Exploring Microorganisms from Plastic-Polluted Sites: Unveiling Plastic Degradation and PHA Production Potential
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection of Samples
2.2. Bacterial Isolation
2.3. Agar-Based Screening Methodology
2.4. 16S rRNA Sequencing
2.5. Phylogenetic Analysis
2.6. Upcycling Capabilities of Selected Isolates
2.6.1. Emulsification Assay
2.6.2. Reactive Extrusion
2.6.3. Bacterial Cultivation for PHA Production
2.6.4. Quantitative and Qualitative Analysis of Derived PHAs
2.6.5. Estimation of TPA Content in Cultivation Media by HPLC Analysis
3. Results and Discussion
3.1. Isolation and Screening of Microbial Strains from Plastic-Polluted Sites
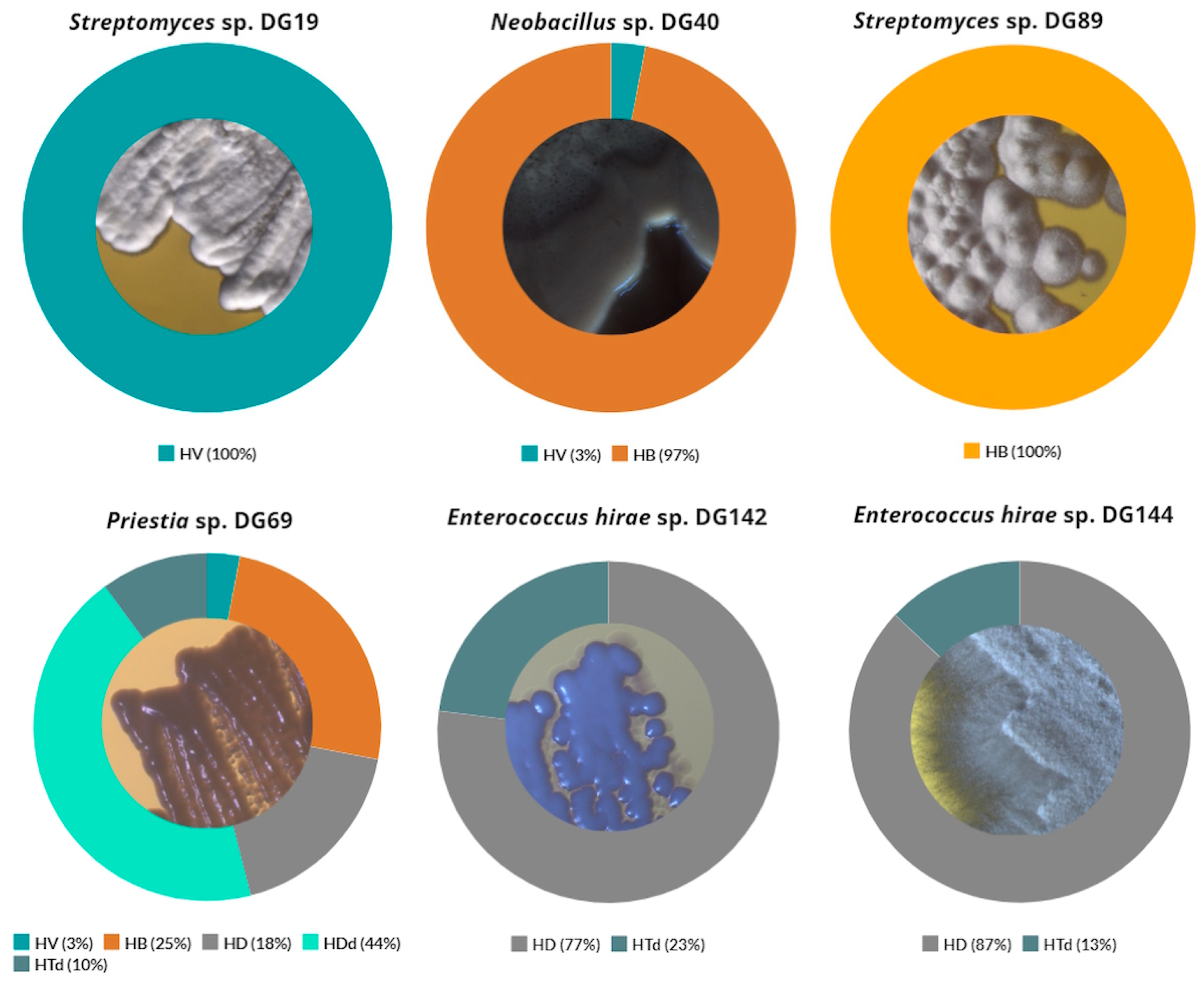
3.2. Most Promising Strains Selection and Characterization
3.3. Phylogenetic Analysis
3.4. Upcycling Capabilities of Isolated Strains
3.4.1. Emulsification Assay
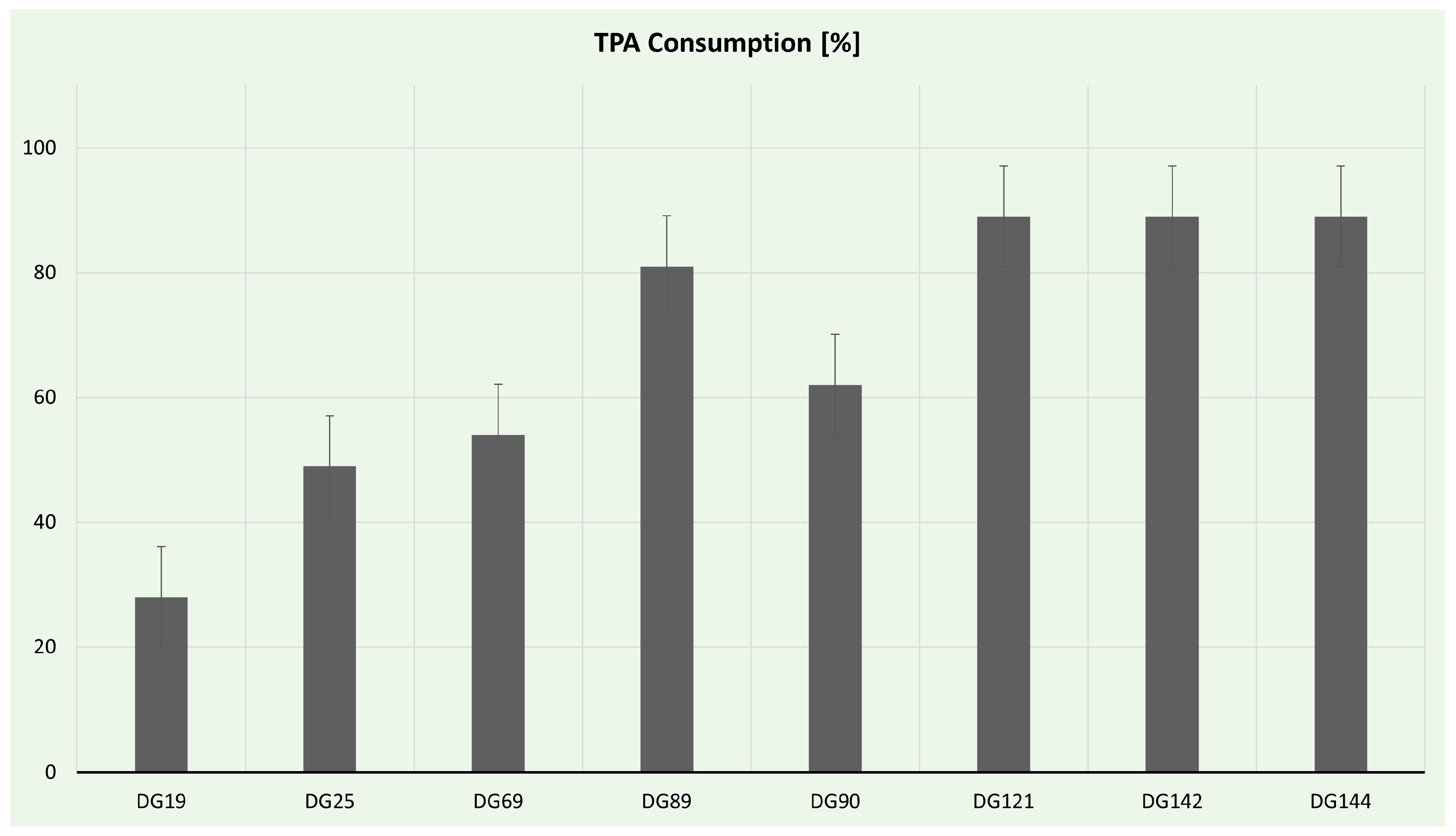
3.4.2. PHA Production
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Purohit, J.; Chattopadhyay, A.; Teli, B. Metagenomic Exploration of Plastic Degrading Microbes for Biotechnological Application. Curr. Genom. 2020, 21, 253–270. [Google Scholar] [CrossRef] [PubMed]
- Gambarini, V.; Pantos, O.; Kingsbury, J.M.; Weaver, L.; Handley, K.M.; Lear, G. Phylogenetic Distribution of Plastic-Degrading Microorganisms. mSystems 2021, 6. [Google Scholar] [CrossRef]
- Thumarat, U.; Nakamura, R.; Kawabata, T.; Suzuki, H.; Kawai, F. Biochemical and genetic analysis of a cutinase-type polyesterase from a thermophilic Thermobifida alba AHK119. Appl. Microbiol. Biotechnol. 2011, 95, 419–430. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.; Oeser, T.; Then, J.; Kühn, N.; Barth, M.; Schmidt, J.; Zimmermann, W. Functional characterization and structural modeling of synthetic polyester-degrading hydrolases from Thermomonospora curvata. AMB Express 2014, 4, 44. [Google Scholar] [CrossRef]
- Tanasupawat, S.; Takehana, T.; Yoshida, S.; Hiraga, K.; Oda, K. Ideonella sakaiensis sp. nov., isolated from a microbial consortium that degrades PET. Int. J. Syst. Evol. Microbiol. 2016, 66, 2813–2818. [Google Scholar] [CrossRef] [PubMed]
- Egmond, M. Fusarium solani pisi cutinase. Biochimie 2000, 82, 1015–1021. [Google Scholar] [CrossRef] [PubMed]
- Ronkvist, Å.M.; Xie, W.; Lu, W.; Gross, R.A. Cutinase-Catalyzed Hydrolysis of Poly(ethylene terephthalate). Macromolecules 2009, 42, 5128–5138. [Google Scholar] [CrossRef]
- Maeda, H.; Yamagata, Y.; Abe, K.; Hasegawa, F.; Machida, M.; Ishioka, R.; Gomi, K.; Nakajima, T. Purification and characterization of a biodegradable plastic-degrading enzyme from Aspergillus oryzae. Appl. Microbiol. Biotechnol. 2005, 67, 778–788. [Google Scholar] [CrossRef]
- Da Costa, J.P.; Nunes, A.R.; Santos, P.S.M.; Girão, A.V.; Duarte, A.C.; Rocha-Santos, T. Degradation of polyethylene microplastics in seawater: Insights into the environmental degradation of polymers. J. Environ. Sci. Health Part A 2018, 53, 866–875. [Google Scholar] [CrossRef]
- Song, L.; Wang, Y.; Tang, W.; Lei, Y. Bacterial community diversity in municipal waste landfill sites. Appl. Microbiol. Biotechnol. 2015, 99, 7745–7756. [Google Scholar] [CrossRef]
- Kumar, R.; Pandit, P.; Kumar, D.; Patel, Z.; Pandya, L.; Kumar, M.; Joshi, C.; Joshi, M. Landfill microbiome harbour plastic degrading genes: A metagenomic study of solid waste dumping site of Gujarat, India. Sci. Total Environ. 2021, 779, 146184. [Google Scholar] [CrossRef] [PubMed]
- Hiraga, K.; Taniguchi, I.; Yoshida, S.; Kimura, Y.; Oda, K. Biodegradation of waste PET. EMBO Rep. 2019, 20, e49365. [Google Scholar] [CrossRef] [PubMed]
- Malik, N.; Lakhawat, S.S.; Kumar, V.; Sharma, V.; Bhatti, J.S.; Sharma, P.K. Recent advances in the omics-based assessment of microbial consortia in the plastisphere environment: Deciphering the dynamic role of hidden players. Process Saf. Environ. Prot. 2023, 176, 207–225. [Google Scholar] [CrossRef]
