Plant Growth-Promoting Bacteria of Soil: Designing of Consortia Beneficial for Crop Production
Abstract
1. Introduction
2. Agrochemistry and Prospects for Using Plant Growth-Promoting Bacteria
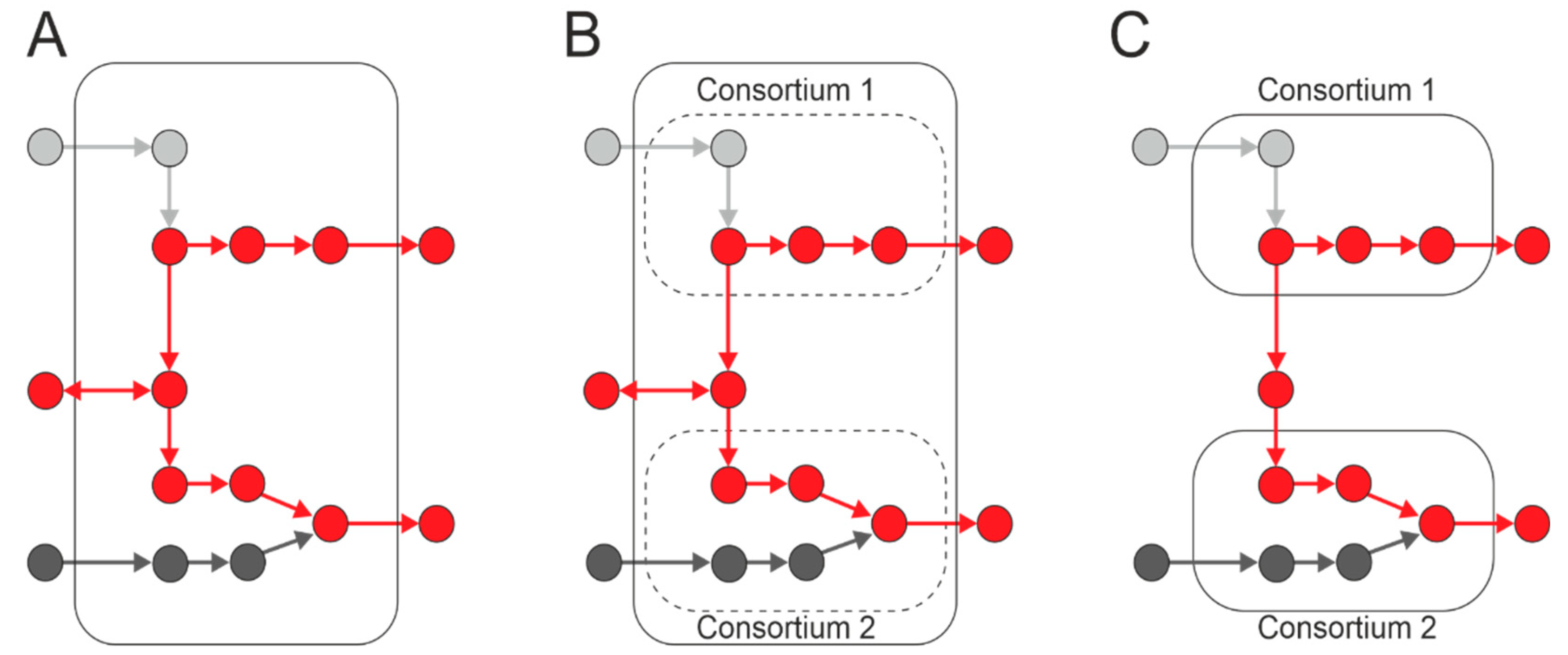
3. Microbial Consortia
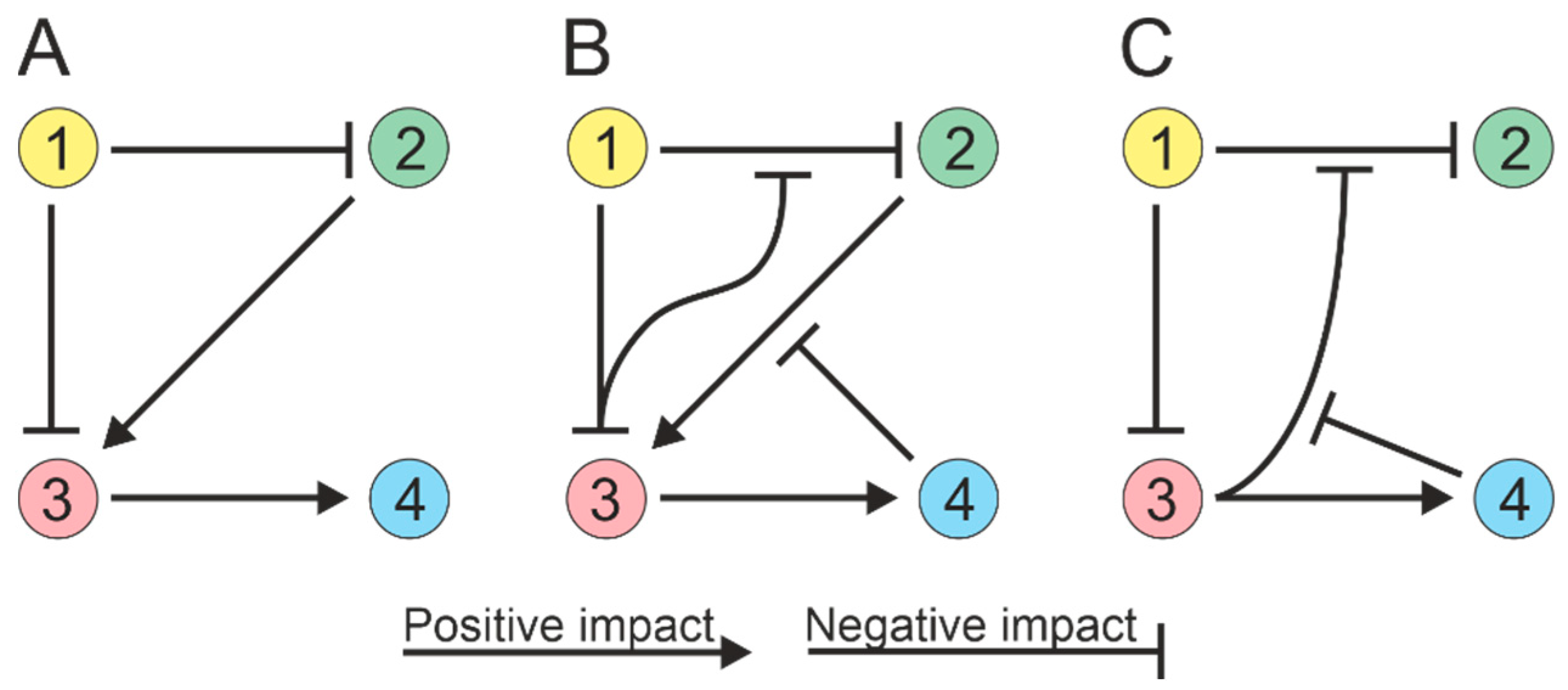
4. Microbial Interactions
5. Modeling Consortia
5.1. The “Bottom-Up” Approach
5.2. The “Top-Down” Approach
5.3. Practical Applications of Synthetic/Artificial Microbial Communities
5.4. Identification of Microbes with Key Characteristics
5.5. Development and Stabilization of Microbial Communities
6. Designing PGPB Consortia That Can Reduce the Response to Abiotic Stress
7. Multi-Omics Approaches for Studying Microorganisms and Their Consortia
8. Analysis of Metabolic Networks of Microbial Communities
Strategies for Design of Microbial Community Networks
9. Economic Impact of PGPB Application
10. Conclusions
Author Contributions
Funding
Informed Consent Statement
Conflicts of Interest
References
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. The Rhizosphere Microbiome: Significance of Plant Beneficial, Plant Pathogenic, and Human Pathogenic Microorganisms. FEMS Microbiol. Rev. 2013, 37, 634–663. [Google Scholar] [CrossRef]
- Yukun, G.; Jianghui, C.; Genzeng, R.; Shilin, W.; Puyuan, Y.; Congpei, Y.; Hongkai, L.; Jinhua, C. Changes in the Root-Associated Bacteria of Sorghum Are Driven by the Combined Effects of Salt and Sorghum Development. Environ. Microbiome 2021, 16, 14. [Google Scholar] [CrossRef]
- Zhang, J.; Cook, J.; Nearing, J.T.; Zhang, J.; Raudonis, R.; Glick, B.R.; Langille, M.G.I.; Cheng, Z. Harnessing the Plant Microbiome to Promote the Growth of Agricultural Crops. Microbiol. Res. 2021, 245, 126690. [Google Scholar] [CrossRef] [PubMed]
- Finkel, O.M.; Castrillo, G.; Herrera Paredes, S.; Salas González, I.; Dangl, J.L. Understanding and Exploiting Plant Beneficial Microbes. Curr. Opin. Plant Biol. 2017, 38, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Benito, P.; Carro, L.; Bacigalupe, R.; Ortúzar, M.; Trujillo, M.E. From Roots to Leaves: The Capacity of Micromonospora to Colonize Different Legume Tissues. Phytobiomes J. 2022, 6, 35–44. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; van Themaat, E.V.L.; Schulze-Lefert, P. Structure and Functions of the Bacterial Microbiota of Plants. Annu. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef] [PubMed]
- De Souza, R.S.C.; Armanhi, J.S.L.; Damasceno, N.D.B.; Imperial, J.; Arruda, P. Genome Sequences of a Plant Beneficial Synthetic Bacterial Community Reveal Genetic Features for Successful Plant Colonization. Front. Microbiol. 2019, 10, 1779. [Google Scholar] [CrossRef]
- de Souza, R.S.C.; Armanhi, J.S.L.; Arruda, P. From Microbiome to Traits: Designing Synthetic Microbial Communities for Improved Crop Resiliency. Front. Plant Sci. 2020, 11, 1179. [Google Scholar] [CrossRef]
- Herrera Paredes, S.; Gao, T.; Law, T.F.; Finkel, O.M.; Mucyn, T.; Teixeira, P.J.P.L.; Salas González, I.; Feltcher, M.E.; Powers, M.J.; Shank, E.A.; et al. Design of Synthetic Bacterial Communities for Predictable Plant Phenotypes. PLoS Biol. 2018, 16, e2003962. [Google Scholar] [CrossRef]
- Levy, A.; Salas Gonzalez, I.; Mittelviefhaus, M.; Clingenpeel, S.; Herrera Paredes, S.; Miao, J.; Wang, K.; Devescovi, G.; Stillman, K.; Monteiro, F.; et al. Genomic Features of Bacterial Adaptation to Plants. Nat. Genet. 2018, 50, 138–150. [Google Scholar] [CrossRef]
- Bernstein, H.C.; Carlson, R.P. Microbial Consortia Engineering for Cellular Factories: In Vitro to In Silico Systems. Comput. Struct. Biotechnol. J. 2012, 3, e201210017. [Google Scholar] [CrossRef]
- Lindemann, S.R.; Bernstein, H.C.; Song, H.-S.; Fredrickson, J.K.; Fields, M.W.; Shou, W.; Johnson, D.R.; Beliaev, A.S. Engineering Microbial Consortia for Controllable Outputs. ISME J. 2016, 10, 2077–2084. [Google Scholar] [CrossRef]
- Song, H.-S.; Cannon, W.; Beliaev, A.; Konopka, A. Mathematical Modeling of Microbial Community Dynamics: A Methodological Review. Processes 2014, 2, 711–752. [Google Scholar] [CrossRef]
- Biggs, M.B.; Medlock, G.L.; Kolling, G.L.; Papin, J.A. Metabolic Network Modeling of Microbial Communities. WIREs Syst. Biol. Med. 2015, 7, 317–334. [Google Scholar] [CrossRef] [PubMed]
- Cardona, C.; Weisenhorn, P.; Henry, C.; Gilbert, J.A. Network-Based Metabolic Analysis and Microbial Community Modeling. Curr. Opin. Microbiol. 2016, 31, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Wanapaisan, P.; Laothamteep, N.; Vejarano, F.; Chakraborty, J.; Shintani, M.; Muangchinda, C.; Morita, T.; Suzuki-Minakuchi, C.; Inoue, K.; Nojiri, H.; et al. Synergistic Degradation of Pyrene by Five Culturable Bacteria in a Mangrove Sediment-Derived Bacterial Consortium. J. Hazard. Mater. 2018, 342, 561–570. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Xue, R.; Zhou, J.; Wen, X.; Shi, Z.; Chen, M.; Xin, F.; Zhang, W.; Dong, W.; Jiang, M. Characterization of Acetamiprid Biodegradation by the Microbial Consortium ACE-3 Enriched From Contaminated Soil. Front. Microbiol. 2020, 11, 1429. [Google Scholar] [CrossRef]
- Wang, D.; Qin, L.; Liu, E.; Chai, G.; Su, Z.; Shan, J.; Yang, Z.; Wang, Z.; Wang, H.; Meng, H.; et al. Biodegradation Performance and Diversity of Enriched Bacterial Consortia Capable of Degrading High-Molecular-Weight Polycyclic Aromatic Hydrocarbons. Environ. Technol. 2021, 43, 4200–4211. [Google Scholar] [CrossRef] [PubMed]
- Eliazer Nelson, A.R.L.; Ravichandran, K.; Antony, U. The Impact of the Green Revolution on Indigenous Crops of India. J. Ethn. Foods 2019, 6, 8. [Google Scholar] [CrossRef]
- Kumar, S.; Dhar, S.; Barthakur, S.; Chandrakala, M.; Kochewad, S.A.; Meena, L.R.; Meena, L.K.; Kumar, S.; Singh, M.; Kumar, D. Effect of Integrated Potassium Management on Soil Biological Properties and Yields of Corn under Corn-Wheat Cropping System. Int. J. Curr. Microbiol. Appl. Sci. 2018, 7, 1855–1866. [Google Scholar] [CrossRef]
