Changing Strategies for the Detection of Bacteria in Platelet Components in Ireland: From Primary and Secondary Culture (2010–2020) to Large Volume Delayed Sampling (2020–2023)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Platelet Products at the IBTS
2.2. Bacterial Screening of Platelets
2.2.1. Pre-November 2020 (Pre-LVDS)
2.2.2. Post-November 2020 (Post-LVDS)
2.2.3. Testing at Expiry
2.3. Processing of Positive BACT/ALERT Bottles
2.4. Product Recall
2.5. Classification of Positive Platelets
- Confirmed Positive refers to a positive signal on BACT/ALERT 3D, a positive subculture from the BACT/ALERT bottle, and a further culture of the same species from the platelet unit, or, for pools, from the pool or from one other component from the donations used in the pool.
- False Positive refers to a positive signal on BACT/ALERT 3D, but no organism detected on subculture or Gram stain of the associated bottles, or on reculture of the unit.
- Indeterminate Positive (adapted from Benjamin et al. [8]): positive subculture from the initial BACT/ALERT bottle but not confirmed on subsequent reculture of remaining components, irrespective of the proportion of the associated components available for testing (i.e., all components may have been available for testing *, or only a proportion of them); or no residual components were available for retesting. * Please note these cases are not considered false positive by our definitions.
3. Results
3.1. Pre-LVDS Period
3.2. Post-LVDS Period
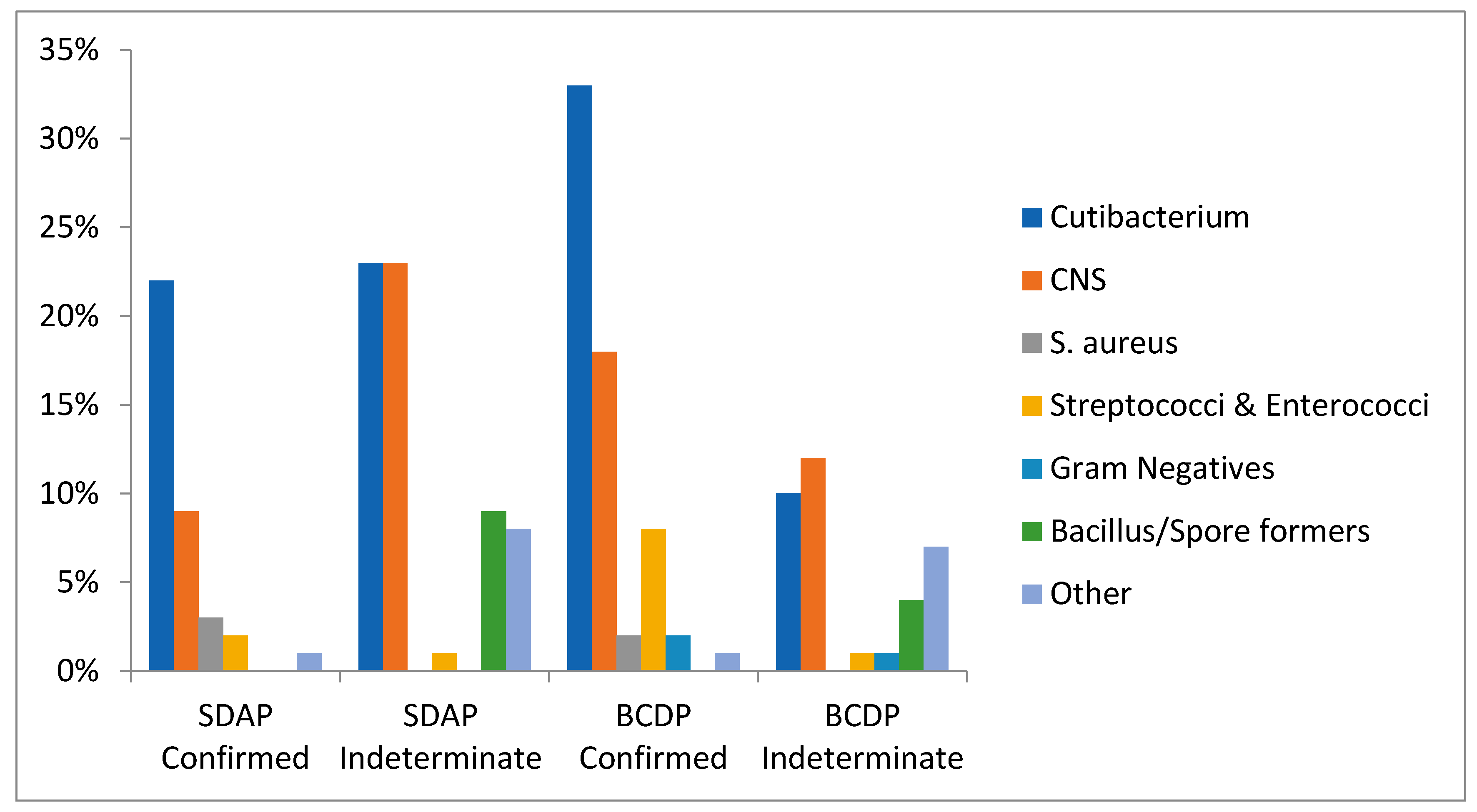
3.3. Organisms Isolated Pre-LVDS
3.4. Co-Components
3.5. Organisms Isolated Post-LVDS
3.6. Positivity According to BACT/ALERT Bottle Type BPA/BPN
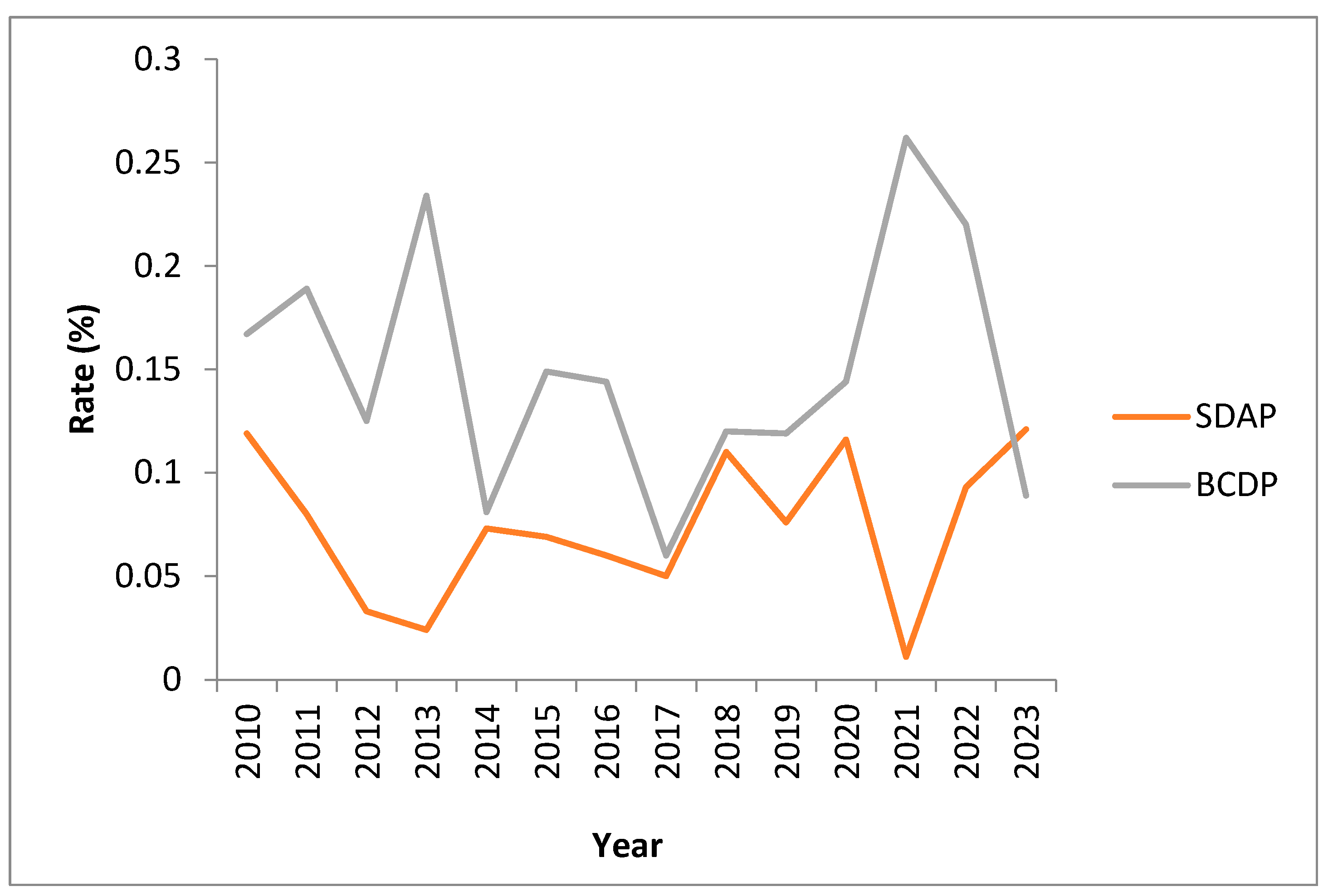
3.7. Positivity Rates over Time
3.8. Residual Risk: Outdate Testing and False Negative BACT/ALERT Screening Results
4. Discussion
4.1. IBTS Data on Bacterial Contamination of Platelets 2010–2020
4.2. Buffy Coat-Derived Platelet data—Cessation of the Second Day-4 Test
4.3. Single-Donor Apheresis Platelets 2010–2020
4.4. Limitations of IBTS LVDS Strategy for Apheresis Platelets (SDAP)
4.5. Close Calls—Rationale for the Value of a 12 h Quarantine
4.6. BCDP Results after Introduction of the Single-Test LVDS Protocol