- Graham Bowditch, T. Penetration of Polyvinyl Chloride and Polypropylene Packaging Films by Ephestia cautella (Lepidoptera: Pyralidae) and Plodia interpunctella (Lepidoptera: Pyralidae) Larvae, and Tribolium confusum (Coleoptera: Tenebrionidae) Adults. J. Econ. Entomol. 1997, 90, 1028–1031. [Google Scholar] [CrossRef]
- Kim, J.-H.; Lee, S.-H.; Lee, B.-M.; Son, K.-H.; Park, H.-Y. Biodegradation Potential of Polyethylene Terephthalate by the Two Insect Gut Symbionts Xanthomonas sp. HY-74 and Bacillus sp. HY-75. Polymers 2023, 15, 3546. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.Q.; Longing, S.; Siebecker, M.G. Consumption and degradation of different consumer plastics by mealworms (Tenebrio molitor): Effects of plastic type, time, and mealworm origin. J. Clean. Prod. 2023, 403, 136842. [Google Scholar] [CrossRef]
- Ong, S.Y.; Heng, K.-S.; Heng, K.-S.; Ong, S.Y.; Sudesh, K. Efficient biosynthesis and recovery of polyhydroxyalkanoate. Malays. J. Microbiol. 2016, 12, 383–398. [Google Scholar]
- Paul-Pont, I.; Ghiglione, J.-F.; Gastaldi, E.; Ter Halle, A.; Huvet, A.; Bruzaud, S.; Lagarde, F.; Galgani, F.; Duflos, G.; George, M.; et al. Discussion about suitable applications for biodegradable plastics regarding their sources, uses and end of life. Waste Manag. 2023, 157, 242–248. [Google Scholar] [CrossRef]
- Nielsen, C.; Rahman, A.; Rehman, A.U.; Walsh, M.K.; Miller, C.D. Food waste conversion to microbial polyhydroxyalkanoates. Microb. Biotechnol. 2017, 10, 1338–1352. [Google Scholar] [CrossRef]
- Surendran, A.; Lakshmanan, M.; Chee, J.Y.; Sulaiman, A.M.; Thuoc, D.V.; Sudesh, K. Can Polyhydroxyalkanoates Be Produced Efficiently From Waste Plant and Animal Oils? Front. Bioeng. Biotechnol. 2020, 8, 169. [Google Scholar] [CrossRef]
- Mannina, G.; Presti, D.; Montiel-Jarillo, G.; Carrera, J.; Suárez-Ojeda, M.E. Recovery of polyhydroxyalkanoates (PHAs) from wastewater: A review. Bioresour. Technol. 2020, 297, 122478. [Google Scholar] [CrossRef]
- Tiso, T.; Narancic, T.; Wei, R.; Pollet, E.; Beagan, N.; Schröder, K.; Honak, A.; Jiang, M.; Kenny, S.T.; Wierckx, N.; et al. Towards bio-upcycling of polyethylene terephthalate. Metab. Eng. 2021, 66, 167–178. [Google Scholar] [CrossRef]
- Johnston, B.; Adamus, G.; Ekere, A.I.; Kowalczuk, M.; Tchuenbou-Magaia, F.; Radecka, I. Bioconversion of Plastic Waste Based on Mass Full Carbon Backbone Polymeric Materials to Value-Added Polyhydroxyalkanoates (PHAs). Bioengineering 2022, 9, 432. [Google Scholar] [CrossRef] [PubMed]
- Wierckx, N.; Prieto, M.A.; Pomposiello, P.; de Lorenzo, V.; O’Connor, K.; Blank, L.M. Plastic waste as a novel substrate for industrial biotechnology. Microb. Biotechnol. 2015, 8, 900–903. [Google Scholar] [CrossRef]
- Johnston, B.; Radecka, I.; Hill, D.; Chiellini, E.; Ilieva, V.I.; Sikorska, W.; Musioł, M.; Zięba, M.; Marek, A.A.; Keddie, D.; et al. The Microbial Production of Polyhydroxyalkanoates from Waste Polystyrene Fragments Attained Using Oxidative Degradation. Polymers 2018, 10, 957. [Google Scholar] [CrossRef] [PubMed]
- Narancic, T.; Salvador, M.; Hughes, G.M.; Beagan, N.; Abdulmutalib, U.; Kenny, S.T.; Wu, H.; Saccomanno, M.; Um, J.; O’Connor, K.E.; et al. Genome analysis of the metabolically versatile Pseudomonas umsongensis GO16: The genetic basis for PET monomer upcycling into polyhydroxyalkanoates. Microb. Biotechnol. 2021, 14, 2463–2480. [Google Scholar] [CrossRef] [PubMed]
- Sousa, A.M.; Machado, I.; Nicolau, A.; Pereira, M.O. Improvements on colony morphology identification towards bacterial profiling. J. Microbiol. Methods 2013, 95, 327–335. [Google Scholar] [CrossRef]
- Breakwell, D.; MacDonald, B.; Woolverton, C.; Smith, K.; Robison, R. Colony Morphology Protocol. Am. Soc. Microbiol. 2007. Available online: https://asm.org/ASM/media/Protocol-Images/Colony-Morphology-Protocol.pdf?ext=.pdf (accessed on 15 March 2023).