- Kumar, S.; Dhar, S.; Barthakur, S.; Rajawat, M.V.S.; Kochewad, S.A.; Kumar, S.; Kumar, D.; Meena, L.R. Farmyard Manure as K-Fertilizer Modulates Soil Biological Activities and Yield of Wheat Using the Integrated Fertilization Approach. Front. Environ. Sci. 2021, 9, 503. [Google Scholar] [CrossRef]
- Fernández-Romero, M.L.; Parras-Alcántara, L.; Lozano-García, B.; Clark, J.M.; Collins, C.D. Soil Quality Assessment Based on Carbon Stratification Index in Different Olive Grove Management Practices in Mediterranean Areas. Catena 2016, 137, 449–458. [Google Scholar] [CrossRef]
- Patra, S.; Mishra, P.; Mahapatra, S.C.; Mithun, S.K. Modelling Impacts of Chemical Fertilizer on Agricultural Production: A Case Study on Hooghly District, West Bengal, India. Model. Earth Syst. Environ. 2016, 2, 1–11. [Google Scholar] [CrossRef]
- Gupta, G.; Dhar, S.; Dass, A.; Sharma, V.K.; Shukla, L.; Singh, R.; Kumar, A.; Kumar, A.; Jinger, D.; Kumar, D.; et al. Assessment of Bio-Inoculants-Mediated Nutrient Management in Terms of Productivity, Profitability and Nutrient Harvest Index of Pigeon Pea–Wheat Cropping System in India. J. Plant Nutr. 2020, 43, 2911–2928. [Google Scholar] [CrossRef]
- Wang, L.; Chai, B. Fate of Antibiotic Resistance Genes and Changes in Bacterial Community with Increasing Breeding Scale of Layer Manure. Front. Microbiol. 2022, 13, 857046. [Google Scholar] [CrossRef] [PubMed]
- Timofeeva, A.M.; Galyamova, M.R.; Sedykh, S.E. Bacterial Siderophores: Classification, Biosynthesis, Perspectives of Use in Agriculture. Plants 2022, 11, 3065. [Google Scholar] [CrossRef]
- Timofeeva, A.; Galyamova, M.; Sedykh, S. Prospects for Using Phosphate-Solubilizing Microorganisms as Natural Fertilizers in Agriculture. Plants 2022, 11, 2119. [Google Scholar] [CrossRef]
- Castaldi, S.; Petrillo, C.; Donadio, G.; Piaz, F.D.; Cimmino, A.; Masi, M.; Evidente, A.; Isticato, R. Plant Growth Promotion Function of Bacillus sp. Strains Isolated from Salt-Pan Rhizosphere and Their Biocontrol Potential against Macrophomina Phaseolina. Int. J. Mol. Sci. 2021, 22, 3324. [Google Scholar] [CrossRef]
- Petrillo, C.; Castaldi, S.; Lanzilli, M.; Selci, M.; Cordone, A.; Giovannelli, D.; Isticato, R. Genomic and Physiological Characterization of Bacilli Isolated From Salt-Pans with Plant Growth Promoting Features. Front. Microbiol. 2021, 12, 715678. [Google Scholar] [CrossRef]
- Kour, D.; Rana, K.L.; Yadav, A.N.; Sheikh, I.; Kumar, V.; Dhaliwal, H.S.; Saxena, A.K. Amelioration of Drought Stress in Foxtail Millet (Setaria italica L.) by P-Solubilizing Drought-Tolerant Microbes with Multifarious Plant Growth Promoting Attributes. Environ. Sustain. 2020, 3, 23–34. [Google Scholar] [CrossRef]
- Khan, N.; Bano, A.; Rahman, M.A.; Guo, J.; Kang, Z.; Babar, M.A. Comparative Physiological and Metabolic Analysis Reveals a Complex Mechanism Involved in Drought Tolerance in Chickpea (Cicer arietinum L.) Induced by PGPR and PGRs. Sci. Rep. 2019, 9, 2097. [Google Scholar] [CrossRef]
- Meenakshi; Annapurna, K.; Govindasamy, V.; Ajit, V.; Choudhary, D.K. Mitigation of Drought Stress in Wheat Crop by Drought Tolerant Endophytic Bacterial Isolates. Vegetos 2019, 32, 486–493. [Google Scholar] [CrossRef]
- Jones, D.L.; Nguyen, C.; Finlay, R.D. Carbon Flow in the Rhizosphere: Carbon Trading at the Soil–Root Interface. Plant Soil 2009, 321, 5–33. [Google Scholar] [CrossRef]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; van der Putten, W.H. Going Back to the Roots: The Microbial Ecology of the Rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef]
- Deng, S.; Wipf, H.M.-L.; Pierroz, G.; Raab, T.K.; Khanna, R.; Coleman-Derr, D. A Plant Growth-Promoting Microbial Soil Amendment Dynamically Alters the Strawberry Root Bacterial Microbiome. Sci. Rep. 2019, 9, 17677. [Google Scholar] [CrossRef]
- Scarlett, K.; Denman, S.; Clark, D.R.; Forster, J.; Vanguelova, E.; Brown, N.; Whitby, C. Relationships between Nitrogen Cycling Microbial Community Abundance and Composition Reveal the Indirect Effect of Soil PH on Oak Decline. ISME J. 2021, 15, 623–635. [Google Scholar] [CrossRef] [PubMed]
- Michalska-Smith, M.; Song, Z.; Spawn-Lee, S.A.; Hansen, Z.A.; Johnson, M.; May, G.; Borer, E.T.; Seabloom, E.W.; Kinkel, L.L. Network Structure of Resource Use and Niche Overlap within the Endophytic Microbiome. ISME J. 2022, 16, 435–446. [Google Scholar] [CrossRef]
- Diffenbaugh, N.S.; Swain, D.L.; Touma, D. Anthropogenic Warming Has Increased Drought Risk in California. Proc. Natl. Acad. Sci. USA 2015, 112, 3931–3936. [Google Scholar] [CrossRef]
- Beaton, D.; Pelletier, P.; Goulet, R.R. Microbial Degradation of Cellulosic Material and Gas Generation: Implications for the Management of Low- and Intermediate-Level Radioactive Waste. Front. Microbiol. 2019, 10, 204. [Google Scholar] [CrossRef]
- Medie, F.M.; Davies, G.J.; Drancourt, M.; Henrissat, B. Genome Analyses Highlight the Different Biological Roles of Cellulases. Nat. Rev. Microbiol. 2012, 10, 227–234. [Google Scholar] [CrossRef]
- Beier, S.; Bertilsson, S. Bacterial Chitin Degradation—Mechanisms and Ecophysiological Strategies. Front. Microbiol. 2013, 4, 149. [Google Scholar] [CrossRef] [PubMed]
- Miyashita, K.; Fujii, T.; Saito, A. Induction and Repression of a Streptomyces Lividans Chitinase Gene Promoter in Response to Various Carbon Sources. Biosci. Biotechnol. Biochem. 2000, 64, 39–43. [Google Scholar] [CrossRef] [PubMed]
- Agaras, B.C.; Scandiani, M.; Luque, A.; Fernández, L.; Farina, F.; Carmona, M.; Gally, M.; Romero, A.; Wall, L.; Valverde, C. Quantification of the Potential Biocontrol and Direct Plant Growth Promotion Abilities Based on Multiple Biological Traits Distinguish Different Groups of Pseudomonas Spp. Isolates. Biol. Control 2015, 90, 173–186. [Google Scholar] [CrossRef]
- Hu, J.; Wei, Z.; Weidner, S.; Friman, V.-P.; Xu, Y.-C.; Shen, Q.-R.; Jousset, A. Probiotic Pseudomonas Communities Enhance Plant Growth and Nutrient Assimilation via Diversity-Mediated Ecosystem Functioning. Soil Biol. Biochem. 2017, 113, 122–129. [Google Scholar] [CrossRef]