4.7. Apheresis Platelet Contamination Rates Post-2020
4.8. Bacterial Contamination and Detection Home and Away
4.9. Septic Transfusion Reactions
5. Limitations
6. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- James, J.D. Bacterial Contamination of Preserved Blood. Vox Sang. 1959, 4, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Bonnel, P.H. Bacterial Contamination of Preserved Blood and Derivatives. Vox Sang. 1961, 6, 60–67. [Google Scholar] [CrossRef]
- Robillard, P.; Delage, G.; Itaj, N.K.; Goldman, M. Use of hemovigilance data to evaluate the effectiveness of diversion and bacterial detection. Transfusion 2011, 51, 1405–1411. [Google Scholar] [CrossRef] [PubMed]
- Kelton, J.G.; Blajchman, M.A. Platelet transfusions. Can. Med. Assoc. J. 1979, 121, 1353–1358. [Google Scholar] [PubMed]
- Wagner, S.J.; Friedman, L.I.; Dodd, R.Y. Transfusion-associated bacterial sepsis. Clin. Microbiol. Rev. 1994, 7, 290–302. [Google Scholar] [CrossRef]
- Pietersz, R.N.I.; Reesink, H.W.; Panzer, S.; Oknaian, S.; Kuperman, S.; Gabriel, C.; Rapaille, A.; Lambermont, M.; Deneys, V.; Sondag, D.; et al. Bacterial contamination in platelet concentrates. Vox Sang. 2014, 106, 256–283. [Google Scholar] [CrossRef]
- McDonald, C.P. Bacterial risk reduction by improved donor arm disinfection, diversion and bacterial screening. Transfus. Med. 2006, 16, 381–396. [Google Scholar] [CrossRef]
- Benjamin, R.J.; McDonald, C.P. The International Experience of Bacterial Screen Testing of Platelet Components with an Automated Microbial Detection System: A Need for Consensus Testing and Reporting Guidelines. Transfus. Med. Rev. 2014, 28, 61–71. [Google Scholar] [CrossRef]
- Horth, R.Z. Fatal Sepsis Associated with Bacterial Contamination of Platelets—Utah and California, August 2017. MMWR Morb. Mortal. Wkly. Rep. 2018, 67, 718–722. Available online: https://www.cdc.gov/mmwr/volumes/67/wr/mm6725a4.htm (accessed on 21 September 2023). [CrossRef]
- Jones, S.A.; Jones, J.M.; Leung, V.; Nakashima, A.K.; Oakeson, K.F.; Smith, A.R.; Hunter, R.; Kim, J.J.; Cumming, M.; McHale, E.; et al. Sepsis Attributed to Bacterial Contamination of Platelets Associated with a Potential Common Source—Multiple States, 2018. MMWR Morb. Mortal. Wkly. Rep. 2019, 68, 519–523. [Google Scholar] [CrossRef]