- Molitor, R.; Bollinger, A.; Kubicki, S.; Loeschcke, A.; Jaeger, K.-E.; Thies, S. Agar plate-based screening methods for the identification of polyester hydrolysis by Pseudomonas species. Microb. Biotechnol. 2020, 13, 274–284. [Google Scholar] [CrossRef]
- Ibrahim MH, A.; Willems, A.; Steinbüchel, A. Isolation and characterization of new poly(3HB)-accumulating star-shaped cell-aggregates-forming thermophilic bacteria. J. Appl. Microbiol. 2010, 109, 1579–1590. [Google Scholar] [CrossRef]
- Álvarez-Barragán, J.; Domínguez-Malfavón, L.; Vargas-Suárez, M.; González-Hernández, R.; Aguilar-Osorio, G.; Loza-Tavera, H. Biodegradative Activities of Selected Environmental Fungi on a Polyester Polyurethane Varnish and Polyether Polyurethane Foams. Appl. Environ. Microbiol. 2016, 82, 5225–5235. [Google Scholar] [CrossRef]
- Nikodinovic, J.; Barrow, K.D.; Chuck, J.-A. High yield preparation of genomic DNA from Streptomyces. Biotechniques 2003, 35, 932–936. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Saltou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Brennan Fournet, M.; Attallah, O.A.; Mojicevic, M.; Chen, Y.; Major, I. High Throughput Mechanochemical Depolymerisation and Purification of Polyethylene Terephthalate and Related Polymers; World Intellectual Property Organization: Geneva, Switzerland, 2022. [Google Scholar]
- Silvestre, S.L.; Araújo, D.; Marques, A.C.; Pires, C.; Matos, M.; Alves, V.D.; Martins, R.F.P.; Freitas, F.; Reis, M.A.M.; Fortunato, E. Microneedle Arrays of Polyhydroxyalkanoate by Laser-Based Micromolding Technique. ACS Appl. Bio Mater. 2020, 3, 5856–5864. [Google Scholar] [CrossRef]
- Rebocho, A.T.; Perreira, J.R.; Neves, L.A.; Alves, V.D.; Sevrin, C.; Grandfils, C.; Freitas, F.; Reis, M.A.M. Preparation and Characterization of Films Based on a Natural P(3HB)/mcl-PHA Blend Obtained through the Co-culture of Cupriavidus Necator and Pseudomonas Citronellolis in Apple Pulp Waste. Bioengineering 2020, 7, 34. [Google Scholar] [CrossRef] [PubMed]
- Bleuven, C.; Landry, C.R. Molecular and cellular bases of adaptation to a changing environment in microorganisms. Proc. R. Soc. B Biol. Sci. 2016, 283, 20161458. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, I. Role of microbiome and biofilm in environmental plastic degradation. Biocatal. Agric. Biotechnol. 2022, 39, 102263. [Google Scholar] [CrossRef]
- Hara, H.; Eltis, L.D.; Davies, J.E.; Mohn, W.W. Transcriptomic Analysis Reveals a Bifurcated Terephthalate Degradation Pathway in Rhodococcus sp. Strain RHA1. J. Bacteriol. 2007, 189, 1641–1647. [Google Scholar] [CrossRef] [PubMed]
- Mooney, A.; Ward, P.G.; O’Connor, K.E. Microbial degradation of styrene: Biochemistry, molecular genetics, and perspectives for biotechnological applications. Appl. Microbiol. Biotechnol. 2006, 72, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Huang, D.; Ji, J.; Völker, C.; Wurm, F.R. Seawater-Degradable Polymers—Fighting the Marine Plastic Pollution. Adv. Sci. 2021, 8, 2001121. [Google Scholar] [CrossRef]
- Mitra, B.; Das, A. The Ability of Insects to Degrade Complex Synthetic Polymers. In Arthropods-New Advances and Perspectives [Working Title]; IntechOpen: London, UK, 2023. [Google Scholar] [CrossRef]
- Kopecká, R.; Kubínová, I.; Sovová, K.; Mravcová, L.; Vítěz, T.; Vítězová, M. Microbial degradation of virgin polyethylene by bacteria isolated from a landfill site. SN Appl. Sci. 2022, 4, 302. [Google Scholar] [CrossRef]
- Geyer, R.; Jambeck, J.R.; Law, K.L. Production, use, and fate of all plastics ever made. Sci. Adv. 2017, 3, e1700782. [Google Scholar] [CrossRef]
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Meier-Kolthoff, J.P.; Klenk, H.P.; Clément, C.; Ouhdouch, Y.; Van Wezel, G.P. Taxonomy, Physiology, and Natural Products of Actinobacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 1–43. [Google Scholar] [CrossRef]
