Impact of Ecological Restoration on the Physicochemical Properties and Bacterial Communities in Alpine Mining Area Soils
Abstract
:1. Introduction
2. Materials and Methods
2.1. Description of the Research Center
2.2. Sample Collection
2.3. Determination of Soil Physical and Chemical Properties
2.4. DNA Extraction and PCR Amplification
2.5. DNA Library Construction and Sequencing
2.6. Data Processing and Statistical Analysis
3. Results
3.1. Variations in Soil Physicochemical Properties across Reclamation Years
3.2. Changes in Heavy Metal Content of Soils with Different Years of Reclamation
3.3. Soil Bacterial Community Composition in Different Reclamation Years
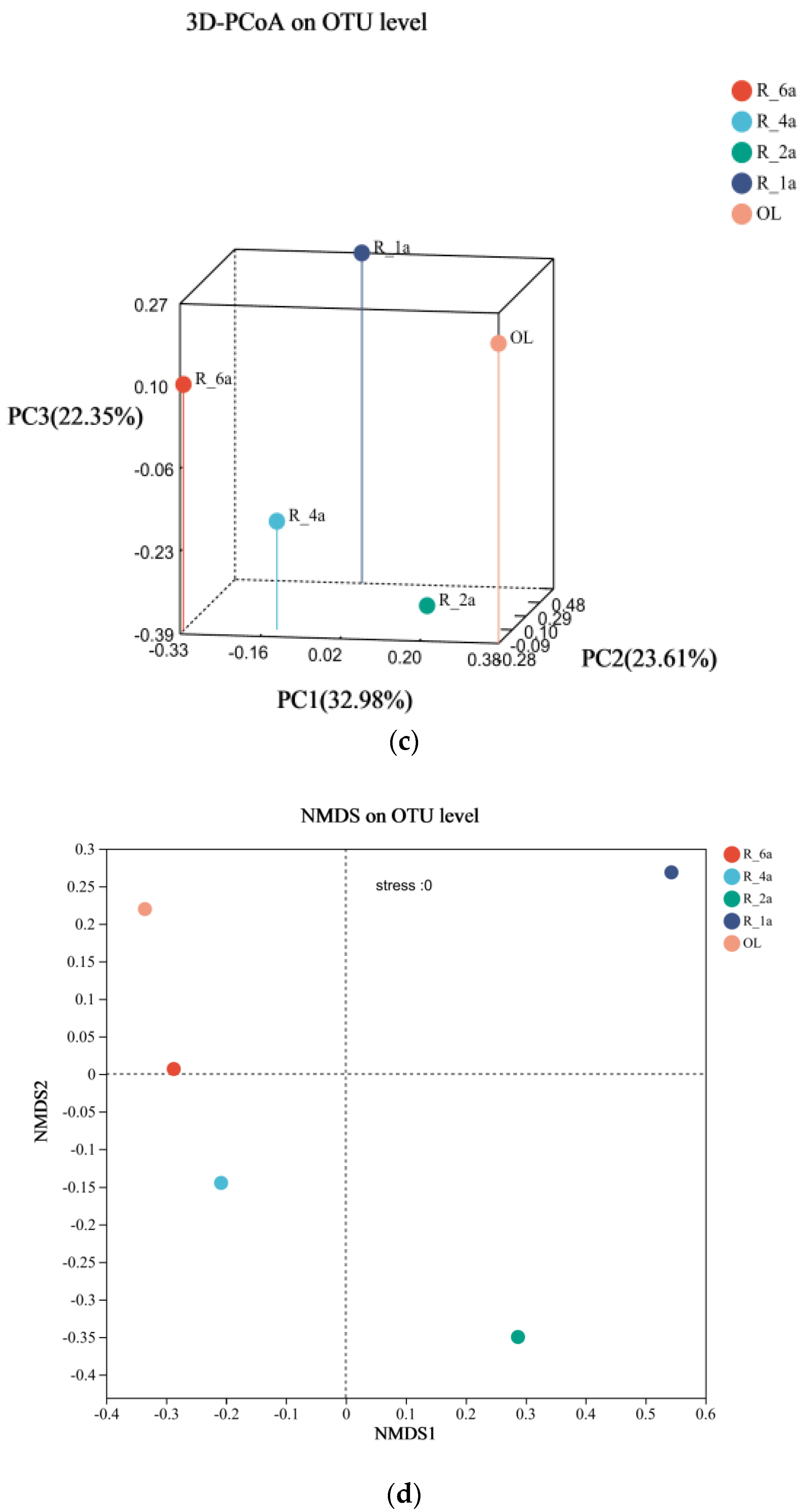
3.4. Diversity and PCA Analysis of Soil Bacterial Communities in Different Reclamation Years
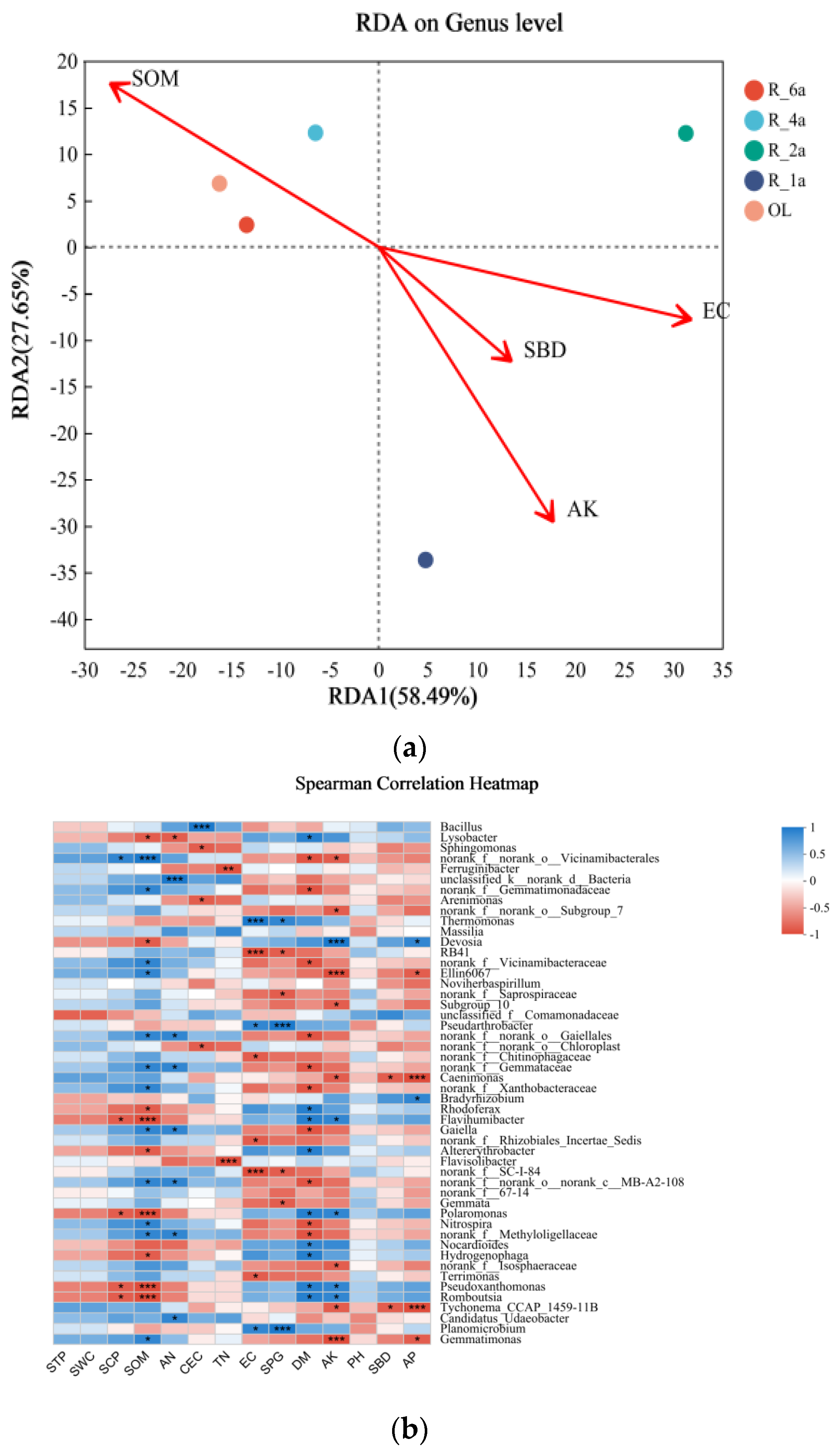
3.5. Correlation between Soil Physicochemical Properties and Microorganisms in Different Reclamation Years
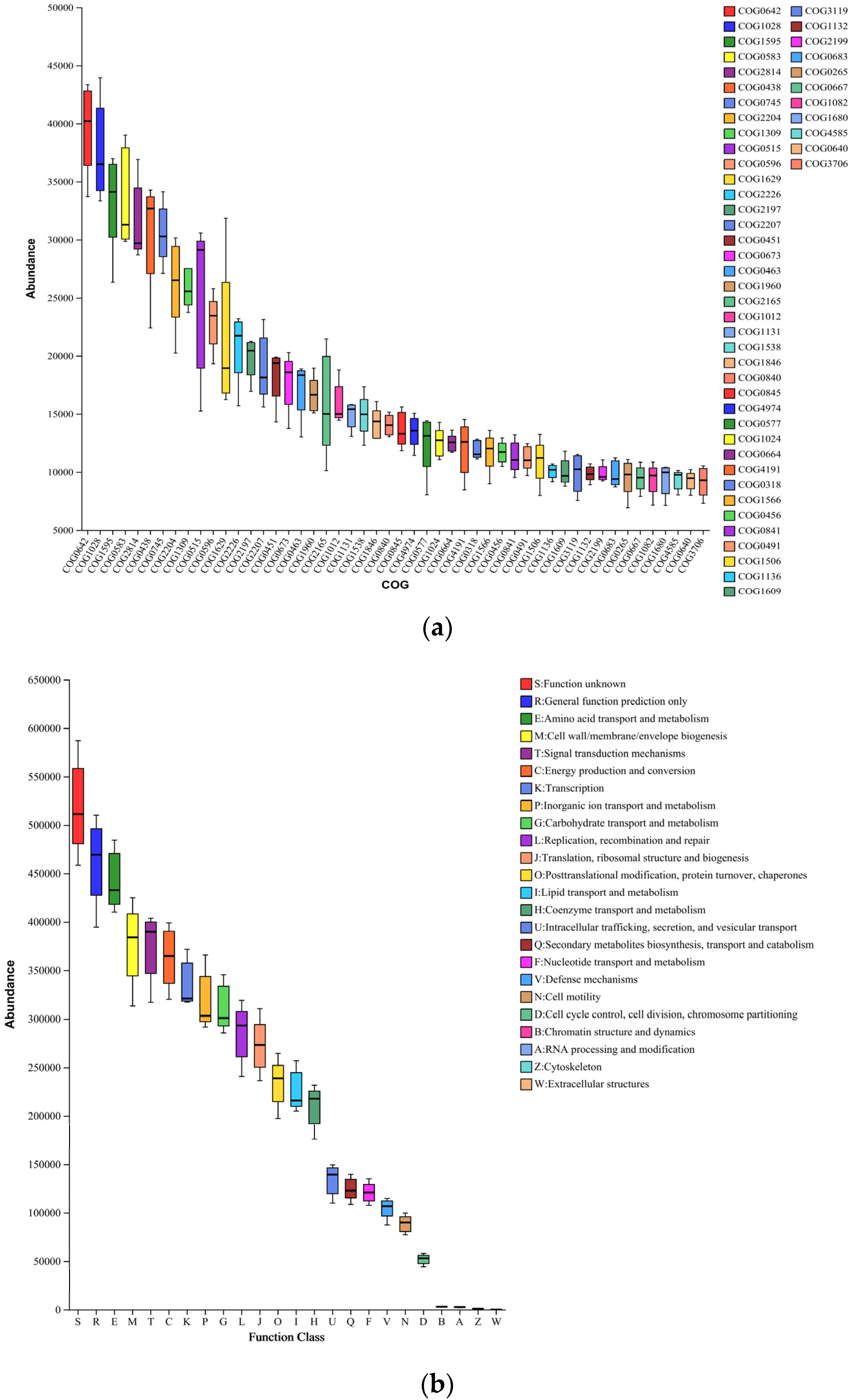
3.6. Microbial Evolution and Potential Functional Shifts in Soils of Different Reclamation Years
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Richter, D.D.J. Humanity’s transformation of Earth’s soil: Pedology’s new frontier. Soil Sci. 2007, 172, 957–967. [Google Scholar] [CrossRef]
- Ly, T.; Wright, J.R.; Weit, N.; McLimans, C.J.; Ulrich, N.; Tokarev, V.; Valkanas, M.M.; Trun, N.; Rummel, S.; Grant, C.J.; et al. Microbial Communities Associated With Passive Acidic Abandoned Coal Mine Remediation. Front. Microbiol. 2019, 10, 1955. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Chandra, A.; Usmani, Z. Impact of coal mining on soil properties and their efficient eco-restoration. Int. J. Energy Technol. Policy 2017, 13, 158. [Google Scholar] [CrossRef]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N. Embracing the unknown: Disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 2017, 15, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Lau, J.A.; Lennon, J.T. Evolutionary ecology of plant–microbe interactions: Soil microbial structure alters selection on plant traits. New Phytol. 2011, 192, 215–224. [Google Scholar] [CrossRef]
- Tao, C.; Li, R.; Xiong, W.; Shen, Z.; Liu, S.; Wang, B.; Ruan, Y.; Geisen, S.; Shen, Q.; Kowalchuk, G.A. Bio-organic fertilizers stimulate indigenous soil Pseudomonas populations to enhance plant disease suppression. Microbiome 2020, 8, 137. [Google Scholar] [CrossRef]