- Nevala-Plagemann, C.; Powers, P.; Mir-Kasimov, M.; Rose, R. A Fatal Case of Septic Shock Secondary to Acinetobacter Bacteremia Acquired from a Platelet Transfusion. Case Rep. Med. 2019, 2019, 3136493. [Google Scholar] [CrossRef] [PubMed]
- Fridey, J.L.; Stramer, S.L.; Nambiar, A.; Moayeri, M.; Bakkour, S.; Langelier, C.; Crawford, E.; Lu, T.; Lanteri, M.C.; Kamm, J.; et al. Sepsis from an apheresis platelet contaminated with Acinetobacter calcoaceticus/baumannii complex bacteria and Staphylococcus saprophyticus after pathogen reduction. Transfusion 2020, 60, 1960–1969. [Google Scholar] [CrossRef] [PubMed]
- Murphy, W.G.; Foley, M.; Doherty, C.; Tierney, G.; Kinsella, A.; Salami, A.; Cadden, E.; Coakley, P. Screening platelet concentrates for bacterial contamination: Low numbers of bacteria and slow growth in contaminated units mandate an alternative approach to product safety. Vox Sang. 2008, 95, 13–19. [Google Scholar] [CrossRef] [PubMed]
- McDonald, C.; Allen, J.; Brailsford, S.; Roy, A.; Ball, J.; Moule, R.; Vasconcelos, M.; Morrison, R.; Pitt, T. Bacterial screening of platelet components by National Health Service Blood and Transplant, an effective risk reduction measure. Transfusion 2017, 57, 1122–1131. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Arcos, S.; Evans, S.; McIntyre, T.; Pang, C.; Yi, Q.L.; DiFranco, C.; Goldman, M. Extension of platelet shelf life with an improved bacterial testing algorithm. Transfusion 2020, 60, 2918–2928. [Google Scholar] [CrossRef] [PubMed]
- Domanović, D.; Cassini, A.; Bekeredjian-Ding, I.; Bokhorst, A.; Bouwknegt, M.; Facco, G.; Galea, G.; Grossi, P.; Jashari, R.; Jungbauer, C.; et al. Prioritizing of bacterial infections transmitted through substances of human origin in Europe. Transfusion 2017, 57, 1311–1317. [Google Scholar] [CrossRef] [PubMed]
- Bloch, E.M.; Marshall, C.E.; Boyd, J.S.; Shifflett, L.; Tobian, A.A.R.; Gehrie, E.A.; Ness, P.M. Implementation of secondary bacterial culture testing of platelets to mitigate residual risk of septic transfusion reactions. Transfusion 2018, 58, 1647–1653. [Google Scholar] [CrossRef]