- Rodríguez-Fonseca, M.F.; Sánchez-Suárez, J.; Valero, M.F.; Ruiz-Balaguera, S.; Díaz, L.E. Streptomyces as Potential Synthetic Polymer Degraders: A Systematic Review. Bioengineering 2021, 8, 154. [Google Scholar] [CrossRef]
- Taddei, A.; Rodríguez, M.J.; Márquez-Vilchez, E.; Castelli, C. Isolation and identification of Streptomyces spp. from Venezuelan soils: Morphological and biochemical studies. I. Microbiol. Res. 2006, 161, 222–231. [Google Scholar] [CrossRef]
- Janakiev, T.; Milošević, K.; Petrović, M.; Miljković, J.; Stanković, N.; Zdravković, D.S.; Dimkić, I. Chironomus riparius Larval Gut Bacteriobiota and Its Potential in Microplastic Degradation. Microb. Ecol. 2023, 86, 1909–1922. [Google Scholar] [CrossRef]
- Bergey, H.D. Bergey’s Manual of Determinative Bacteriology; Williams & Wilkins Co.: Philadelphia, PA, USA, 1975. [Google Scholar]
- Vary, P.S.; Biedendieck, R.; Fuerch, T.; Meinhardt, F.; Rohde, M.; Deckwer, W.-D.; Jahn, D. Bacillus megaterium—From simple soil bacterium to industrial protein production host. Appl. Microbiol. Biotechnol. 2007, 76, 957–967. [Google Scholar] [CrossRef] [PubMed]
- Meng, T.K.; Kassim, A.S.B.M.; Razak, A.H.B.A.; Fauzi, N.A.B.M. Bacillus megaterium: A Potential and an Efficient Bio-Degrader of Polystyrene. Braz. Arch. Biol. Technol. 2021, 64, e21190321. [Google Scholar] [CrossRef]
- Biedendieck, R.; Knuuti, T.; Moore, S.J.; Jahn, D. The “beauty in the beast”—The multiple uses of Priestia megaterium in biotechnology. Appl. Microbiol. Biotechnol. 2021, 105, 5719–5737. [Google Scholar] [CrossRef]
- Zerhouni, K.; Abbouni, B.; Kanoun, K.; Larbi Daouadji, K.; Tifrit, A.; Benahmed, M.; Chaouche, T.M. Isolation and identification of low density polythene-degrading bacteria from soil of North West of Algeria. South Asian J. Exp. Biol. 2019, 8, 76–82. [Google Scholar] [CrossRef]
- Puglisi, E.; Romaniello, F.; Galletti, S.; Boccaleri, E.; Frache, A.; Cocconcelli, P.S. Selective bacterial colonization processes on polyethylene waste samples in an abandoned landfill site. Sci. Rep. 2019, 9, 14138. [Google Scholar] [CrossRef] [PubMed]
- Saikia, S.S.; Borah, B.K.; Baruah, G.; Rokozeno; Deka, M.K. Characterization of the gut microbes of greater wax moth (Galleria mellonella Linnaeus) shows presence of potential polymer degraders. Folia Microbiol. 2022, 67, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Kag, S.; Kumar, P.; Kataria, R. Potato Peel Waste as an Economic Feedstock for PHA Production by Bacillus circulans. Appl. Biochem. Biotechnol. Appl. Biochem. Biotechnol. 2023, 1–15. [Google Scholar] [CrossRef]
- Phukon, P.; Saikia, J.P.; Konwar, B.K. Bio-plastic (P-3HB-co-3HV) from Bacillus circulans (MTCC 8167) and its biodegradation. Colloids Surf B Biointerfaces 2012, 92, 30–34. [Google Scholar] [CrossRef]
- Nakkabi, A.; Noureddine, E.; Moulay, S.; Ibnsouda, K.S.; Mohammed, F.F. Biodegradation of Poly (Ethylene Terephthalate) by Bacillus subtilis. Int. J. Recent. Adv. Multidiscip. Res. 2015, 2, 1060–1062. [Google Scholar]
- Benavides Fernández, C.D.; Guzmán Castillo, M.P.; Quijano Pérez, S.A.; Carvajal Rodríguez, L.V. Microbial degradation of polyethylene terephthalate: A systematic review. SN Appl. Sci. 2022, 4, 263. [Google Scholar] [CrossRef]
- Li, W.; Jia, M.-X.; Deng, J.; Wang, J.-H.; Lin, Q.-L.; Liu, C.; Wang, S.-S.; Tang, J.-X.; Zeng, X.-X.; Ma, L.; et al. Isolation, genetic identification and degradation characteristics of COD-degrading bacterial strain in slaughter wastewater. Saudi J. Biol. Sci. 2018, 25, 1800–1805. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Li, S.; Dang, G.; Jia, R.; Chen, S.; Deng, X.; Liu, G.; Beckers, Y.; Cai, H. Screening and characterization of Bacillus velezensis LB-Y-1 toward selection as a potential probiotic for poultry with multi-enzyme production property. Front. Microbiol. 2023, 14, 1143265. [Google Scholar] [CrossRef] [PubMed]