- Yuan, J.; Wen, T.; Zhang, H.; Zhao, M.; Penton, C.R.; Thomashow, L.S.; Shen, Q. Predicting disease occurrence with high accuracy based on soil macroecological patterns of Fusarium wilt. ISME J. 2020, 14, 2936–2950. [Google Scholar] [CrossRef]
- Bakker, P.A.; Berendsen, R.L.; Van Pelt, J.A.; Vismans, G.; Yu, K.; Li, E.; Van Bentum, S.; Poppeliers, S.W.; Gil, J.J.S.; Zhang, H.; et al. The Soil-Borne Identity and Microbiome-Assisted Agriculture: Looking Back to the Future. Mol. Plant 2020, 13, 1394–1401. [Google Scholar] [CrossRef]
- French, E.; Kaplan, I.; Iyer-Pascuzzi, A.; Nakatsu, C.H.; Enders, L. Emerging strategies for precision microbiome management in diverse agroecosystems. Nat. Plants 2021, 7, 256–267. [Google Scholar] [CrossRef]
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. The rhizosphere microbiome: Significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 2013, 37, 634–663. [Google Scholar] [CrossRef] [PubMed]
- Ezeokoli, O.T.; Mashigo, S.K.; Paterson, D.G.; Bezuidenhout, C.C.; Adeleke, R.A. Microbial community structure and relationship with physicochemical properties of soil stockpiles in selected South African opencast coal mines. Soil Sci. Plant Nutr. 2019, 65, 332–341. [Google Scholar] [CrossRef]
- Vuko, M.; Cania, B.; Vogel, C.; Kublik, S.; Schloter, M.; Schulz, S. Shifts in reclamation management strategies shape the role of exopolysaccharide and lipopolysaccharide-producing bacteria during soil formation. Microb. Biotechnol. 2020, 13, 584–598. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, L.; Wen, H.; Zhou, T.; Zhang, T.; Gao, X. 454 Pyrosequencing Analysis of Bacterial Diversity Revealed by a Comparative Study of Soils from Mining Subsidence and Reclamation Areas. J. Microbiol. Biotechnol. 2014, 24, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Čížková, B.; Woś, B.; Pietrzykowski, M.; Frouz, J. Development of soil chemical and microbial properties in reclaimed and unreclaimed grasslands in heaps after opencast lignite mining. Ecol. Eng. 2018, 123, 103–111. [Google Scholar] [CrossRef]
- Rampelotto, P.H.; Ferreira, A.d.S.; Barboza, A.D.M.; Roesch, L.F.W. Changes in Diversity, Abundance, and Structure of Soil Bacterial Communities in Brazilian Savanna Under Different Land Use Systems. Microb. Ecol. 2013, 66, 593–607. [Google Scholar] [CrossRef] [PubMed]