- Assainar, S.K.; Abbott, L.K.; Mickan, B.S.; Whiteley, A.S.; Siddique, K.H.M.; Solaiman, Z.M. Response of Wheat to a Multiple Species Microbial Inoculant Compared to Fertilizer Application. Front. Plant Sci. 2018, 9, 1601. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Yang, T.; Friman, V.-P.; Xu, Y.; Shen, Q.; Jousset, A. Trophic Network Architecture of Root-Associated Bacterial Communities Determines Pathogen Invasion and Plant Health. Nat. Commun. 2015, 6, 8413. [Google Scholar] [CrossRef]
- Hu, J.; Wei, Z.; Friman, V.-P.; Gu, S.; Wang, X.; Eisenhauer, N.; Yang, T.; Ma, J.; Shen, Q.; Xu, Y.; et al. Probiotic Diversity Enhances Rhizosphere Microbiome Function and Plant Disease Suppression. MBio 2016, 7, e01790-16. [Google Scholar] [CrossRef]
- Castro-Sowinski, S.; Herschkovitz, Y.; Okon, Y.; Jurkevitch, E. Effects of Inoculation with Plant Growth-Promoting Rhizobacteria on Resident Rhizosphere Microorganisms. FEMS Microbiol. Lett. 2007, 276, 1–11. [Google Scholar] [CrossRef]
- Xiong, W.; Guo, S.; Jousset, A.; Zhao, Q.; Wu, H.; Li, R.; Kowalchuk, G.A.; Shen, Q. Bio-Fertilizer Application Induces Soil Suppressiveness against Fusarium Wilt Disease by Reshaping the Soil Microbiome. Soil Biol. Biochem. 2017, 114, 238–247. [Google Scholar] [CrossRef]
- Delgado-Baquerizo, M.; Maestre, F.T.; Reich, P.B.; Jeffries, T.C.; Gaitan, J.J.; Encinar, D.; Berdugo, M.; Campbell, C.D.; Singh, B.K. Microbial Diversity Drives Multifunctionality in Terrestrial Ecosystems. Nat. Commun. 2016, 7, 10541. [Google Scholar] [CrossRef]
- Mueller, U.G.; Sachs, J.L. Engineering Microbiomes to Improve Plant and Animal Health. Trends Microbiol. 2015, 23, 606–617. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Yu, Z.; Yu, X.; Xue, Y.; Huang, B.; Yang, J. Invasion by Cordgrass Increases Microbial Diversity and Alters Community Composition in a Mangrove Nature Reserve. Front. Microbiol. 2017, 8, 2503. [Google Scholar] [CrossRef]
- Gu, Y.; Wei, Z.; Wang, X.; Friman, V.-P.; Huang, J.; Wang, X.; Mei, X.; Xu, Y.; Shen, Q.; Jousset, A. Pathogen Invasion Indirectly Changes the Composition of Soil Microbiome via Shifts in Root Exudation Profile. Biol. Fertil. Soils 2016, 52, 997–1005. [Google Scholar] [CrossRef]
- Stringlis, I.A.; Yu, K.; Feussner, K.; de Jonge, R.; Van Bentum, S.; Van Verk, M.C.; Berendsen, R.L.; Bakker, P.A.H.M.; Feussner, I.; Pieterse, C.M.J. MYB72-Dependent Coumarin Exudation Shapes Root Microbiome Assembly to Promote Plant Health. Proc. Natl. Acad. Sci. USA 2018, 115, E5213–E5222. [Google Scholar] [CrossRef] [PubMed]
- Vannier, N.; Agler, M.; Hacquard, S. Microbiota-Mediated Disease Resistance in Plants. PLoS Pathog. 2019, 15, e1007740. [Google Scholar] [CrossRef]
- Mallon, C.A.; Le Roux, X.; van Doorn, G.S.; Dini-Andreote, F.; Poly, F.; Salles, J.F. The Impact of Failure: Unsuccessful Bacterial Invasions Steer the Soil Microbial Community Away from the Invader’s Niche. ISME J. 2018, 12, 728–741. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, H.K.; Appidi, M.R.; Villalobos Solis, M.I.; Wang, J.; Carper, D.L.; Burdick, L.; Pelletier, D.A.; Doktycz, M.J.; Hettich, R.L.; Abraham, P.E. Metaproteomics Reveals Insights into Microbial Structure, Interactions, and Dynamic Regulation in Defined Communities as They Respond to Environmental Disturbance. BMC Microbiol. 2021, 21, 308. [Google Scholar] [CrossRef]
- Garbeva, P.; Silby, M.W.; Raaijmakers, J.M.; Levy, S.B.; de Boer, W. Transcriptional and Antagonistic Responses of Pseudomonas Fluorescens Pf0-1 to Phylogenetically Different Bacterial Competitors. ISME J. 2011, 5, 973–985. [Google Scholar] [CrossRef]
- Traxler, M.F.; Watrous, J.D.; Alexandrov, T.; Dorrestein, P.C.; Kolter, R. Interspecies Interactions Stimulate Diversification of the Streptomyces Coelicolor Secreted Metabolome. MBio 2013, 4, e00459-13. [Google Scholar] [CrossRef]
- Tyc, O.; van den Berg, M.; Gerards, S.; van Veen, J.A.; Raaijmakers, J.M.; de Boer, W.; Garbeva, P. Impact of Interspecific Interactions on Antimicrobial Activity among Soil Bacteria. Front. Microbiol. 2014, 5, 567. [Google Scholar] [CrossRef]
- Garbeva, P.; de Boer, W. Inter-Specific Interactions between Carbon-Limited Soil Bacteria Affect Behavior and Gene Expression. Microb. Ecol. 2009, 58, 36–46. [Google Scholar] [CrossRef]
- Tracanna, V.; Ossowicki, A.; Petrus, M.L.C.; Overduin, S.; Terlouw, B.R.; Lund, G.; Robinson, S.L.; Warris, S.; Schijlen, E.G.W.M.; van Wezel, G.P.; et al. Dissecting Disease-Suppressive Rhizosphere Microbiomes by Functional Amplicon Sequencing and 10× Metagenomics. mSystems 2021, 6, e01116-20. [Google Scholar] [CrossRef] [PubMed]
- Briones, A.; Raskin, L. Diversity and Dynamics of Microbial Communities in Engineered Environments and Their Implications for Process Stability. Curr. Opin. Biotechnol. 2003, 14, 270–276. [Google Scholar] [CrossRef] [PubMed]
- Eiteman, M.A.; Lee, S.A.; Altman, R.; Altman, E. A Substrate-Selective Co-Fermentation Strategy with Escherichia Coli Produces Lactate by Simultaneously Consuming Xylose and Glucose. Biotechnol. Bioeng. 2009, 102, 822–827. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; Ward, T.E.; Regan, J.M. Electricity Production from Cellulose in a Microbial Fuel Cell Using a Defined Binary Culture. Environ. Sci. Technol. 2007, 41, 4781–4786. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, H.C.; Paulson, S.D.; Carlson, R.P. Synthetic Escherichia Coli Consortia Engineered for Syntrophy Demonstrate Enhanced Biomass Productivity. J. Biotechnol. 2012, 157, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Milucka, J.; Ferdelman, T.G.; Polerecky, L.; Franzke, D.; Wegener, G.; Schmid, M.; Lieberwirth, I.; Wagner, M.; Widdel, F.; Kuypers, M.M.M. Zero-Valent Sulphur Is a Key Intermediate in Marine Methane Oxidation. Nature 2012, 491, 541–546. [Google Scholar] [CrossRef] [PubMed]
- Kolenbrander, P.E. Multispecies Communities: Interspecies Interactions Influence Growth on Saliva as Sole Nutritional Source. Int. J. Oral Sci. 2011, 3, 49–54. [Google Scholar] [CrossRef]