- Gui, Z.; Liu, G.; Liu, X.; Cai, R.; Liu, R.; Sun, C. A Deep-Sea Bacterium Is Capable of Degrading Polyurethane. Microbiol. Spectr. 2023, 11, e0007323. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Chen, C.; Shen, Q.; Rosen, B.P.; Zhao, F.-J. Genetically Engineering Bacillus subtilis with a Heat-Resistant Arsenite Methyltransferase for Bioremediation of Arsenic-Contaminated Organic Waste. Appl. Environ. Microbiol. 2015, 81, 6718–6724. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.; Zimmermann, W. Microbial enzymes for the recycling of recalcitrant petroleum-based plastics: How far are we? Microb. Biotechnol. 2017, 10, 1308–1322. [Google Scholar] [CrossRef] [PubMed]
- Yao, Z.; Seong, H.J.; Jang, Y.-S. Degradation of low density polyethylene by Bacillus species. Appl. Biol. Chem. 2022, 65, 84. [Google Scholar] [CrossRef]
- Nakkabi, A.; Sadiki, M.; Fahim, M.; Ittobane, N.; IbnsoudaKoraichi, S.; Barkai, H.; El Abed, S. Biodegradation of Poly(ester urethane)s by Bacillus subtilis. Int. J. Environ. Res. 2015, 9, 157–162. [Google Scholar]
- Mukherjee, S.; Roy Chowdhuri, U.; Kundu, P.P. Bio-degradation of polyethylene waste by simultaneous use of two bacteria: Bacillus licheniformis for production of bio-surfactant and Lysinibacillus fusiformis for bio-degradation. RSC Adv. 2016, 6, 2982–2992. [Google Scholar] [CrossRef]
- Arena, M.; Abbate, C.; Fukushima, K.; Gennari, M. Degradation of poly (lactic acid) and nanocomposites by Bacillus licheniformis. Environ. Sci. Pollut. Res. 2011, 18, 865–870. [Google Scholar] [CrossRef]
- Jain, K.; Bhunia, H.; Reddy, M.S. Degradation of polypropylene-poly-L-lactide blends by Bacillus isolates: A microcosm and field evaluation. Bioremediation J. 2022, 26, 64–75. [Google Scholar] [CrossRef]
- Wang, Y.; Hu, T.; Zhang, W.; Lin, J.; Wang, Z.; Lyu, S.; Tong, H. Biodegradation of polylactic acid by a mesophilic bacteria Bacillus safensis. Chemosphere 2023, 318, 137991. [Google Scholar] [CrossRef]
- Abdullah, A.; Waqas, M.; Haris, M.; Asim, N.; Islam, H.U.; Khan, A.; Khattak, H.; Ali, S. Biodegradable Potential of Bacillus amyloliquefaciens and Bacillus safensis Using Low Density Polyethylene Thermoplastic (LDPE) Substrate. Eur. J. Environ. Public Health 2021, 5, em0069. [Google Scholar] [CrossRef] [PubMed]
- Oulidi, O.; Nakkabi, A.; Bouymajane, A.; Elaraaj, I.; Filali, F.R.; Fahim, M.; El Moualij, N. Biodegradation of polyamide 6 by Lysinibacillus sp, Alcaligene faecalis and Enterococcus faecalis. Clean. Chem. Eng. 2022, 3, 100054. [Google Scholar] [CrossRef]
- Hchaichi, I.; Bandini, F.; Spini, G.; Banni, M.; Cocconcelli, P.S.; Puglisi, E. Enterococcus faecalis and Vibrio harveyi colonize low-density polyethylene and biodegradable plastics under marine conditions. FEMS Microbiol. Lett. 2020, 367, fnaa125. [Google Scholar] [CrossRef] [PubMed]
- Nur, C.; Guven, O. Poly--hydroxybutyrate (PHB) production from domestic wastewater using Enterobacter aerogenes 12Bi strain. Afr. J. Microbiol. Res. 2011, 5, 690–702. [Google Scholar]
- Bhuwal, A.K.; Singh, G.; Aggarwal, N.K.; Goyal, V.; Yadav, A. Isolation and Screening of Polyhydroxyalkanoates Producing Bacteria from Pulp, Paper, and Cardboard Industry Wastes. Int. J. Biomater. 2013, 2013, 752821. [Google Scholar] [CrossRef]
- Obradors, N.; Aguilar, J. Efficient biodegradation of high-molecular-weight polyethylene glycols by pure cultures of Pseudomonas stutzeri. Appl. Environ. Microbiol. 1991, 57, 2383–2388. [Google Scholar] [CrossRef]
- Liu, P.; Zhang, T.; Zhen, Y.; Li, Q.; Liang, Q.; Qi, Q. Screening and genome analysis of a Pseudomonas stutzeri that degrades PET monomer terephthalate. Acta Microbiol. Sin. 2022, 62, 200–212. [Google Scholar]
- Hooda, S.; Annu; Mondal, P. Insights into the degradation of high-density polyethylene microplastics using microbial strains: Effect of process parameters, degradation kinetics and modeling. Waste Manag. 2023, 164, 143–153. [Google Scholar] [CrossRef]