- Yin, C.; Jones, K.L.; Peterson, D.E.; Garrett, K.A.; Hulbert, S.H.; Paulitz, T.C. Members of soil bacterial communities sensitive to tillage and crop rotation. Soil Biol. Biochem. 2010, 42, 2111–2118. [Google Scholar] [CrossRef]
- Ai, C.; Zhang, S.; Zhang, X.; Guo, D.; Zhou, W.; Huang, S. Distinct responses of soil bacterial and fungal communities to changes in fertilization regime and crop rotation. Geoderma 2018, 319, 156–166. [Google Scholar] [CrossRef]
- Van der Heijen, M.G.A. The unseen majority: Soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems (vol 11, pg 296, 2008). Ecol. Lett. 2008, 11, 651. [Google Scholar] [CrossRef]
- Fan, X.; Wang, Y.; Niu, F.; Li, W.; Wu, X.; Ding, Z.; Pang, W.; Lin, Z. Environmental Characteristics of High Ice-Content Permafrost on the Qinghai–Tibetan Plateau. Remote Sens. 2023, 15, 4496. [Google Scholar] [CrossRef]
- Karami, A.; Homaee, M.; Afzalinia, S.; Ruhipour, H.; Basirat, S. Organic resource management: Impacts on soil aggregate stability and other soil physico-chemical properties. Agric. Ecosyst. Environ. 2012, 148, 22–28. [Google Scholar] [CrossRef]
- Weisburg, W.; Barns, S.; Pelletier, D.; Lane, D. 16S ribosomal DNA amplification for phylogenetic study. J. Bacter. 1991, 173, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Souza, T.P.; Marques, S.C.; Santos, D.M.d.S.e.; Dias, E.S. Analysis of thermophilic fungal populations during phase II of composting for the cultivation of Agaricus subrufescens. World J. Microbiol. Biotechnol. 2014, 30, 2419–2425. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Lozupone, C.; Knight, R. UniFrac: A New Phylogenetic Method for Comparing Microbial Communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Kumar, S.; Maiti, S.K.; Chaudhuri, S. Soil development in 2–21 years old coalmine reclaimed spoil with trees: A case study from Sonepur-Bazari opencast project, Raniganj Coalfield, India. Ecol. Eng. 2015, 84, 311–324. [Google Scholar] [CrossRef]
- Feng, Y.; Guo, Z.; Zhong, L.; Zhao, F.; Zhang, J.; Lin, X. Balanced Fertilization Decreases Environmental Filtering on Soil Bacterial Community Assemblage in North China. Front. Microbiol. 2017, 8, 2376. [Google Scholar] [CrossRef]
- Yan, Y.; Klinkhamer, P.G.; van Veen, J.A.; Kuramae, E.E. Environmental filtering: A case of bacterial community assembly in soil. Soil Biol. Biochem. 2019, 136, 107531. [Google Scholar] [CrossRef]
