer, S.; Ruggiero, C.E. Petrobactin is the Primary Siderophore Synthesized by Bacillus anthracis Str. Sterne under Conditions of Iron Starvation. Biometals 2005, 18, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Miethke, M.; Klotz, O.; Linne, U.; May, J.J.; Beckering, C.L.; Marahiel, M.A. Ferri-bacillibactin uptake and hydrolysis in Bacillus subtilis. Mol. Microbiol. 2006, 61, 1413–1427. [Google Scholar] [CrossRef] [PubMed]
- Santos, S.; Neto, I.F.F.; Machado, M.D.; Soares, H.M.V.M.; Soares, E.V. Siderophore Production by Bacillus megaterium: Effect of Growth Phase and Cultural Conditions. Appl. Biochem. Biotechnol. 2013, 172, 549–560. [Google Scholar] [CrossRef]
- Ahmed, E.; Holmström, S.J.M. Siderophores in environmental research: Roles and applications. Microb. Biotechnol. 2014, 7, 196–208. [Google Scholar] [CrossRef]
- Nagoba, B.; Vedpathak, D. Medical Applications of Siderophores. Electron. J. Gen. Med. 2011, 8, 229–235. [Google Scholar] [CrossRef]
- Hotta, K.; Kim, C.-Y.; Fox, D.T.; Koppisch, A.T. Siderophore-mediated iron acquisition in Bacillus anthracis and related strains. Microbiology 2010, 156, 1918–1925. [Google Scholar] [CrossRef]
- Miethke, M.; Schmidt, S.; Marahiel, M.A. The Major Facilitator Superfamily-Type Transporter YmfE and the Multidrug-Efflux Activator Mta Mediate Bacillibactin Secretion in Bacillus subtilis. J. Bacteriol. 2008, 190, 5143–5152. [Google Scholar] [CrossRef]
- Ollinger, J.; Song, K.-B.; Antelmann, H.; Hecker, M.; Helmann, J.D. Role of the Fur Regulon in Iron Transport in Bacillus subtilis. J. Bacteriol. 2006, 188, 3664–3673. [Google Scholar] [CrossRef]
- Jacques, J.-F.; Jang, S.; Prévost, K.; Desnoyers, G.; Desmarais, M.; Imlay, J.; Massé, E. RyhB small RNA modulates the free intracellular iron pool and is essential for normal growth during iron limitation in Escherichia coli. Mol. Microbiol. 2006, 62, 1181–1190. [Google Scholar] [CrossRef]
- Smaldone, G.T.; Revelles, O.; Gaballa, A.; Sauer, U.; Antelmann, H.; Helmann, J. A Global Investigation of the Bacillus subtilis Iron-Sparing Response Identifies Major Changes in Metabolism. J. Bacteriol. 2012, 194, 2594–2605. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.E.; Huang, W.; Brewer, L.K.; Nguyen, A.T.; Kane, M.A.; Wilks, A.; Oglesby-Sherrouse, A.G. Proteomic analysis of the Pseudomonas aeruginosa iron starvation response reveals PrrF small regulatory RNA-dependent iron regulation of twitching motility, amino acid metabolism, and zinc homeostasis proteins. J. Bacteriol. 2019, 201, e00754-18. [Google Scholar] [CrossRef] [PubMed]
- Pelley, J.W. Glycolysis and pyruvate oxidation. In Elsevier’s Integrated Review Biochemistry, 2nd ed.; Pelley, J.W., Ed.; Saunders: Philadelphia, PA, USA, 2012; pp. 49–55. [Google Scholar]
- Schenk, G.; Duggleby, R.G.; Nixon, P.F. Properties and functions of the thiamin diphosphate dependent enzyme trans-ketolase. Int. J. Biochem. Cell Biol. 1998, 30, 1297–1318. [Google Scholar] [CrossRef]
- Veech, R.L. Tricarboxylic Acid Cycle. In Encyclopedia of Biological Chemistry, 2nd ed.; Lennarz, W.J., Lane, M.D., Eds.; Academic Press: Pittsburgh, PA, USA, 2013; pp. 436–440. [Google Scholar]
- Chand, S.; Mahajan, R.V.; Prasad, J.P.; Sahoo, D.K.; Mihooliya, K.N.; Dhar, M.S.; Sharma, G. A comprehensive review on microbial l-asparaginase: Bioprocessing, characterization, and industrial applications. Biotechnol. Appl. Biochem. 2020, 67, 619–647. [Google Scholar] [CrossRef] [PubMed]
- Cooper, A.J.L.; Shurubor, Y.I.; Dorai, T.; Pinto, J.T.; Isakova, E.P.; Deryabina, Y.I.; Denton, T.T.; Krasnikov, B.F. ω-Amidase: An underappreciated, but important enzyme in l-glutamine and l-asparagine metabolism; relevance to sulfur and nitrogen metabolism, tumor biology and hyperammonemic diseases. Amino Acids 2015, 48, 1–20. [Google Scholar] [CrossRef]
- Jaisson, S.; Veiga-da-Cunha, M.; Van Schaftingen, E. Molecular identification of ω-amidase, the enzyme that is functionally coupled with glutamine transaminases, as the putative tumor suppressor Nit2. Biochimie 2009, 91, 1066–1071. [Google Scholar] [CrossRef]