- Ghoul, M.; Mitri, S. The Ecology and Evolution of Microbial Competition. Trends Microbiol. 2016, 24, 833–845. [Google Scholar] [CrossRef]
- Jagmann, N.; Brachvogel, H.-P.; Philipp, B. Parasitic Growth of Pseudomonas Aeruginosa in Co-Culture with the Chitinolytic Bacterium Aeromonas Hydrophila. Environ. Microbiol. 2010, 12, 1787–1802. [Google Scholar] [CrossRef]
- Poisot, T.; Stouffer, D.B.; Gravel, D. Beyond Species: Why Ecological Interaction Networks Vary through Space and Time. Oikos 2015, 124, 243–251. [Google Scholar] [CrossRef]
- Wootton, J.T. Indirect Effects in Complex Ecosystems: Recent Progress and Future Challenges. J. Sea Res. 2002, 48, 157–172. [Google Scholar] [CrossRef]
- Kelsic, E.D.; Zhao, J.; Vetsigian, K.; Kishony, R. Counteraction of Antibiotic Production and Degradation Stabilizes Microbial Communities. Nature 2015, 521, 516–519. [Google Scholar] [CrossRef]
- Abrudan, M.I.; Smakman, F.; Grimbergen, A.J.; Westhoff, S.; Miller, E.L.; van Wezel, G.P.; Rozen, D.E. Socially Mediated Induction and Suppression of Antibiosis during Bacterial Coexistence. Proc. Natl. Acad. Sci. USA 2015, 112, 11054–11059. [Google Scholar] [CrossRef] [PubMed]
- Lozano, G.L.; Bravo, J.I.; Garavito Diago, M.F.; Park, H.B.; Hurley, A.; Peterson, S.B.; Stabb, E.V.; Crawford, J.M.; Broderick, N.A.; Handelsman, J. Introducing THOR, a Model Microbiome for Genetic Dissection of Community Behavior. MBio 2019, 10, e02846-18. [Google Scholar] [CrossRef] [PubMed]
- Zengler, K.; Hofmockel, K.; Baliga, N.S.; Behie, S.W.; Bernstein, H.C.; Brown, J.B.; Dinneny, J.R.; Floge, S.A.; Forry, S.P.; Hess, M.; et al. EcoFABs: Advancing Microbiome Science through Standardized Fabricated Ecosystems. Nat. Methods 2019, 16, 567–571. [Google Scholar] [CrossRef]
- Kong, Z.; Hart, M.; Liu, H. Paving the Way From the Lab to the Field: Using Synthetic Microbial Consortia to Produce High-Quality Crops. Front. Plant Sci. 2018, 9, 1467. [Google Scholar] [CrossRef]
- Rodríguez Amor, D.; Dal Bello, M. Bottom-Up Approaches to Synthetic Cooperation in Microbial Communities. Life 2019, 9, 22. [Google Scholar] [CrossRef]
- Smercina, D.N.; Bailey, V.L.; Hofmockel, K.S. Micro on a Macroscale: Relating Microbial-Scale Soil Processes to Global Ecosystem Function. FEMS Microbiol. Ecol. 2021, 97, fiab091. [Google Scholar] [CrossRef]
- Irabor, A.; Mmbaga, M.T. Evaluation of Selected Bacterial Endophytes for Biocontrol Potential against Phytophthora Blight of Bell Pepper (Capsicum annuum L.). J. Plant Pathol. Microbiol. 2017, 8, 31–34. [Google Scholar] [CrossRef]
- Yahya, M.; Islam, E.U.; Rasul, M.; Farooq, I.; Mahreen, N.; Tawab, A.; Irfan, M.; Rajput, L.; Amin, I.; Yasmin, S. Differential Root Exudation and Architecture for Improved Growth of Wheat Mediated by Phosphate Solubilizing Bacteria. Front. Microbiol. 2021, 12, 744094. [Google Scholar] [CrossRef] [PubMed]
- Hays, S.G.; Patrick, W.G.; Ziesack, M.; Oxman, N.; Silver, P.A. Better Together: Engineering and Application of Microbial Symbioses. Curr. Opin. Biotechnol. 2015, 36, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Mee, M.T.; Collins, J.J.; Church, G.M.; Wang, H.H. Syntrophic Exchange in Synthetic Microbial Communities. Proc. Natl. Acad. Sci. USA 2014, 111, E2149–E2156. [Google Scholar] [CrossRef] [PubMed]
- Bairey, E.; Kelsic, E.D.; Kishony, R. High-Order Species Interactions Shape Ecosystem Diversity. Nat. Commun. 2016, 7, 12285. [Google Scholar] [CrossRef] [PubMed]
- Duncker, K.E.; Holmes, Z.A.; You, L. Engineered Microbial Consortia: Strategies and Applications. Microb. Cell Fact. 2021, 20, 211. [Google Scholar] [CrossRef] [PubMed]
- Zegeye, E.K.; Brislawn, C.J.; Farris, Y.; Fansler, S.J.; Hofmockel, K.S.; Jansson, J.K.; Wright, A.T.; Graham, E.B.; Naylor, D.; McClure, R.S.; et al. Selection, Succession, and Stabilization of Soil Microbial Consortia. mSystems 2019, 4, e00055-19. [Google Scholar] [CrossRef] [PubMed]
- McClure, R.; Naylor, D.; Farris, Y.; Davison, M.; Fansler, S.J.; Hofmockel, K.S.; Jansson, J.K. Development and Analysis of a Stable, Reduced Complexity Model Soil Microbiome. Front. Microbiol. 2020, 11, 1987. [Google Scholar] [CrossRef] [PubMed]
- Großkopf, T.; Soyer, O.S. Synthetic Microbial Communities. Curr. Opin. Microbiol. 2014, 18, 72–77. [Google Scholar] [CrossRef]
- Gilmore, S.P.; Lankiewicz, T.S.; Wilken, S.E.; Brown, J.L.; Sexton, J.A.; Henske, J.K.; Theodorou, M.K.; Valentine, D.L.; O’Malley, M.A. Top-Down Enrichment Guides in Formation of Synthetic Microbial Consortia for Biomass Degradation. ACS Synth. Biol. 2019, 8, 2174–2185. [Google Scholar] [CrossRef]
- Tecon, R.; Mitri, S.; Ciccarese, D.; Or, D.; van der Meer, J.R.; Johnson, D.R. Bridging the Holistic-Reductionist Divide in Microbial Ecology. mSystems 2019, 4, e00265-18. [Google Scholar] [CrossRef]
- Qin, Y.; Druzhinina, I.S.; Pan, X.; Yuan, Z. Microbially Mediated Plant Salt Tolerance and Microbiome-Based Solutions for Saline Agriculture. Biotechnol. Adv. 2016, 34, 1245–1259. [Google Scholar] [CrossRef]
- Niu, B.; Paulson, J.N.; Zheng, X.; Kolter, R. Simplified and Representative Bacterial Community of Maize Roots. Proc. Natl. Acad. Sci. USA 2017, 114, E2450–E2459. [Google Scholar] [CrossRef]
- Kaminsky, L.M.; Trexler, R.V.; Malik, R.J.; Hockett, K.L.; Bell, T.H. The Inherent Conflicts in Developing Soil Microbial Inoculants. Trends Biotechnol. 2019, 37, 140–151. [Google Scholar] [CrossRef]
- Chiu, H.-C.; Levy, R.; Borenstein, E. Emergent Biosynthetic Capacity in Simple Microbial Communities. PLoS Comput. Biol. 2014, 10, e1003695. [Google Scholar] [CrossRef]
- Burke, C.; Steinberg, P.; Rusch, D.; Kjelleberg, S.; Thomas, T. Bacterial Community Assembly Based on Functional Genes Rather than Species. Proc. Natl. Acad. Sci. USA 2011, 108, 14288–14293. [Google Scholar] [CrossRef]
- Tsolakidou, M.-D.; Stringlis, I.A.; Fanega-Sleziak, N.; Papageorgiou, S.; Tsalakou, A.; Pantelides, I.S. Rhizosphere-Enriched Microbes as a Pool to Design Synthetic Communities for Reproducible Beneficial Outputs. FEMS Microbiol. Ecol. 2019, 95, fiz138. [Google Scholar] [CrossRef]