- Huang, X.; Zhou, X.; Zhang, J.; Cai, Z. Highly connected taxa located in the microbial network are prevalent in the rhizosphere soil of healthy plant. Biol. Fertil. Soils 2019, 55, 299–312. [Google Scholar] [CrossRef]
- Pascale, A.; Proietti, S.; Pantelides, I.S.; Stringlis, I.A. Modulation of the Root Microbiome by Plant Molecules: The Basis for Targeted Disease Suppression and Plant Growth Promotion. Front. Plant Sci. 2020, 10, 1741. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Jia, Z.; Sun, Q.; Zhan, J.; Yang, Y.; Wang, D. Ecological restoration alters microbial communities in mine tailings profiles. Sci. Rep. 2016, 6, 25193. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Xiao, E.; Häggblom, M.; Krumins, V.; Dong, Y.; Sun, X.; Li, F.; Wang, Q.; Li, B.; Yan, B. Bacterial Survival Strategies in an Alkaline Tailing Site and the Physiological Mechanisms of Dominant Phylotypes As Revealed by Metagenomic Analyses. Environ. Sci. Technol. 2018, 52, 13370–13380. [Google Scholar] [CrossRef] [PubMed]
- Caravaca, F.; Alguacil, M.; Figueroa, D.; Barea, J.; Roldán, A. Re-establishment of Retama sphaerocarpa as a target species for reclamation of soil physical and biological properties in a semi-arid Mediterranean area. For. Ecol. Manag. 2003, 182, 49–58. [Google Scholar] [CrossRef]
- Li, Y.; Li, Z.; Jiang, Y.; Weng, B.; Lin, W. Variations of rhizosphere bacterial communities in tea (Camellia sinensis L.) continuous cropping soil by high-throughput pyrosequencing approach. J. Appl. Microbiol. 2016, 121, 787–799. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, K.; Wemheuer, B.; Korolkow, V.; Wemheuer, F.; Nacke, H.; Schöning, I.; Schrumpf, M.; Daniel, R. Driving forces of soil bacterial community structure, diversity, and function in temperate grasslands and forests. Sci. Rep. 2016, 6, 33696. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Noya, Y.E.; Gómez-Acata, S.; Montoya-Ciriaco, N.; Rojas-Valdez, A.; Suárez-Arriaga, M.C.; Valenzuela-Encinas, C.; Jiménez-Bueno, N.; Verhulst, N.; Govaerts, B.; Dendooven, L. Relative impacts of tillage, residue management and crop-rotation on soil bacterial communities in a semi-arid agroecosystem. Soil Biol. Biochem. 2013, 65, 86–95. [Google Scholar] [CrossRef]
- Miralles, I.; Ortega, R.; Almendros, G.; Sánchez-Marañón, M.; Soriano, M. Soil quality and organic carbon ratios in mountain agroecosystems of South-east Spain. Geoderma 2009, 150, 120–128. [Google Scholar] [CrossRef]