- Uefuji, M.; Kasuya, K.; Doi, Y. Enzymatic degradation of poly[(R)-3-hydroxybutyrate]: Secretion and properties of PHB depolymerase from Pseudomonas stutzeri. Polym. Degrad. Stab. 1997, 58, 275–281. [Google Scholar] [CrossRef]
- Drzyzga, O.; Prieto, A. Plastic waste management, a matter for the ‘community’. Microb. Biotechnol. 2019, 12, 66–68. [Google Scholar] [CrossRef] [PubMed]
- Fields, R.D.; Rodriguez, F.; Finn, R.K. Microbial degradation of polyesters: Polycaprolactone degraded by P. pullulans. J. Appl. Polym. Sci. 1974, 18, 3571–3579. [Google Scholar] [CrossRef]
- Shah, Z.; Krumholz, L.; Aktas, D.F.; Hasan, F.; Khattak, M.; Shah, A.A. Degradation of polyester polyurethane by a newly isolated soil bacterium, Bacillus subtilis strain MZA-75. Biodegradation 2013, 24, 865–877. [Google Scholar] [CrossRef] [PubMed]
- Shah, Z.; Hasan, F.; Krumholz, L.; Aktas, D.F.; Shah, A.A. Degradation of polyester polyurethane by newly isolated Pseudomonas aeruginosa strain MZA-85 and analysis of degradation products by GC–MS. Int. Biodeterior. Biodegradation 2013, 77, 114–122. [Google Scholar] [CrossRef]
- Sriyapai, P.; Chansiri, K.; Sriyapai, T. Isolation and Characterization of Polyester-Based Plastics-Degrading Bacteria from Compost Soils. Microbiology 2018, 87, 290–300. [Google Scholar] [CrossRef]
- Liu, G.; Zhong, H.; Yang, X.; Liu, Y.; Shao, B.; Liu, Z. Advances in applications of rhamnolipids biosurfactant in environmental remediation: A review. Biotechnol. Bioeng. 2018, 115, 796–814. [Google Scholar] [CrossRef]
- López-Prieto, A.; Rodríguez-López, L.; Rincón-Fontán, M.; Cruz, J.M.; Moldes, A.B. Characterization of extracellular and cell bound biosurfactants produced by Aneurinibacillus aneurinilyticus isolated from commercial corn steep liquor. Microbiol. Res. 2021, 242, 126614. [Google Scholar] [CrossRef]
- Hajfarajollah, H.; Mokhtarani, B.; Tohidi, A.; Bazsefidpar, S.; Akbari Noghabi, K. Overproduction of lipopeptide biosurfactant by Aneurinibacillus thermoaerophilus HAK01 in various fed-batch modes under thermophilic conditions. RSC Adv. 2019, 9, 30419–30427. [Google Scholar] [CrossRef]
- Manikkasundaram, V.; Baskaran, A.; Kaari, M.; Angamuthu, V.; Venugopal, G.; Manikkam, R. Production and characterization of glycolipid biosurfactant from Streptomyces enissocaesilis HRB1 and its evaluation for biomedical and bioremediation applications. J. Surfactants Deterg. 2023, 26, 491–503. [Google Scholar] [CrossRef]
- Zambry, N.S.; Rusly, N.S.; Awang, M.S.; Md Noh, N.A.; Yahya AR, M. Production of lipopeptide biosurfactant in batch and fed-batch Streptomyces sp. PBD-410L cultures growing on palm oil. Bioprocess. Biosyst. Eng. 2021, 44, 1577–1592. [Google Scholar] [CrossRef]
- Mgbechidinma, C.L.; Akan, O.D.; Zhang, C.; Huang, M.; Linus, N.; Zhu, H.; Wakil, S.M. Integration of green economy concepts for sustainable biosurfactant production—A review. Bioresour. Technol. 2022, 364, 128021. [Google Scholar] [CrossRef]
- Bugnicourt, E.; Cinelli, P.; Lazzeri, A.; Alvarez, V. Polyhydroxyalkanoate (PHA): Review of synthesis, characteristics, processing and potential applications in packaging. Express Polym. Lett. 2014, 8, 791–808. [Google Scholar] [CrossRef]
- Akiyama, M.; Tsuge, T.; Doi, Y. Environmental life cycle comparison of polyhydroxyalkanoates produced from renewable carbon resources by bacterial fermentation. Polym. Degrad. Stab. 2003, 80, 183–194. [Google Scholar] [CrossRef]
- Kingsly, J.S.; Chathalingath, N.; Parthiban, S.A.; Sivakumar, D.; Sambugan, S.; Senniyappan, V.; Sekar, D.V.; Jasmine, A.H.; Gunasekar, G. Utilization of sugarcane molasses as the main carbon source for the production of polyhydroxyalkanoates from Enterobacter cloacae. Energy Nexus 2022, 6, 100071. [Google Scholar] [CrossRef]
- Obruca, S.; Benesova, P.; Marsalek, L.; Marova, I. Use of Lignocellulosic Materials for PHA Production. Chem. Biochem. Eng. Q. 2015, 29, 135–144. [Google Scholar] [CrossRef]