- Thomloudi, E.-E.; Tsalgatidou, P.C.; Douka, D.; Spantidos, T.-N.; Dimou, M.; Venieraki, A.; Katinakis, P. Multistrain versus Single-Strain Plant Growth Promoting Microbial Inoculants—The Compatibility Issue. Hell. Plant Prot. J. 2019, 12, 61–77. [Google Scholar] [CrossRef]
- Yang, N.; Nesme, J.; Røder, H.L.; Li, X.; Zuo, Z.; Petersen, M.; Burmølle, M.; Sørensen, S.J. Emergent Bacterial Community Properties Induce Enhanced Drought Tolerance in Arabidopsis. NPJ Biofilms Microbiomes 2021, 7, 82. [Google Scholar] [CrossRef] [PubMed]
- Chaiya, L.; Gavinlertvatana, P.; Teaumroong, N.; Pathom-aree, W.; Chaiyasen, A.; Sungthong, R.; Lumyong, S. Enhancing Teak (Tectona grandis) Seedling Growth by Rhizosphere Microbes: A Sustainable Way to Optimize Agroforestry. Microorganisms 2021, 9, 1990. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, R.L.; Vismans, G.; Yu, K.; Song, Y.; de Jonge, R.; Burgman, W.P.; Burmølle, M.; Herschend, J.; Bakker, P.A.H.M.; Pieterse, C.M.J. Disease-Induced Assemblage of a Plant-Beneficial Bacterial Consortium. ISME J. 2018, 12, 1496–1507. [Google Scholar] [CrossRef] [PubMed]
- Goswami, S.K.; Kashyap, P.L.; Awasthi, S. Deciphering Rhizosphere Microbiome for the Development of Novel Bacterial Consortium and Its Evaluation for Salt Stress Management in Solanaceous Crops in India. Indian Phytopathol. 2019, 72, 479–488. [Google Scholar] [CrossRef]
- Wang, C.-J.; Yang, W.; Wang, C.; Gu, C.; Niu, D.-D.; Liu, H.-X.; Wang, Y.-P.; Guo, J.-H. Induction of Drought Tolerance in Cucumber Plants by a Consortium of Three Plant Growth-Promoting Rhizobacterium Strains. PLoS ONE 2012, 7, e52565. [Google Scholar] [CrossRef] [PubMed]
- Saikia, J.; Sarma, R.K.; Dhandia, R.; Yadav, A.; Bharali, R.; Gupta, V.K.; Saikia, R. Alleviation of Drought Stress in Pulse Crops with ACC Deaminase Producing Rhizobacteria Isolated from Acidic Soil of Northeast India. Sci. Rep. 2018, 8, 3560. [Google Scholar] [CrossRef]
- Minchev, Z.; Kostenko, O.; Soler, R.; Pozo, M.J. Microbial Consortia for Effective Biocontrol of Root and Foliar Diseases in Tomato. Front. Plant Sci. 2021, 12, 756368. [Google Scholar] [CrossRef] [PubMed]
- Grandel, N.E.; Reyes Gamas, K.; Bennett, M.R. Control of Synthetic Microbial Consortia in Time, Space, and Composition. Trends Microbiol. 2021, 29, 1095–1105. [Google Scholar] [CrossRef]
- Ortiz, A.; Vega, N.M.; Ratzke, C.; Gore, J. Interspecies Bacterial Competition Regulates Community Assembly in the C. elegans Intestine. ISME J. 2021, 15, 2131–2145. [Google Scholar] [CrossRef]
- Tanentzap, A.J.; Brandt, A.J.; Smissen, R.D.; Heenan, P.B.; Fukami, T.; Lee, W.G. When Do Plant Radiations Influence Community Assembly? The Importance of Historical Contingency in the Race for Niche Space. N. Phytol. 2015, 207, 468–479. [Google Scholar] [CrossRef]
- Nadeem, S.M.; Ahmad, M.; Zahir, Z.A.; Javaid, A.; Ashraf, M. The Role of Mycorrhizae and Plant Growth Promoting Rhizobacteria (PGPR) in Improving Crop Productivity under Stressful Environments. Biotechnol. Adv. 2014, 32, 429–448. [Google Scholar] [CrossRef]
- Zimmer, S.; Messmer, M.; Haase, T.; Piepho, H.-P.; Mindermann, A.; Schulz, H.; Habekuß, A.; Ordon, F.; Wilbois, K.-P.; Heß, J. Effects of Soybean Variety and Bradyrhizobium Strains on Yield, Protein Content and Biological Nitrogen Fixation under Cool Growing Conditions in Germany. Eur. J. Agron. 2016, 72, 38–46. [Google Scholar] [CrossRef]
- Armanhi, J.S.L.; De Souza, R.S.C.; Damasceno, N.D.B.; De Araujo, L.M.; Imperial, J.; Arruda, P. A Community-Based Culture Collection for Targeting Novel Plant Growth-Promoting Bacteria from the Sugarcane Microbiome. Front. Plant Sci. 2018, 8, 2191. [Google Scholar] [CrossRef]
- Toju, H.; Peay, K.G.; Yamamichi, M.; Narisawa, K.; Hiruma, K.; Naito, K.; Fukuda, S.; Ushio, M.; Nakaoka, S.; Onoda, Y.; et al. Core Microbiomes for Sustainable Agroecosystems. Nat. Plants 2018, 4, 247–257. [Google Scholar] [CrossRef]
- Vorholt, J.A.; Vogel, C.; Carlström, C.I.; Müller, D.B. Establishing Causality: Opportunities of Synthetic Communities for Plant Microbiome Research. Cell Host Microbe 2017, 22, 142–155. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Boedicker, J.Q.; Choi, J.W.; Ismagilov, R.F. Defined Spatial Structure Stabilizes a Synthetic Multispecies Bacterial Community. Proc. Natl. Acad. Sci. USA 2008, 105, 18188–18193. [Google Scholar] [CrossRef] [PubMed]
- Minty, J.J.; Singer, M.E.; Scholz, S.A.; Bae, C.-H.; Ahn, J.-H.; Foster, C.E.; Liao, J.C.; Lin, X.N. Design and Characterization of Synthetic Fungal-Bacterial Consortia for Direct Production of Isobutanol from Cellulosic Biomass. Proc. Natl. Acad. Sci. USA 2013, 110, 14592–14597. [Google Scholar] [CrossRef]
- Fahad, S.; Bajwa, A.A.; Nazir, U.; Anjum, S.A.; Farooq, A.; Zohaib, A.; Sadia, S.; Nasim, W.; Adkins, S.; Saud, S.; et al. Crop Production under Drought and Heat Stress: Plant Responses and Management Options. Front. Plant Sci. 2017, 8, 1147. [Google Scholar] [CrossRef]
- Niu, X.; Song, L.; Xiao, Y.; Ge, W. Drought-Tolerant Plant Growth-Promoting Rhizobacteria Associated with Foxtail Millet in a Semi-Arid Agroecosystem and Their Potential in Alleviating Drought Stress. Front. Microbiol. 2018, 8, 2580. [Google Scholar] [CrossRef]
- Paredes-Páliz, K.; Rodríguez-Vázquez, R.; Duarte, B.; Caviedes, M.A.; Mateos-Naranjo, E.; Redondo-Gómez, S.; Caçador, M.I.; Rodríguez-Llorente, I.D.; Pajuelo, E. Investigating the Mechanisms Underlying Phytoprotection by Plant Growth-promoting Rhizobacteria in Spartina Densiflora under Metal Stress. Plant Biol. 2018, 20, 497–506. [Google Scholar] [CrossRef]
- Kasim, W.A.; Gaafar, R.M.; Abou-Ali, R.M.; Omar, M.N.; Hewait, H.M. Effect of Biofilm Forming Plant Growth Promoting Rhizobacteria on Salinity Tolerance in Barley. Ann. Agric. Sci. 2016, 61, 217–227. [Google Scholar] [CrossRef]