- Brevik, E.C.; Hartemink, A.E. Early soil knowledge and the birth and development of soil science. CATENA 2010, 83, 23–33. [Google Scholar] [CrossRef]
- Bossolani, J.W.; Crusciol, C.A.; Leite, M.F.; Merloti, L.F.; Moretti, L.G.; Pascoaloto, I.M.; Kuramae, E.E. Modulation of the soil microbiome by long-term Ca-based soil amendments boosts soil organic carbon and physicochemical quality in a tropical no-till crop rotation system. Soil Biol. Biochem. 2021, 156, 108188. [Google Scholar] [CrossRef]
- Wang, R.; Zhang, H.; Sun, L.; Qi, G.; Chen, S.; Zhao, X. Microbial community composition is related to soil biological and chemical properties and bacterial wilt outbreak. Sci. Rep. 2017, 7, 343. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.; Ren, H.; Li, S.; Leng, X.; Yao, X. Soil Bacterial Community Structure and Co-occurrence Pattern during Vegetation Restoration in Karst Rocky Desertification Area. Front. Microbiol. 2017, 8, 2377. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Wang, X.; Carrión, V.J.; Yin, S.; Yue, Z.; Liao, Y.; Dong, Y.; Li, X. Acidic amelioration of soil amendments improves soil health by impacting rhizosphere microbial assemblies. Soil Biol. Biochem. 2022, 167, 108599. [Google Scholar] [CrossRef]
- Banning, N.C.; Gleeson, D.B.; Grigg, A.H.; Grant, C.D.; Andersen, G.L.; Brodie, E.L.; Murphy, D.V. Soil Microbial Community Successional Patterns during Forest Ecosystem Restoration. Appl. Environ. Microbiol. 2011, 77, 6158–6164. [Google Scholar] [CrossRef] [PubMed]
- Rocha, U.N.; Van Overbeek, L.; Van Elsas, J.D. Exploration of hitherto-uncultured bacteria from the rhizosphere. FEMS Microbiol. Ecol. 2009, 69, 313–328. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, C.C.; Kishi, L.T.; Lopes, E.M.; Omori, W.P.; de Souza, J.A.M.; Alves, L.M.C.; Lemos, E.G.d.M. Bacterial communities in mining soils and surrounding areas under regeneration process in a former ore mine. Braz. J. Microbiol. 2018, 49, 489–502. [Google Scholar] [CrossRef] [PubMed]
- Gremion, F.; Chatzinotas, A.; Harms, H. Comparative 16S rDNA and 16S rRNA sequence analysis indicates that Actinobacteria might be a dominant part of the metabolically active bacteria in heavy metal-contaminated bulk and rhizosphere soil. Environ. Microbiol. 2003, 5, 896–907. [Google Scholar] [CrossRef] [PubMed]