- Hassan, M.A.; Yee, L.-N.; Yee, P.L.; Ariffin, H.; Raha, A.R.; Shirai, Y.; Sudesh, K. Sustainable production of polyhydroxyalkanoates from renewable oil-palm biomass. Biomass Bioenergy 2013, 50, 1–9. [Google Scholar] [CrossRef]
- Asyraf, M.; Ishak, M.; Syamsir, A.; Nurazzi, N.; Sabaruddin, F.; Shazleen, S.; Norrrahim, M.; Rafidah, M.; Ilyas, R.; Rashid, M.Z.A.; et al. Mechanical properties of oil palm fibre-reinforced polymer composites: A review. J. Mater. Res. Technol. 2022, 17, 33–65. [Google Scholar] [CrossRef]
- Chanasit, W.; Hodgson, B.; Sudesh, K.; Umsakul, K. Efficient production of polyhydroxyalkanoates (PHAs) from Pseudomonas mendocina PSU using a biodiesel liquid waste (BLW) as the sole carbon source. Biosci. Biotechnol. Biochem. 2016, 80, 1440–1450. [Google Scholar] [CrossRef]
- Van Thuoc, D.; My, D.N.; Loan, T.T.; Sudesh, K. Utilization of waste fish oil and glycerol as carbon sources for polyhydroxyalkanoate production by Salinivibrio sp. M318. Int. J. Biol. Macromol. 2019, 141, 885–892. [Google Scholar] [CrossRef]
- Esmael, M.E.; Ibrahim, M.I.; Aldhumri, S.A.; Bayoumi, R.A.; Matsuo, K.; Khattab, A.M. Lipid-membranes interaction, structural assessment, and sustainable production of polyhydroxyalkanoate by Priestia filamentosa AZU-A6 from sugarcane molasses. Int. J. Biol. Macromol. 2023, 242, 124721. [Google Scholar] [CrossRef]
- Jung, H.J.; Kim, S.H.; Shin, N.; Oh, S.-J.; Hwang, J.H.; Kim, H.J.; Kim, Y.-H.; Bhatia, S.K.; Jeon, J.-M.; Yoon, J.-J.; et al. Polyhydroxybutyrate (PHB) production from sugar cane molasses and tap water without sterilization using novel strain, Priestia sp. YH4. Int. J. Biol. Macromol. 2023, 250, 126152. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, S.; Chinnadurai, G.S.; Perumal, P. Polyhydroxybutyrate by Streptomyces sp.: Production and characterization. Int. J. Biol. Macromol. 2017, 104, 1165–1171. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Fox, B.G.; Takasuka, T.E. Consolidated bioprocessing of plant biomass to polyhydroxyalkanoate by co-culture of Streptomyces sp. SirexAA-E and Priestia megaterium. Bioresour. Technol. 2023, 376, 128934. [Google Scholar] [CrossRef]
- Koller, M. Switching from petro-plastics to microbial polyhydroxyalkanoates (PHA): The biotechnological escape route of choice out of the plastic predicament? Eurobiotech J. 2019, 3, 32–44. [Google Scholar] [CrossRef]
- Kenny, S.T.; Runic, J.N.; Kaminsky, W.; Woods, T.; Babu, R.P.; Keely, C.M.; Blau, W.; O’connor, K.E. Up-Cycling of PET (Polyethylene Terephthalate) to the Biodegradable Plastic PHA (Polyhydroxyalkanoate). Environ. Sci. Technol. 2008, 42, 7696–7701. [Google Scholar] [CrossRef]
- Urtuvia, V.; Villegas, P.; González, M.; Seeger, M. Bacterial production of the biodegradable plastics polyhydroxyalkanoates. Int. J. Biol. Macromol. 2014, 70, 208–213. [Google Scholar] [CrossRef]
- Spasic, J.; Mandic, M.; Djokic, L.; Nikodinovic-Runic, J. Streptomyces spp. in the biocatalysis toolbox. Appl. Microbiol. Biotechnol. 2018, 102, 3513–3536. [Google Scholar] [CrossRef] [PubMed]
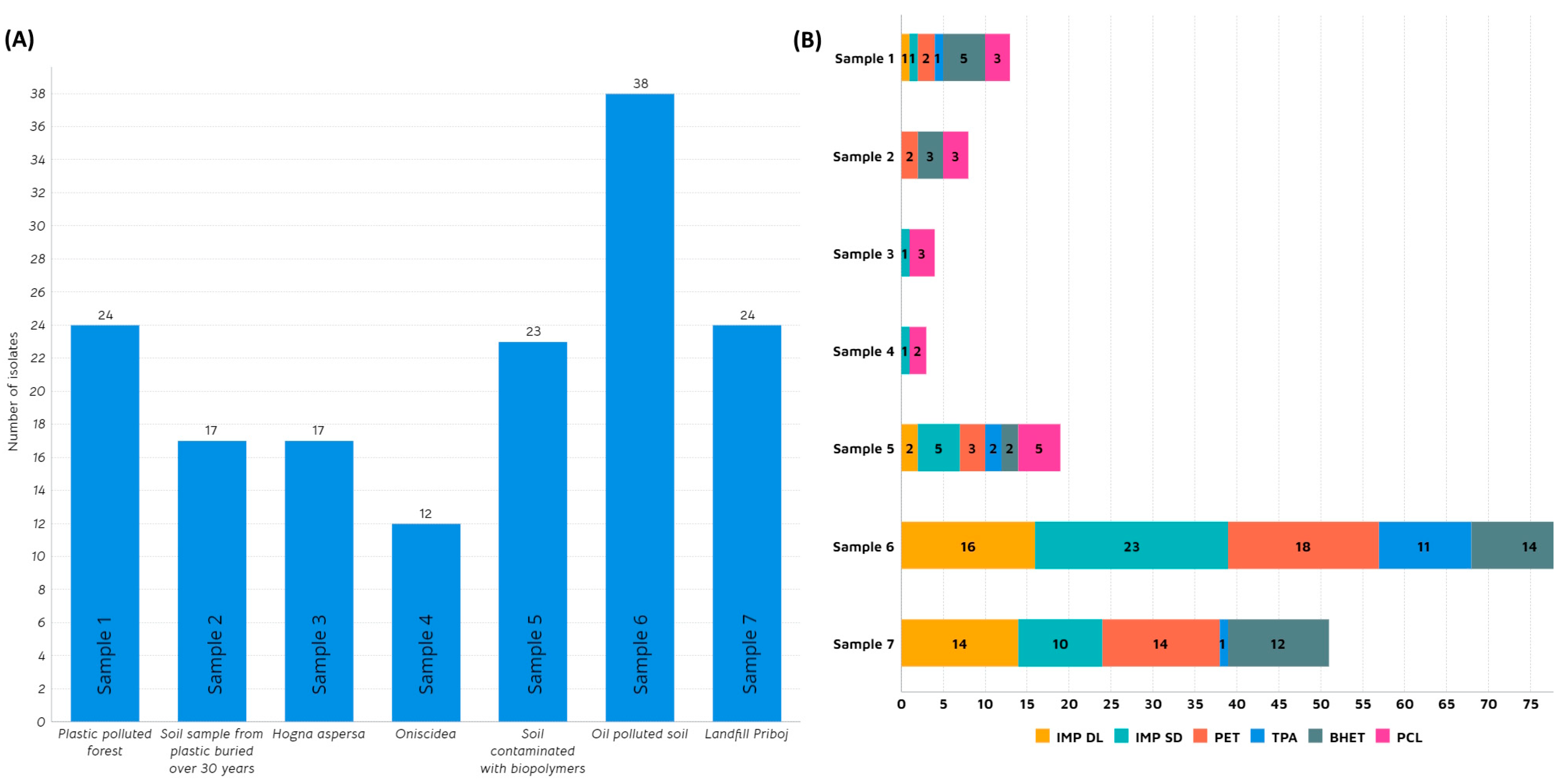
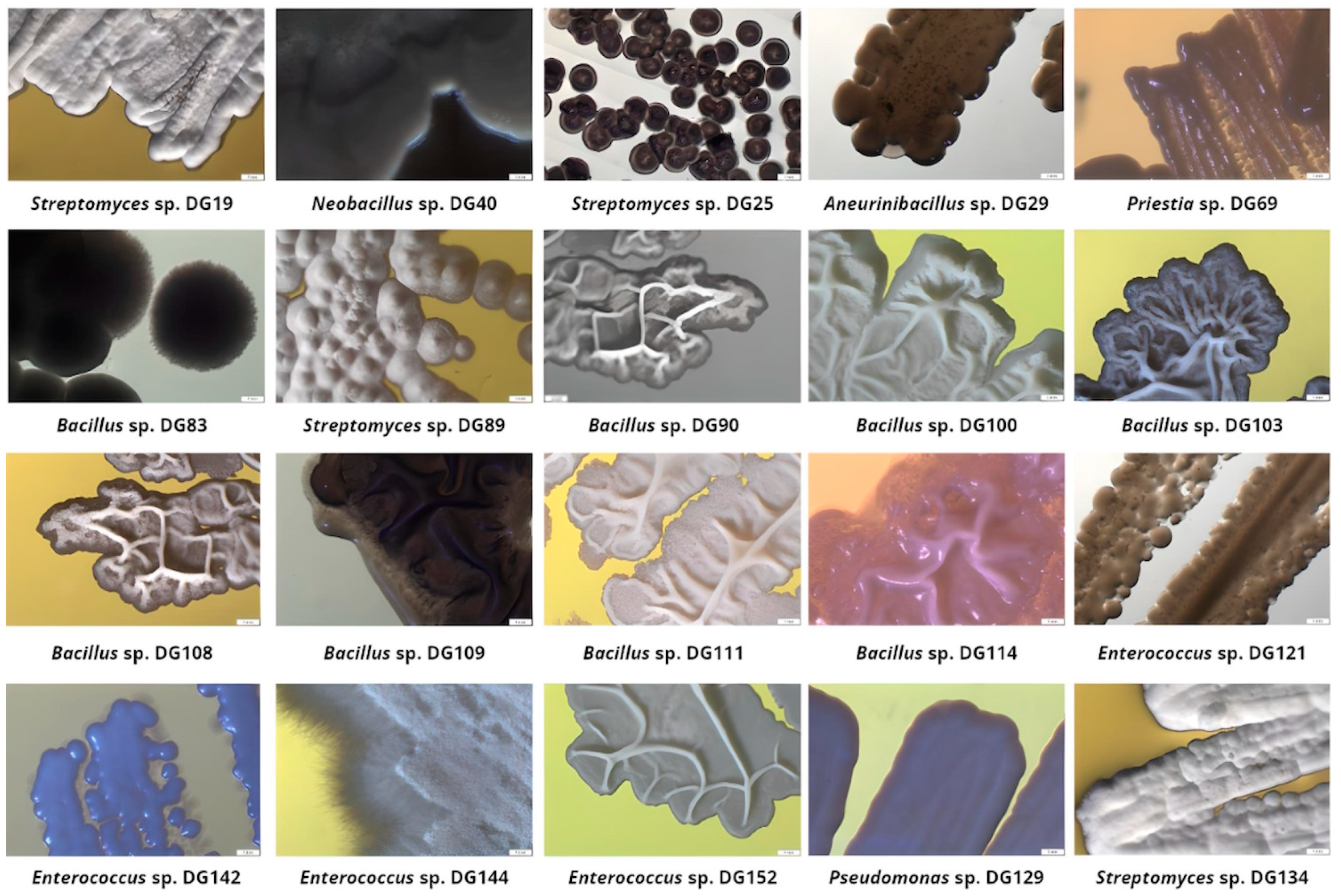
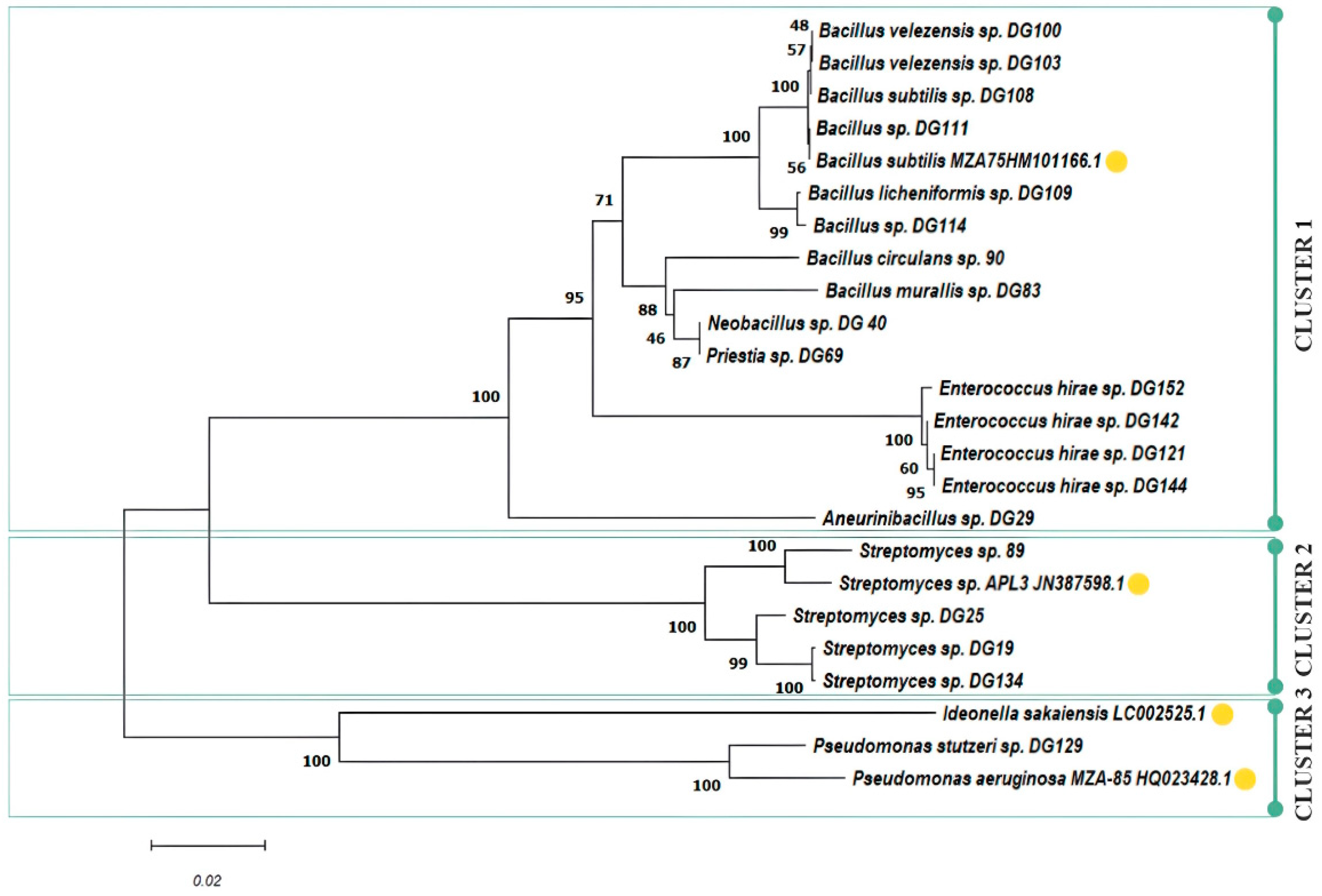

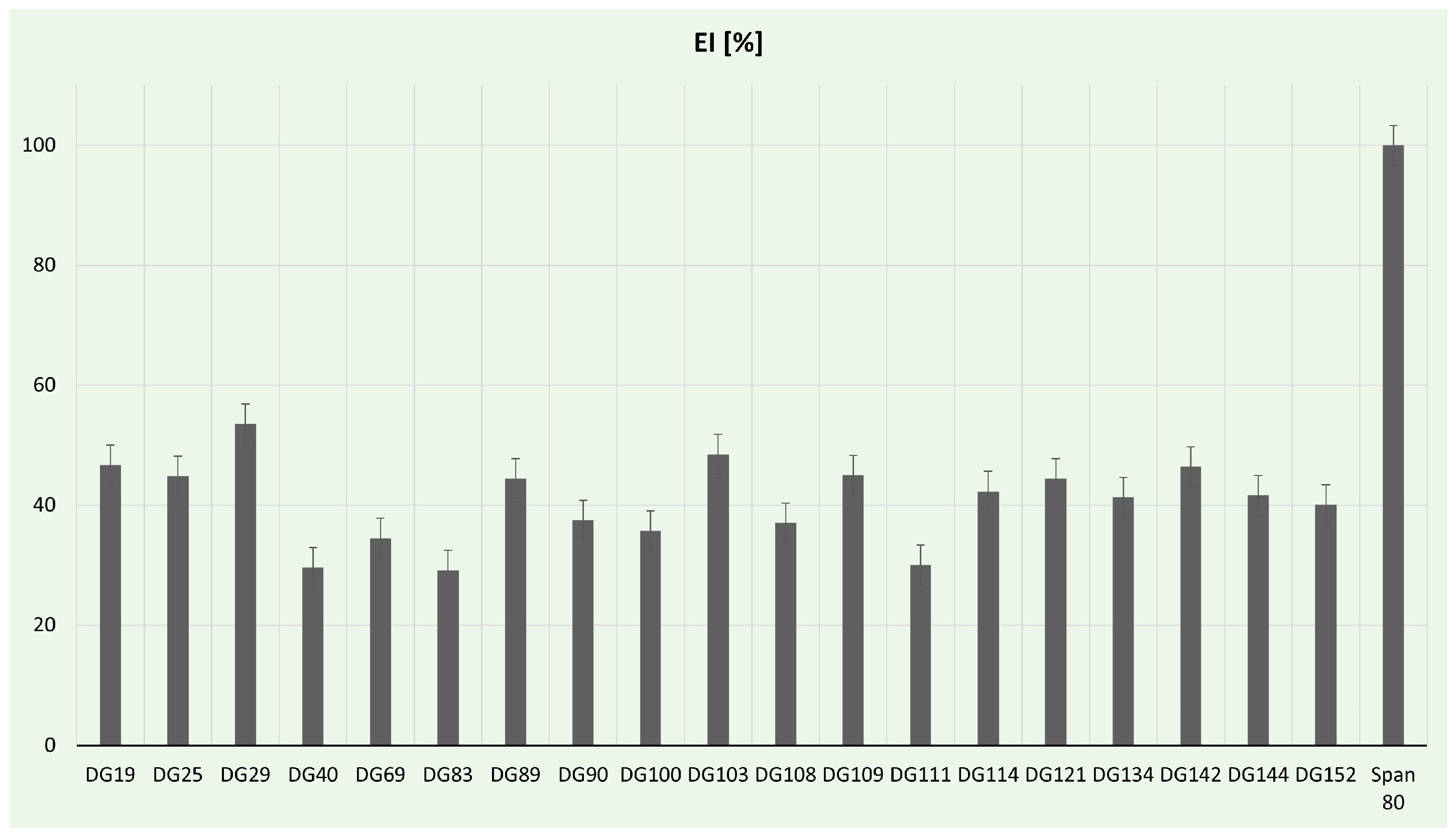
| Sample ID | Collection Date | Description/Criteria | Location | Coordinates | Average Temperature [°C] |
|---|---|---|---|---|---|
| S1 | 20 September 2020 | Forest polluted with plastic waste | Athlone, Ireland | 53.41991° N 7.88762° W | 9–17 |
| S2 | 1 September 2020 | Soil sample with plastic buried 30 years ago | Athlone, Ireland | 53.41991° N 8.62351° W | 9–17 |
| S3 | 20 October 2020 | Hogna aspersa | Athlone, Ireland | 53.4239° N 7.9407° W | 7–14 |
| S4 | 20 October 2020 | Oniscidea | Athlone, Ireland | 53.4239° N 7.9407° W | 7–14 |
| S5 | 28 October 2020 | Soil contaminated with biopolymers | Athlone, Ireland | 53.41991° N 7.88762° W | 7–14 |
| S6 | 10 October 2021 | Landfill | Priboj, Serbia | 43.5668° N 19.5330° E | 5–20 |