- Cloutier, M.; De Korte, D.; ISBT Transfusion-Transmitted Infectious Diseases Working Party, Subgroup on Bacteria. Residual risks of bacterial contamination for pathogen-reduced platelet components. Vox Sang. 2022, 117, 879–886. [Google Scholar] [CrossRef]
- Liu, H.; Wang, X. Pathogen reduction technology for blood component: A promising solution for prevention of emerging infectious disease and bacterial contamination in blood transfusion services. J. Photochem. Photobiol. 2021, 8, 100079. [Google Scholar] [CrossRef]
- Prax, M.; Bekeredjian-Ding, I.; Krut, O. Microbiological Screening of Platelet Concentrates in Europe. Transfus. Med. Hemother. 2019, 46, 76–86. [Google Scholar] [CrossRef]
- de Korte, D.; Marcelis, J.H. Platelet concentrates: Reducing the risk of transfusion-transmitted bacterial infections. Int. J. Clin. Transfus. Med. 2014, 2, 29–37. [Google Scholar] [CrossRef]
- Doyle, B.; (IBTS, NBC, Dublin, Ireland). Platelet Stock Report 2022. Irish Blood Transfusion Service. Personal Communication, 2023. [Google Scholar]
- Walker, B.S.; Schmidt, R.L.; Fisher, M.A.; White, S.K.; Blaylock, R.C.; Metcalf, R.A. The comparative safety of bacterial risk control strategies for platelet components: A simulation study. Transfusion 2020, 60, 1723–1731. [Google Scholar] [CrossRef] [PubMed]
- Delage, G.; Bernier, F. Bacterial culture of platelets with the large volume delayed sampling approach: A narrative review. Ann. Blood 2021, 6. Available online: https://aob.amegroups.org/article/view/6453 (accessed on 30 September 2023). [CrossRef]
- McDonald, C.P.; Hartley, S.; Orchard, K.; Hughes, G.; Brett, M.M.; Hewitt, P.E.; Barbara, J.A. Fatal Clostridium perfringens sepsis from a pooled platelet transfusion. Transfus. Med. 1998, 8, 19–22. [Google Scholar] [CrossRef]
- Center for Biologics Evaluation and Research. Bacterial Risk Control Strategies for Blood Collection Establishments and Transfusion Services to Enhance the Safety and Availability of Platelets for Transfusion. FDA. 2022. Available online: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/bacterial-risk-control-strategies-blood-collection-establishments-and-transfusion-services-enhance (accessed on 26 October 2023).