- Xu, N.; Yang, L.; Fan, Y.; Yang, J.; Yue, D.; Liang, Y.; Price, B.; Cohen, S.; Huang, T. YouTube-VOS: Sequence-to-Sequence Video Object Segmentation; Springer: Berlin, Germany, 2018; pp. 603–619. [Google Scholar]
- Shaheen, R.; Svensson, B.; Andersson, M.A.; Christiansson, A.; Salkinoja-Salonen, M. Persistence Strategies of Bacillus Cereus Spores Isolated from Dairy Silo Tanks. Food Microbiol. 2010, 27, 347–355. [Google Scholar] [CrossRef]
- Poli, A.; Anzelmo, G.; Nicolaus, B. Bacterial Exopolysaccharides from Extreme Marine Habitats: Production, Characterization and Biological Activities. Mar. Drugs 2010, 8, 1779–1802. [Google Scholar] [CrossRef]
- Chen, Y.; Yan, F.; Chai, Y.; Liu, H.; Kolter, R.; Losick, R.; Guo, J. Biocontrol of Tomato Wilt Disease by B Acillus Subtilis Isolates from Natural Environments Depends on Conserved Genes Mediating Biofilm Formation. Environ. Microbiol. 2013, 15, 848–864. [Google Scholar] [CrossRef] [PubMed]
- Nunkaew, T.; Kantachote, D.; Nitoda, T.; Kanzaki, H.; Ritchie, R.J. Characterization of Exopolymeric Substances from Selected Rhodopseudomonas Palustris Strains and Their Ability to Adsorb Sodium Ions. Carbohydr. Polym. 2015, 115, 334–341. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, M.; Hasnain, S.; Berge, O.; Mahmood, T. Inoculating Wheat Seedlings with Exopolysaccharide-Producing Bacteria Restricts Sodium Uptake and Stimulates Plant Growth under Salt Stress. Biol. Fertil. Soils 2004, 40, 157–162. [Google Scholar] [CrossRef]
- Zippel, B.; Neu, T.R. Characterization of Glycoconjugates of Extracellular Polymeric Substances in Tufa-Associated Biofilms by Using Fluorescence Lectin-Binding Analysis. Appl. Environ. Microbiol. 2011, 77, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Redmile-Gordon, M.A.; Brookes, P.C.; Evershed, R.P.; Goulding, K.W.T.; Hirsch, P.R. Measuring the Soil-Microbial Interface: Extraction of Extracellular Polymeric Substances (EPS) from Soil Biofilms. Soil Biol. Biochem. 2014, 72, 163–171. [Google Scholar] [CrossRef]
- Zörb, C.; Geilfus, C.-M.; Dietz, K.-J. Salinity and Crop Yield. Plant Biol. 2019, 21, 31–38. [Google Scholar] [CrossRef]
- Kumar, A.; Singh, S.; Mukherjee, A.; Rastogi, R.P.; Verma, J.P. Salt-Tolerant Plant Growth-Promoting Bacillus Pumilus Strain JPVS11 to Enhance Plant Growth Attributes of Rice and Improve Soil Health under Salinity Stress. Microbiol. Res. 2021, 242, 126616. [Google Scholar] [CrossRef]
- Kumar Arora, N.; Fatima, T.; Mishra, J.; Mishra, I.; Verma, S.; Verma, R.; Verma, M.; Bhattacharya, A.; Verma, P.; Mishra, P.; et al. Halo-Tolerant Plant Growth Promoting Rhizobacteria for Improving Productivity and Remediation of Saline Soils. J. Adv. Res. 2020, 26, 69–82. [Google Scholar] [CrossRef]
- Numan, M.; Bashir, S.; Khan, Y.; Mumtaz, R.; Shinwari, Z.K.; Khan, A.L.; Khan, A.; AL-Harrasi, A. Plant Growth Promoting Bacteria as an Alternative Strategy for Salt Tolerance in Plants: A Review. Microbiol. Res. 2018, 209, 21–32. [Google Scholar] [CrossRef]
- Kasassi, A.; Rakimbei, P.; Karagiannidis, A.; Zabaniotou, A.; Tsiouvaras, K.; Nastis, A.; Tzafeiropoulou, K. Soil Contamination by Heavy Metals: Measurements from a Closed Unlined Landfill. Bioresour. Technol. 2008, 99, 8578–8584. [Google Scholar] [CrossRef]
- Lwin, C.S.; Seo, B.-H.; Kim, H.-U.; Owens, G.; Kim, K.-R. Application of Soil Amendments to Contaminated Soils for Heavy Metal Immobilization and Improved Soil Quality—A Critical Review. Soil Sci. Plant Nutr. 2018, 64, 156–167. [Google Scholar] [CrossRef]
- Xu, X.; Xu, M.; Zhao, Q.; Xia, Y.; Chen, C.; Shen, Z. Complete Genome Sequence of Cd(II)-Resistant Arthrobacter sp. PGP41, a Plant Growth-Promoting Bacterium with Potential in Microbe-Assisted Phytoremediation. Curr. Microbiol. 2018, 75, 1231–1239. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Xu, M.; Wei, Q.; Tang, M.; Guan, L.; Lou, L.; Xu, X.; Hu, Z.; Chen, Y.; Shen, Z.; et al. Promotion of Growth and Phytoextraction of Cadmium and Lead in Solanum Nigrum L. Mediated by Plant-Growth-Promoting Rhizobacteria. Ecotoxicol. Environ. Saf. 2020, 205, 111333. [Google Scholar] [CrossRef] [PubMed]
- Masmoudi, F.; Abdelmalek, N.; Tounsi, S.; Dunlap, C.A.; Trigui, M. Abiotic Stress Resistance, Plant Growth Promotion and Antifungal Potential of Halotolerant Bacteria from a Tunisian Solar Saltern. Microbiol. Res. 2019, 229, 126331. [Google Scholar] [CrossRef] [PubMed]
- Turan, M.; Gulluce, M.; Şahin, F. Effects of Plant-Growth-Promoting Rhizobacteria on Yield, Growth, and Some Physiological Characteristics of Wheat and Barley Plants. Commun. Soil Sci. Plant Anal. 2012, 43, 1658–1673. [Google Scholar] [CrossRef]
- Mishra, P.K.; Bisht, S.C.; Ruwari, P.; Selvakumar, G.; Joshi, G.K.; Bisht, J.K.; Bhatt, J.C.; Gupta, H.S. Alleviation of Cold Stress in Inoculated Wheat (Triticum aestivum L.) Seedlings with Psychrotolerant Pseudomonads from NW Himalayas. Arch. Microbiol. 2011, 193, 497–513. [Google Scholar] [CrossRef]
- Wu, H.; Gu, Q.; Xie, Y.; Lou, Z.; Xue, P.; Fang, L.; Yu, C.; Jia, D.; Huang, G.; Zhu, B.; et al. Cold-adapted Bacilli Isolated from the Qinghai–Tibetan Plateau Are Able to Promote Plant Growth in Extreme Environments. Environ. Microbiol. 2019, 21, 3505–3526. [Google Scholar] [CrossRef] [PubMed]
- Calvo, P.; Ormeño-Orrillo, E.; Martínez-Romero, E.; Zúñiga, D. Characterization of Bacillus Isolates of Potato Rhizosphere from Andean Soils of Peru and Their Potential PGPR Characteristics. Braz. J. Microbiol. 2010, 41, 899–906. [Google Scholar] [CrossRef]
- Ghyselinck, J.; Velivelli, S.L.S.; Heylen, K.; O’Herlihy, E.; Franco, J.; Rojas, M.; De Vos, P.; Prestwich, B.D. Bioprospecting in Potato Fields in the Central Andean Highlands: Screening of Rhizobacteria for Plant Growth-Promoting Properties. Syst. Appl. Microbiol. 2013, 36, 116–127. [Google Scholar] [CrossRef]
- Knief, C.; Delmotte, N.; Chaffron, S.; Stark, M.; Innerebner, G.; Wassmann, R.; von Mering, C.; Vorholt, J.A. Metaproteogenomic Analysis of Microbial Communities in the Phyllosphere and Rhizosphere of Rice. ISME J. 2012, 6, 1378–1390. [Google Scholar] [CrossRef]