- Ceja-Navarro, J.A.; Rivera-Orduña, F.N.; Patiño-Zúñiga, L.; Vila-Sanjurjo, A.; Crossa, J.; Govaerts, B.; Dendooven, L. Phylogenetic and Multivariate Analyses To Determine the Effects of Different Tillage and Residue Management Practices on Soil Bacterial Communities. Appl. Environ. Microbiol. 2010, 76, 3685–3691. [Google Scholar] [CrossRef]
- Lamb, E.G.; Kennedy, N.; Siciliano, S.D. Effects of plant species richness and evenness on soil microbial community diversity and function. Plant Soil 2011, 338, 483–495. [Google Scholar] [CrossRef]
- Vikram, S.; Guerrero, L.D.; Makhalanyane, T.P.; Le, P.T.; Seely, M.; Cowan, D.A. Metagenomic analysis provides insights into functional capacity in a hyperarid desert soil niche community. Environ. Microbiol. 2016, 18, 1875–1888. [Google Scholar] [CrossRef]
- Zhang, F.H.; Yang, H.C.; Gale, W.J.; Cheng, Z.B.; Yan, J.H. Temporal changes in soil organic carbon and aggregate-associated organic carbon after reclamation of abandoned, salinized farmland. J. Agric. Sci. 2017, 155, 205–215. [Google Scholar] [CrossRef]
- Miralles, I.; Trasar-Cepeda, C.; Leirós, M.C.; Gil-Sotres, F. Labile carbon in biological soil crusts in the Tabernas desert, SE Spain. Soil Biol. Biochem. 2013, 58, 1–8. [Google Scholar] [CrossRef]
- Delgado-Baquerizo, M.; Trivedi, P.; Trivedi, C.; Eldridge, D.J.; Reich, P.B.; Jeffries, T.C.; Singh, B.K. Microbial richness and composition independently drive soil multifunctionality. Funct. Ecol. 2017, 31, 2330–2343. [Google Scholar] [CrossRef]
- Yamada, T.; Sekiguchi, Y.; Imachi, H.; Kamagata, Y.; Ohashi, A.; Harada, H. Diversity, localization, and physiological properties of fila-mentous microbes belonging to Chloroflexi subphylum I in mesophilic and thermophilic methanogenic sludge granules. Appl. Environ. Microbiol. 2005, 71, 7493–7503. [Google Scholar] [CrossRef] [PubMed]
- Kuramae, E.E.; Yergeau, E.; Wong, L.C.; Pijl, A.S.; Veen, J.A.; Kowalchuk, G.A. Soil characteristics more strongly influence soil bacterial communities than land-use type. FEMS Microbiol. Ecol. 2012, 79, 12–24. [Google Scholar] [CrossRef]
- Wang, C.; Liu, S.; Zhang, Y.; Liu, B.; He, F.; Xu, D.; Zhou, Q.; Wu, Z. Bacterial Communities and Their Predicted Functions Explain the Sediment Nitrogen Changes Along with Submerged Macrophyte Restoration. Microb. Ecol. 2018, 76, 625–636. [Google Scholar] [CrossRef]