- Hu, X.; Quinn, P.J.; Wang, Z.; Han, G.; Wang, X. Genetic Modification and Bioprocess Optimization for S-Adenosyl-L-methionine Biosynthesis. Subcell. Biochem. 2012, 64, 327–341. [Google Scholar] [CrossRef]
- Qin, Z.; Yan, Q.; Ma, Q.; Jiang, Z. Crystal structure and characterization of a novel L-serine ammonia-lyase from Rhizomucor miehe. Biochem. Biophys. Res. Commun. 2015, 466, 431–437. [Google Scholar] [CrossRef]
- Lin, C.J.; Wang, M.C. Microbial metabolites regulate host lipid metabolism through NR5A-Hedgehog signalling. Nat. Cell Biol. 2017, 19, 550–557. [Google Scholar] [CrossRef]
- Ohshima, N.; Yamashita, S.; Takahashi, N.; Kuroishi, C.; Shiro, Y.; Takio, K. Escherichia coli cytosolic glycerophosphodiester phosphodiesterase (UgpQ) requires Mg2+, Co2+, or Mn2+ for its enzyme activity. J. Bacteriol. 2008, 190, 1219–1223. [Google Scholar] [CrossRef]
- Brzoska, P.; Boos, W. The ugp-encoded glycerophosphoryldiester phosphodiesterase, a transport-related enzyme of Escherichia coli. FEMS Microbiol. Rev. 1989, 5, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Zhu, L.; Guo, J.; Xiao, X.; Ma, Z.; Wang, J. Bacillus subtilis STU6 ameliorates iron deficiency in tomato by enhancement of polyamine-mediated iron remobilization. J. Agric. Food Chem. 2019, 67, 320–330. [Google Scholar] [CrossRef] [PubMed]




| Accession | Description | FC (Fe−/Fe+) | p Value | Regulate |
|---|---|---|---|---|
| Carbohydrate metabolism | ||||
| gene1651 | pyruvate dehydrogenase E1 component beta subunit (PDHB) | 0.718 | 0.006 | down |
| gene4021 | aconitase (ACO) | 0.747 | 0.036 | down |
| gene2203 | phosphoglycerate kinase (PGK) | 0.749 | 0.030 | down |
| gene2204 | type I glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | 0.660 | 0.008 | down |
| gene4010 | transketolase (TKT) | 0.367 | 0.017 | down |
| gene2263 | beta-phosphoglucomutase (PGMB) | 0.501 | 0.028 | down |
| Amino acid metabolism | ||||
| gene1700 | Asparaginase (ANS) | 0.572 | 0.007 | down |
| gene0508 | ω-amidase | 0.816 | 0.022 | down |
| gene0981 | methionine adenosyltransferase (MAT) | 0.723 | 0.022 | down |
| gene0743 | L-serine ammonia-lyase, iron-sulfur-dependent subunit beta (SDH) | 0.729 | 0.038 | down |
| Lipid metabolism | ||||
| gene3591 | glycerophosphodiester phosphodiesterase (GDPD) | 0.810 | 0.039 | down |
| gene0063 | glycerol-3-phosphate dehydrogenase (GPD) | 0.725 | 0.044 | down |
| Fe-containing proteins | ||||
| gene2592 | heme-dependent peroxidase HemQ | 0.549 | 0.028 | down |
| gene1752 | ferredoxin | 0.348 | 0.040 | down |
| gene0858 | 2′,3′-cyclic-nucleotide 2′-phosphodiesterase (CNP) | 0.537 | 0.002 | down |
| gene2067 | Fe-S cluster assembly protein SufD | 0.741 | 0.039 | down |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yue, Z.; Liu, Y.; Chen, Y.; Chen, C.; Zhang, J.; He, L.; Ma, K. Comprehensive Genomics and Proteomics Analysis Reveals the Multiple Response Strategies of Endophytic Bacillus sp. WR13 to Iron Limitation. Microorganisms 2023, 11, 367. https://doi.org/10.3390/microorganisms11020367
Yue Z, Liu Y, Chen Y, Chen C, Zhang J, He L, Ma K. Comprehensive Genomics and Proteomics Analysis Reveals the Multiple Response Strategies of Endophytic Bacillus sp. WR13 to Iron Limitation. Microorganisms. 2023; 11(2):367. https://doi.org/10.3390/microorganisms11020367
Chicago/Turabian StyleYue, Zonghao, Yongchuang Liu, Yanjuan Chen, Can Chen, Ju Zhang, Le He, and Keshi Ma. 2023. "Comprehensive Genomics and Proteomics Analysis Reveals the Multiple Response Strategies of Endophytic Bacillus sp. WR13 to Iron Limitation" Microorganisms 11, no. 2: 367. https://doi.org/10.3390/microorganisms11020367
APA StyleYue, Z., Liu, Y., Chen, Y., Chen, C., Zhang, J., He, L., & Ma, K. (2023). Comprehensive Genomics and Proteomics Analysis Reveals the Multiple Response Strategies of Endophytic Bacillus sp. WR13 to Iron Limitation. Microorganisms, 11(2), 367. https://doi.org/10.3390/microorganisms11020367