- Sessitsch, A.; Hardoim, P.; Döring, J.; Weilharter, A.; Krause, A.; Woyke, T.; Mitter, B.; Hauberg-Lotte, L.; Friedrich, F.; Rahalkar, M.; et al. Functional Characteristics of an Endophyte Community Colonizing Rice Roots as Revealed by Metagenomic Analysis. Mol. Plant-Microbe Interact. 2012, 25, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Bakker, P.A.H.M.; Berendsen, R.L.; Doornbos, R.F.; Wintermans, P.C.A.; Pieterse, C.M.J. The Rhizosphere Revisited: Root Microbiomics. Front. Plant Sci. 2013, 4, 165. [Google Scholar] [CrossRef] [PubMed]
- Forchetti, G.; Masciarelli, O.; Alemano, S.; Alvarez, D.; Abdala, G. Endophytic Bacteria in Sunflower (Helianthus annuus L.): Isolation, Characterization, and Production of Jasmonates and Abscisic Acid in Culture Medium. Appl. Microbiol. Biotechnol. 2007, 76, 1145–1152. [Google Scholar] [CrossRef]
- Lakshmanan, V.; Selvaraj, G.; Bais, H.P. Functional Soil Microbiome: Belowground Solutions to an Aboveground Problem. Plant Physiol. 2014, 166, 689–700. [Google Scholar] [CrossRef]
- Schlaeppi, K.; Bulgarelli, D. The Plant Microbiome at Work. Mol. Plant-Microbe Interact. 2015, 28, 212–217. [Google Scholar] [CrossRef]
- Jansson, J.K.; Hofmockel, K.S. The Soil Microbiome—From Metagenomics to Metaphenomics. Curr. Opin. Microbiol. 2018, 43, 162–168. [Google Scholar] [CrossRef]
- Yang, T.; Adams, J.M.; Shi, Y.; He, J.; Jing, X.; Chen, L.; Tedersoo, L.; Chu, H. Soil Fungal Diversity in Natural Grasslands of the Tibetan Plateau: Associations with Plant Diversity and Productivity. N. Phytol. 2017, 215, 756–765. [Google Scholar] [CrossRef]
- Song, H.-K.; Shi, Y.; Yang, T.; Chu, H.; He, J.-S.; Kim, H.; Jablonski, P.; Adams, J.M. Environmental Filtering of Bacterial Functional Diversity along an Aridity Gradient. Sci. Rep. 2019, 9, 866. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; He, Z.; Yang, Y.; Deng, Y.; Tringe, S.G.; Alvarez-Cohen, L. High-Throughput Metagenomic Technologies for Complex Microbial Community Analysis: Open and Closed Formats. MBio 2015, 6, e02288-44. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Garrido-Oter, R.; Münch, P.C.; Weiman, A.; Dröge, J.; Pan, Y.; McHardy, A.C.; Schulze-Lefert, P. Structure and Function of the Bacterial Root Microbiota in Wild and Domesticated Barley. Cell Host Microbe 2015, 17, 392–403. [Google Scholar] [CrossRef]
- Peng, J.; Wegner, C.-E.; Bei, Q.; Liu, P.; Liesack, W. Metatranscriptomics Reveals a Differential Temperature Effect on the Structural and Functional Organization of the Anaerobic Food Web in Rice Field Soil. Microbiome 2018, 6, 169. [Google Scholar] [CrossRef] [PubMed]
- Roy Chowdhury, T.; Lee, J.-Y.; Bottos, E.M.; Brislawn, C.J.; White, R.A.; Bramer, L.M.; Brown, J.; Zucker, J.D.; Kim, Y.-M.; Jumpponen, A.; et al. Metaphenomic Responses of a Native Prairie Soil Microbiome to Moisture Perturbations. mSystems 2019, 4, e00061-19. [Google Scholar] [CrossRef] [PubMed]
- Guttman, D.S.; McHardy, A.C.; Schulze-Lefert, P. Microbial Genome-Enabled Insights into Plant–Microorganism Interactions. Nat. Rev. Genet. 2014, 15, 797–813. [Google Scholar] [CrossRef] [PubMed]
- Callister, S.J.; Fillmore, T.L.; Nicora, C.D.; Shaw, J.B.; Purvine, S.O.; Orton, D.J.; White, R.A.; Moore, R.J.; Burnet, M.C.; Nakayasu, E.S.; et al. Addressing the Challenge of Soil Metaproteome Complexity by Improving Metaproteome Depth of Coverage through Two-Dimensional Liquid Chromatography. Soil Biol. Biochem. 2018, 125, 290–299. [Google Scholar] [CrossRef]
- Hultman, J.; Waldrop, M.P.; Mackelprang, R.; David, M.M.; McFarland, J.; Blazewicz, S.J.; Harden, J.; Turetsky, M.R.; McGuire, A.D.; Shah, M.B.; et al. Multi-Omics of Permafrost, Active Layer and Thermokarst Bog Soil Microbiomes. Nature 2015, 521, 208–212. [Google Scholar] [CrossRef]
- Kleiner, M.; Thorson, E.; Sharp, C.E.; Dong, X.; Liu, D.; Li, C.; Strous, M. Assessing Species Biomass Contributions in Microbial Communities via Metaproteomics. Nat. Commun. 2017, 8, 1558. [Google Scholar] [CrossRef]
- Neurohr, G.E.; Amon, A. Relevance and Regulation of Cell Density. Trends Cell Biol. 2020, 30, 213–225. [Google Scholar] [CrossRef]
- Milo, R. What Is the Total Number of Protein Molecules per Cell Volume? A Call to Rethink Some Published Values. BioEssays 2013, 35, 1050–1055. [Google Scholar] [CrossRef]
- Kleiner, M. Metaproteomics: Much More than Measuring Gene Expression in Microbial Communities. mSystems 2019, 4, e00115-19. [Google Scholar] [CrossRef]
- Han, X.; He, L.; Xin, L.; Shan, B.; Ma, B. PeaksPTM: Mass Spectrometry-Based Identification of Peptides with Unspecified Modifications. J. Proteome Res. 2011, 10, 2930–2936. [Google Scholar] [CrossRef]
- Macek, B.; Forchhammer, K.; Hardouin, J.; Weber-Ban, E.; Grangeasse, C.; Mijakovic, I. Protein Post-Translational Modifications in Bacteria. Nat. Rev. Microbiol. 2019, 17, 651–664. [Google Scholar] [CrossRef] [PubMed]
- Mijakovic, I.; Grangeasse, C.; Turgay, K. Exploring the Diversity of Protein Modifications: Special Bacterial Phosphorylation Systems. FEMS Microbiol. Rev. 2016, 40, 398–417. [Google Scholar] [CrossRef]
- Li, Z.; Wang, Y.; Yao, Q.; Justice, N.B.; Ahn, T.-H.; Xu, D.; Hettich, R.L.; Banfield, J.F.; Pan, C. Diverse and Divergent Protein Post-Translational Modifications in Two Growth Stages of a Natural Microbial Community. Nat. Commun. 2014, 5, 4405. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Sun, J.; Cao, H.; Tian, R.; Cai, L.; Ding, W.; Qian, P.-Y. Post-Translational Modifications Are Enriched within Protein Functional Groups Important to Bacterial Adaptation within a Deep-Sea Hydrothermal Vent Environment. Microbiome 2016, 4, 49. [Google Scholar] [CrossRef] [PubMed]
- Erce, M.A.; Pang, C.N.I.; Hart-Smith, G.; Wilkins, M.R. The Methylproteome and the Intracellular Methylation Network. Proteomics 2012, 12, 564–586. [Google Scholar] [CrossRef]
- van Staalduinen, L.M.; Jia, Z. Post-Translational Hydroxylation by 2OG/Fe(II)-Dependent Oxygenases as a Novel Regulatory Mechanism in Bacteria. Front. Microbiol. 2015, 5, 798. [Google Scholar] [CrossRef][Green Version]