| S7 | 20 January 2021 | Oil polluted soil | Poza Rica, Mexico | 20.5271° N 97.4629° W | 17–25 |
| Origin | Isolate | IMP-DL | IMP-SD | PET | TPA | BHET |
|---|---|---|---|---|---|---|
| S1 | 19 | |||||
| S2 | 25 | |||||
| S2 | 29 | |||||
| S1 | 40 | |||||
| S4 | 69 | |||||
| S5 | 83 | |||||
| S5 | 89 | |||||
| S5 | 90 | |||||
| S7 | 100 | |||||
| S7 | 103 | |||||
| S7 | 108 | |||||
| S7 | 109 | |||||
| S7 | 111 | |||||
| S7 | 114 | |||||
| S6 | 121 | |||||
| S6 | 129 | |||||
| S6 | 134 | |||||
| S6 | 142 | |||||
| S6 | 144 | |||||
| S6 | 152 | |||||
 | ||||||
| Strain ID | Growth [ΔOD600nm] | ΔCDW [g/L] | Fluorescence | PHA [%] | Residual Cell Dry Weight [g/L] |
|---|---|---|---|---|---|
| DG19 | (1) | 1.39 ± 0.06 | +/− | 0.32 ± 0.1 | 1.337 |
| DG25 | (1) | 0.08 ± 0.1 | +/− | n.d. | 0.08 |
| DG29 | (1) | 0.62 ± 0.02 | − | n.d. | 0.62 |
| DG40 | 0.19 | 0.44 ± 0.13 | + | 3.34 ± 0.51 | 0.425 |
| DG69 | 0.45 | 1.06 ± 0.04 | +/− | 4.14 ± 0.55 | 1.016 |
| DG83 | 0.72 | 0.23 ± 0.11 | + | n.d. | 0.23 |
| DG89 | (1) | 0.36 ± 0.08 | +/− | 0.11 ± 0.06 | 0.359 |
| DG90 | 0.23 | 0.13 ± 0.06 | +/− | n.d. | 0.13 |
| DG97 | (1) | 0.43 ± 0.21 | − | n.d. | 0.43 |
| DG100 | 0.38 | 0 | + | n.d. | 0 |
| DG103 | 0.98 | 0 | − | n.d. | 0 |
| DG108 | 0.66 | 0.12 ± 0.02 | − | n.d. | 0.12 |
| DG109 | 0.51 | 0.11 ± 0.02 | − | n.d. | 0.11 |
| DG111 | 0.70 | 0 | +/− | n.d. | 0 |
| DG114 | 0.90 | 0 | + | n.d. | 0 |
| DG121 | 0.42 | 0.52 ± 0.1 | − | n.d. | 0.52 |
| DG134 | (1) | 0.9 ± 0.19 | − | n.d. | 0.9 |
| DG142 | 0.94 | 0.27 ± 0.04 | +/− | 1.26 ± 0.13 | 0.266 |
| DG144 | 1.73 | 0.65 ± 0.1 | +/− | 2.80 ± 0.56 | 0.632 |
| DG152 | 1.35 | 0.72 ± 0.06 | − | n.d. | 0.72 |
| Isolate | 16S rRNA Sequence [bp] | BLAST Analysis Results | Plastic Degradation Ability | Emulsification Index [%] | PHA Production [%] |
|---|---|---|---|---|---|
| Source: Plastic polluted forest | |||||
| DG19 | 1375 | Streptomyces sp. | PU, PET, BHET | 46.66 | 0.32 ± 0.1 |
| DG40 | 1204 | Neobacillus sp. | PET, BHET | 29.62 | 3.34 ± 0.51 |
| Source: Soil contaminated with biopolymers | |||||
| DG89 | 1378 | Streptomyces sp. | PU, PET | 44.44 | 0.11 ± 0.06 |
| Source: Soil sample from plastic buried over 30 years | |||||
| DG69 | 1178 | Priestia sp. | PU, PET | 34.48 | 4.14 ± 0.55 |
| Source: Landfill Priboj | |||||
| DG142 | 1296 | Enterococcus sp. | PU, PET, TPA | 46.42 | 1.26 ± 0.13 |
| DG144 | 1367 | Enterococcus sp. | PU, PET, TPA | 41.66 | 2.80 ± 0.56 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Herrera, D.A.G.; Mojicevic, M.; Pantelic, B.; Joshi, A.; Collins, C.; Batista, M.; Torres, C.; Freitas, F.; Murray, P.; Nikodinovic-Runic, J.; et al. Exploring Microorganisms from Plastic-Polluted Sites: Unveiling Plastic Degradation and PHA Production Potential. Microorganisms 2023, 11, 2914. https://doi.org/10.3390/microorganisms11122914
Herrera DAG, Mojicevic M, Pantelic B, Joshi A, Collins C, Batista M, Torres C, Freitas F, Murray P, Nikodinovic-Runic J, et al. Exploring Microorganisms from Plastic-Polluted Sites: Unveiling Plastic Degradation and PHA Production Potential. Microorganisms. 2023; 11(12):2914. https://doi.org/10.3390/microorganisms11122914
Chicago/Turabian StyleHerrera, Diana A. Garza, Marija Mojicevic, Brana Pantelic, Akanksha Joshi, Catherine Collins, Maria Batista, Cristiana Torres, Filomena Freitas, Patrick Murray, Jasmina Nikodinovic-Runic, and et al. 2023. "Exploring Microorganisms from Plastic-Polluted Sites: Unveiling Plastic Degradation and PHA Production Potential" Microorganisms 11, no. 12: 2914. https://doi.org/10.3390/microorganisms11122914
APA StyleHerrera, D. A. G., Mojicevic, M., Pantelic, B., Joshi, A., Collins, C., Batista, M., Torres, C., Freitas, F., Murray, P., Nikodinovic-Runic, J., & Brennan Fournet, M. (2023). Exploring Microorganisms from Plastic-Polluted Sites: Unveiling Plastic Degradation and PHA Production Potential. Microorganisms, 11(12), 2914. https://doi.org/10.3390/microorganisms11122914