- Goldfarb, K.C.; Karaoz, U.; Hanson, C.A.; Santee, C.A.; Bradford, M.A.; Treseder, K.K.; Wallenstein, M.D.; Brodie, E.L. Differential Growth Responses of Soil Bacterial Taxa to Carbon Substrates of Varying Chemical Recalcitrance. Front. Microbiol. 2011, 2, 94. [Google Scholar] [CrossRef]

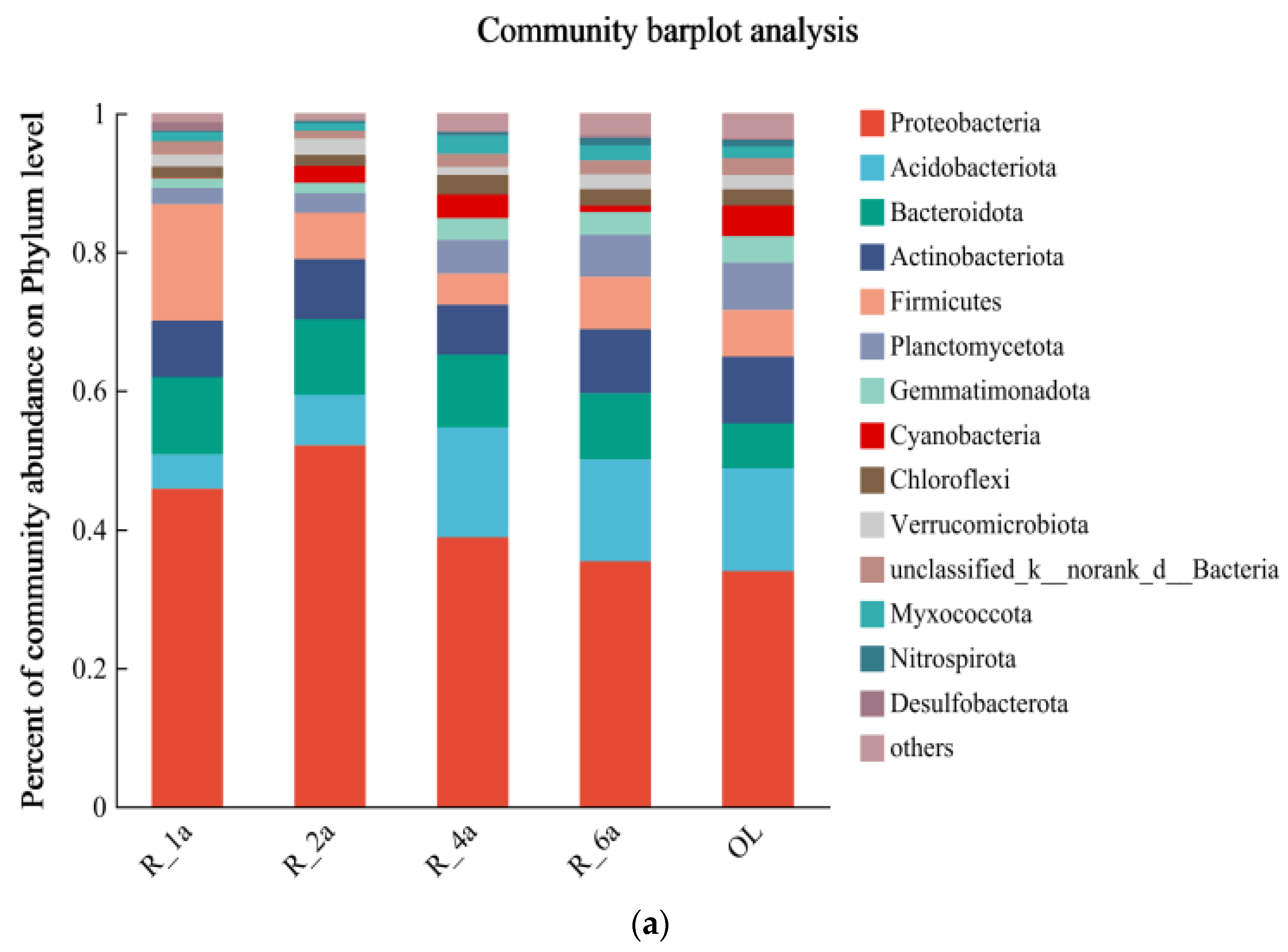
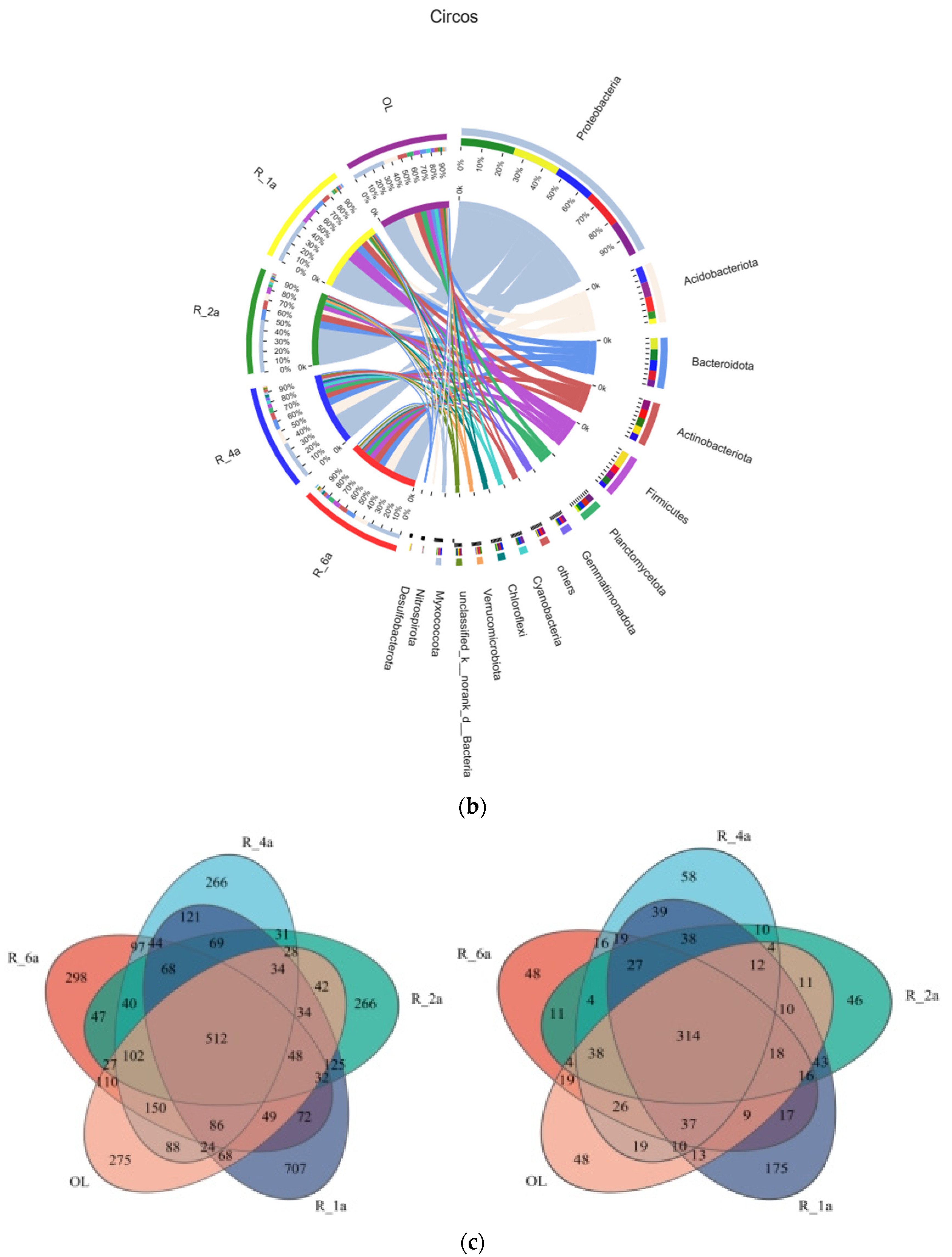
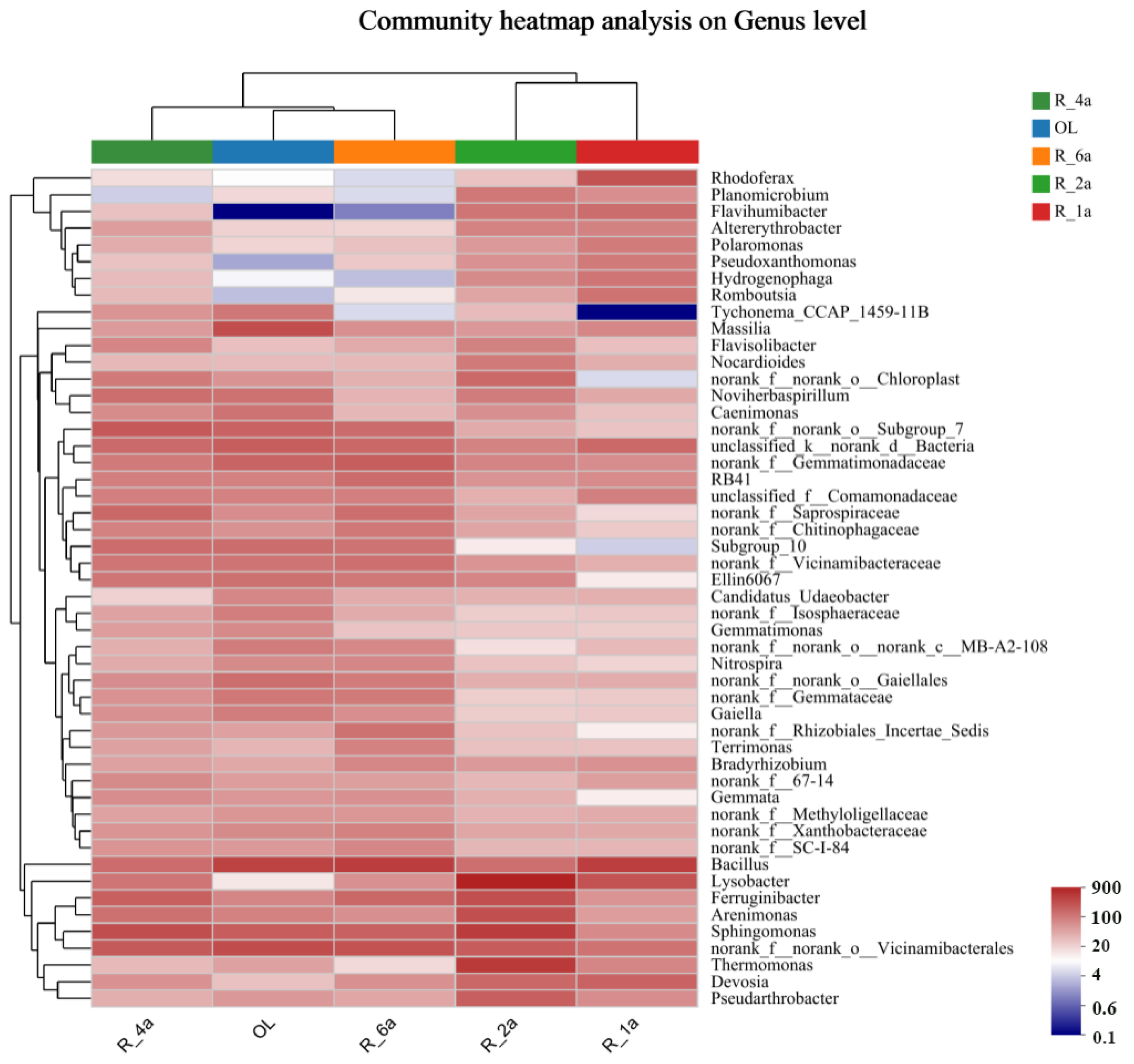
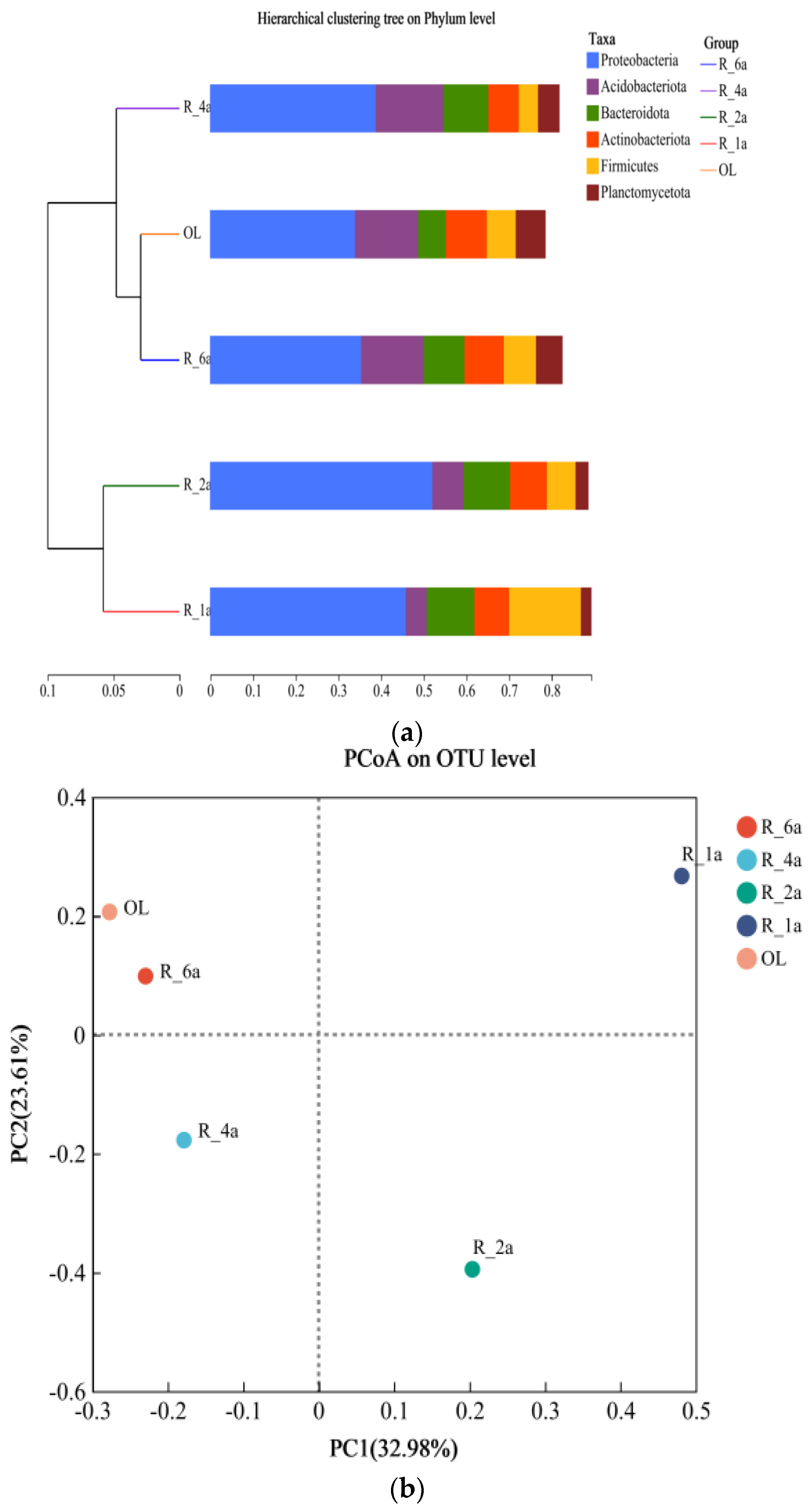


| Plot Number | Plot Name | Reclamation Period | Types of Reclaimed Vegetation | Aspect of Slope |
|---|---|---|---|---|
| R_1a(S1) | 4242BP | 1 | Elymus nutans, Poa crymophila cv Qinghai, Elymus sibiricus L., Festuca sinensis, Poa pratensis L., Floret alkali grass | West |
| R_2a(S2) | 4386BP | 2 | Elymus nutans, Poa crymophila cv Qinghai, Elymus sibiricus L., Festuca sinensis, Poa pratensis L., Floret alkali grass, ryegrass | west |
| R_4a(S3) | 4230BP | 4 | Elymus nutans, Poa crymophila cv Qinghai, Elymus sibiricus L., Festuca sinensis, rape flower | west |
| R_6a(S4) | 4420BP | 6 | Elymus nutans, Poa crymophila cv Qinghai, Elymus sibiricus L., Festuca sinensis | west |
| OL(S5) | 4242YT | — | Elymus nutans, Poa crymophila cv Qinghai, Elymus sibiricus L., Festuca sinensis, Kobresia pygmaea, Kobresia humilis | west |
| Treatment | Unit | R_1a | R_2a | R_4a | R_6a | OL |
|---|---|---|---|---|---|---|
| pH | / | 7.91 ± 0.01 ab | 7.55 ± 0.55 b | 8.00 ± 0.03 a | 7.91 ± 0.06 ab | 7.04 ± 0.11 c |
| EC | μs·cm−1 | 205.97 ± 19.07 b | 272.33 ± 29.33 a | 92.97 ± 2.47 d | 77.00 ± 60.00 d | 124.50 ± 2.50 c |
| CEC | cmol·kg−1 | 18.43 ± 2.84 ab | 12.30 ± 1.18 b | 13.17 ± 0.02 b | 27.98 ± 6.96 a | 16.75 ± 0.04 ab |
| SBD | g·cm−3 | 1.66 ± 0.07 a | 1.56 ± 0.06 a | 1.60 ± 0.01 a | 1.61 ± 0.02 a | 1.16 ± 0.11 b |
| SPG | g·cm−3 | 2.65 ± 0.02 a | 2.67 ± 0.01 a | 2.59 ± 0.04 a | 2.62 ± 0.02 a | 2.63 ± 0.01 a |
| STP | % | 37.39 ± 0.02 b | 41.67 ± 0.02 b | 38.35 ± 0.10 b | 38.45 ± 0.80 b | 55.72 ± 0.04 a |
| SCP | % | 23.63 ± 0.09 b | 30.50 ± 0.01 ab | 26.50 ± 0.02 ab | 33.50 ± 0.04 ab | 42.13 ± 0.03 a |
| SWC | % | 19.46 ± 1.80 b | 21.42 ± 1.20 b | 19.75 ± 0.80 b | 19.88 ± 0.60 b | 38.17 ± 1.10 a |
| TN | mg·kg−1 | 1220.00 ± 58.8 a | 1020.00 ± 49.98 b | 1150.00 ± 56.35 ab | 1200 ± 58.80 a | 1220.00 ± 60.00 a |
| DM | % | 99.00 ± 2.60 a | 99.00 ± 1.80 a | 98.70 ± 2.50 a | 98.20 ± 3.20 a | 98.20 ± 3.60 a |