- Thiele, I.; Palsson, B.Ø. A Protocol for Generating a High-Quality Genome-Scale Metabolic Reconstruction. Nat. Protoc. 2010, 5, 93–121. [Google Scholar] [CrossRef]
- Shoaie, S.; Ghaffari, P.; Kovatcheva-Datchary, P.; Mardinoglu, A.; Sen, P.; Pujos-Guillot, E.; de Wouters, T.; Juste, C.; Rizkalla, S.; Chilloux, J.; et al. Quantifying Diet-Induced Metabolic Changes of the Human Gut Microbiome. Cell Metab. 2015, 22, 320–331. [Google Scholar] [CrossRef]
- Duarte, N.C.; Becker, S.A.; Jamshidi, N.; Thiele, I.; Mo, M.L.; Vo, T.D.; Srivas, R.; Palsson, B.Ø. Global Reconstruction of the Human Metabolic Network Based on Genomic and Bibliomic Data. Proc. Natl. Acad. Sci. USA 2007, 104, 1777–1782. [Google Scholar] [CrossRef]
- Gianchandani, E.P.; Oberhardt, M.A.; Burgard, A.P.; Maranas, C.D.; Papin, J.A. Predicting Biological System Objectives de Novo from Internal State Measurements. BMC Bioinform. 2008, 9, 43. [Google Scholar] [CrossRef]
- Lander, E.S.; Waterman, M.S. Genomic Mapping by Fingerprinting Random Clones: A Mathematical Analysis. Genomics 1988, 2, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Papin, J.A.; Palsson, B.O. The JAK-STAT Signaling Network in the Human B-Cell: An Extreme Signaling Pathway Analysis. Biophys. J. 2004, 87, 37–46. [Google Scholar] [CrossRef][Green Version]
- Li, F.; Thiele, I.; Jamshidi, N.; Palsson, B.Ø. Identification of Potential Pathway Mediation Targets in Toll-like Receptor Signaling. PLoS Comput. Biol. 2009, 5, e1000292. [Google Scholar] [CrossRef]
- Thiele, I.; Jamshidi, N.; Fleming, R.M.T.; Palsson, B.Ø. Genome-Scale Reconstruction of Escherichia Coli’s Transcriptional and Translational Machinery: A Knowledge Base, Its Mathematical Formulation, and Its Functional Characterization. PLoS Comput. Biol. 2009, 5, e1000312. [Google Scholar] [CrossRef] [PubMed]
- Abubucker, S.; Segata, N.; Goll, J.; Schubert, A.M.; Izard, J.; Cantarel, B.L.; Rodriguez-Mueller, B.; Zucker, J.; Thiagarajan, M.; Henrissat, B.; et al. Metabolic Reconstruction for Metagenomic Data and Its Application to the Human Microbiome. PLoS Comput. Biol. 2012, 8, e1002358. [Google Scholar] [CrossRef] [PubMed]
- Faria, J.P.; Khazaei, T.; Edirisinghe, J.N.; Weisenhorn, P.; Seaver, S.M.D.; Conrad, N.; Harris, N.; DeJongh, M.; Henry, C.S. Constructing and Analyzing Metabolic Flux Models of Microbial Communities; Springer: Heidelberg, Germany, 2016; pp. 247–273. [Google Scholar]
- Henry, C.S.; Bernstein, H.C.; Weisenhorn, P.; Taylor, R.C.; Lee, J.; Zucker, J.; Song, H. Microbial Community Metabolic Modeling: A Community Data-Driven Network Reconstruction. J. Cell. Physiol. 2016, 231, 2339–2345. [Google Scholar] [CrossRef]
- Seaver, S.M.D.; Bradbury, L.M.T.; Frelin, O.; Zarecki, R.; Ruppin, E.; Hanson, A.D.; Henry, C.S. Improved Evidence-Based Genome-Scale Metabolic Models for Maize Leaf, Embryo, and Endosperm. Front. Plant Sci. 2015, 6, 142. [Google Scholar] [CrossRef]
- Available online: https://www.fortunebusinessinsights.com/industry-reports/agricultural-microbial-market-100412 (accessed on 30 October 2023).
- Basu, A.; Prasad, P.; Das, S.N.; Kalam, S.; Sayyed, R.Z.; Reddy, M.S.; El Enshasy, H. Plant Growth Promoting Rhizobacteria (PGPR) as Green Bioinoculants: Recent Developments, Constraints, and Prospects. Sustainability 2021, 13, 1140. [Google Scholar] [CrossRef]
- Soumare, A.; Diedhiou, A.G.; Thuita, M.; Hafidi, M.; Ouhdouch, Y.; Gopalakrishnan, S.; Kouisni, L. Exploiting Biological Nitrogen Fixation: A Route Towards a Sustainable Agriculture. Plants 2020, 9, 1011. [Google Scholar] [CrossRef]
- Tang, A.; Haruna, A.O.; Majid, N.M.A.; Jalloh, M.B. Potential PGPR Properties of Cellulolytic, Nitrogen-Fixing, Phosphate-Solubilizing Bacteria in Rehabilitated Tropical Forest Soil. Microorganisms 2020, 8, 442. [Google Scholar] [CrossRef]


| Type of Bacterial Biofertilizer | Species (Genus) of Bacteria |
|---|---|
| Nitrogen fixation | Acetobacter diazotrophicus, Acetobacter sp., Azoarcus sp., Azospirillum brasilense, Azospirillum lipoferum, Azospirillum sp., Azotobacter chroococcum 2, Azotobacter vinelandii, Azotobacter sp., Bacllius megaterium 1, Bacillus subtilis 1,4, Bradyrhizobium elkanii, Bradyrhizobium japonicum, Delftia acidovorans, Mesorhizobium ciceri, Penicillium bilaii, Paenibacillus polymyxa, Rhizobium japonicum, Rhizobium sp., Rhizophagus irregularis 1 |
| Phosphate solubilization | Bacillus megaterium 1, Bacillus mucilaginosus, Bacillus polymyxa, Bacillus subtilis 1, Bacillus spp., Frateuria aurantia, Glomus intraradices, Pseudomonas fluorescens 3, Pseudomonas striata, Rhizophagus irregularis 1 |
| Solubilization of Potassium and Zinc | Frateuria aurantia, Thiobacillus thiooxidans |
| Stimulation of plant growth | Azotobacter chroococcum 2, Pseudomonas azotoformans, Pseudomonas fluorescens 3 |
| Biocontrolling | Bacillus subtilis 4, Brevibacillus laterosporus, Paenibacillus chitinolyticus, Pseudomonas chlororaphis |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Timofeeva, A.M.; Galyamova, M.R.; Sedykh, S.E. Plant Growth-Promoting Bacteria of Soil: Designing of Consortia Beneficial for Crop Production. Microorganisms 2023, 11, 2864. https://doi.org/10.3390/microorganisms11122864
Timofeeva AM, Galyamova MR, Sedykh SE. Plant Growth-Promoting Bacteria of Soil: Designing of Consortia Beneficial for Crop Production. Microorganisms. 2023; 11(12):2864. https://doi.org/10.3390/microorganisms11122864
Chicago/Turabian StyleTimofeeva, Anna M., Maria R. Galyamova, and Sergey E. Sedykh. 2023. "Plant Growth-Promoting Bacteria of Soil: Designing of Consortia Beneficial for Crop Production" Microorganisms 11, no. 12: 2864. https://doi.org/10.3390/microorganisms11122864
APA StyleTimofeeva, A. M., Galyamova, M. R., & Sedykh, S. E. (2023). Plant Growth-Promoting Bacteria of Soil: Designing of Consortia Beneficial for Crop Production. Microorganisms, 11(12), 2864. https://doi.org/10.3390/microorganisms11122864