| SOM | g·kg−1 | 17.70 ± 0.50 c | 21.70 ± 0.50 b | 31.40 ± 0.30 ab | 41.40 ± 0.20 a | 43.20 ± 0.60 a |
| HN | mg·kg−1 | 81.10 ± 4.87 b | 78.40 ± 4.70 b | 81.00 ± 4.86 b | 174.00 ± 10.44 ab | 243.00 ± 14.58 a |
| AP | mg·kg−1 | 19.60 ± 0.80 a | 11.20 ± 0.90 ab | 3.20 ± 0.70 b | 12.40 ± 0.50 a | 1.90 ± 0.70 b |
| AK | mg·kg−1 | 150.00 ± 4.00 b | 93.00 ± 4.00 ab | 58.00 ± 2.00 a | 60.00 ± 50.00 a | 50.00 ± 50.00 a |
| Treatment | unit | R_1a | R_2a | R_4a | R_6a | OL |
|---|---|---|---|---|---|---|
| Cu | mg·kg−1 | 28.00 ± 0.84 b | 87.00 ± 2.61 ab | 57.00 ± 1.71 a | 50.00 ± 1.50 a | 39.00 ± 1.17 c |
| Cr | mg·kg−1 | 78.00 ± 0.70 b | 167.00 ± 1.50 a | 292.00 ± 2.63 ab | 174.00 ± 1.57 a | 166.00 ± 1.49 a |
| Ni | mg·kg−1 | 52.00 ± 0.62 c | 131.00 ± 1.57 ab | 429.00 ± 5.2 b | 259.00 ± 3.11 a | 122.00 ± 1.46 c |
| Zn | mg·kg−1 | 76.00 ± 1.30 a | 99.00 ± 1.10 a | 87.00 ± 1.20 b | 84.00 ± 1.30 a | 93.00 ± 1.10 ab |
| Pb | mg·kg−1 | 14.00 ± 0.41 a | 24.00 ± 0.70 a | 14.00 ± 0.41 a | 18.00 ± 0.52 a | 16.00 ± 0.46 a |
| Cd | mg·kg−1 | 0.05 ± 0.02 a | 0.05 ± 0.02 a | 0.07 ± 0.01 a | 0.03 ± 0.01 b | 0.04 ± 0.01 b |
| As | mg·kg−1 | 17.60 ± 0.51 a | 32.00 ± 0.19 ab | 12.40 ± 0.07 a | 23.60 ± 0.14 a | 14.20 ± 0.41 a |
| Hg | mg·kg−1 | 0.10 ± 0.003 a | 0.43 ± 0.001 b | 0.113 ± 0.002 ab | 0.054 ± 0.001 b | 0.05 ± 0.002 b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kong, L.; Zhang, L.; Wang, Y.; Huang, Z. Impact of Ecological Restoration on the Physicochemical Properties and Bacterial Communities in Alpine Mining Area Soils. Microorganisms 2024, 12, 41. https://doi.org/10.3390/microorganisms12010041
Kong L, Zhang L, Wang Y, Huang Z. Impact of Ecological Restoration on the Physicochemical Properties and Bacterial Communities in Alpine Mining Area Soils. Microorganisms. 2024; 12(1):41. https://doi.org/10.3390/microorganisms12010041
Chicago/Turabian StyleKong, Lingjian, Lin Zhang, Yingnan Wang, and Zhanbin Huang. 2024. "Impact of Ecological Restoration on the Physicochemical Properties and Bacterial Communities in Alpine Mining Area Soils" Microorganisms 12, no. 1: 41. https://doi.org/10.3390/microorganisms12010041
APA StyleKong, L., Zhang, L., Wang, Y., & Huang, Z. (2024). Impact of Ecological Restoration on the Physicochemical Properties and Bacterial Communities in Alpine Mining Area Soils. Microorganisms, 12(1), 41. https://doi.org/10.3390/microorganisms12010041