- Kamel, H.; Townsend, M.; Bravo, M.; Vassallo, R.R. Improved yield of minimal proportional sample volume platelet bacterial culture. Transfusion 2017, 57, 2413–2419. [Google Scholar] [CrossRef]
- Kamel, H.; Ramirez-Arcos, S.; McDonald, C.; ISBT Transfusion-Transmitted Infectious Disease Bacterial Working Party Bacterial Subgroup. The international experience of bacterial screen testing of platelet components with automated microbial detection systems: An update. Vox Sang. 2022, 117, 647–655. [Google Scholar] [CrossRef]
- Kozakai, M.; Matsumoto, M.; Takakura, A.; Furuta, R.A.; Matsubayashi, K.; Goto, N.; Satake, M. Two cases of Streptococcus dysgalactiae subspecies equisimilis infection transmitted through transfusion of platelet concentrate derived from separate blood donations by the same donor. Vox Sang. 2023, 118, 582–586. [Google Scholar] [CrossRef]
- Gori, M.; Bolzoni, L.; Scaltriti, E.; Andriani, L.; Marano, V.; Morabito, F.; Fappani, C.; Cereda, D.; Giompapa, E.; Chianese, R.; et al. Listeria monocytogenes Transmission from Donated Blood to Platelet Transfusion Recipient, Italy. Emerg. Infect. Dis. 2023, 29, 2108–2111. Available online: https://wwwnc.cdc.gov/eid/article/29/10/23-0746_article (accessed on 27 September 2023). [CrossRef]
- Vollmer, T.; Dabisch-Ruthe, M.; Weinstock, M.; Knabbe, C.; Dreier, J. Late sampling for automated culture to extend the platelet shelf life to 5 days in Germany. Transfusion 2018, 58, 1654–1664. [Google Scholar] [CrossRef]
- Walker, B.S.; White, S.K.; Schmidt, R.L.; Metcalf, R.A. Residual bacterial detection rates after primary culture as determined by secondary culture and rapid testing in platelet components: A systematic review and meta-analysis. Transfusion 2020, 60, 2029–2037. [Google Scholar] [CrossRef]
- Thyer, J.; Perkowska-Guse, Z.; Ismay, S.L.; Keller, A.J.; Chan, H.T.; Dennington, P.M.; Bell, B.; Kotsiou, G.; Pink, J.M. Bacterial testing of platelets—Has it prevented transfusion-transmitted bacterial infections in Australia? Vox Sang. 2018, 113, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Kundrapu, S.; Srivastava, S.; Good, C.E.; Lazarus, H.M.; Maitta, R.W.; Jacobs, M.R. Bacterial contamination and septic transfusion reaction rates associated with platelet components before and after introduction of primary culture: Experience at a US Academic Medical Center 1991 through 2017. Transfusion 2020, 60, 974–985. [Google Scholar] [CrossRef] [PubMed]
- Center for Biologics Evaluation and Research. Important Information for Blood Establishments and Transfusion Services Regarding Bacterial Contamination of Platelets for Transfusion. FDA. 2022. Available online: https://www.fda.gov/vaccines-blood-biologics/safety-availability-biologics/important-information-blood-establishments-and-transfusion-services-regarding-bacterial-0 (accessed on 30 September 2023).
- Department of Agriculture, Food and Marine. Ireland’s National Action Plan for Antimicrobial Resistance 2017–2020 (iNAP). 2019. Available online: https://www.gov.ie/en/publication/babe6-irelands-national-action-plan-for-antimicrobial-resistance-2017-2020-inap/ (accessed on 25 September 2023).
- Department of Health. Antimicrobial Resistance (AMR). 2019. Available online: https://www.gov.ie/en/collection/8f0dd5-antimicrobial-resistance-amr/?referrer=http://www.health.gov.ie/national-patient-safety-office/patient-safety-surveillance/antimicrobial-resistance-amr/ (accessed on 30 September 2023).
- Hong, H.; Xiao, W.; Lazarus, H.M.; Good, C.E.; Maitta, R.W.; Jacobs, M.R. Detection of septic transfusion reactions to platelet transfusions by active and passive surveillance. Blood 2016, 127, 496–502. [Google Scholar] [CrossRef]
- Jacobs, M.R.; Good, C.E.; Lazarus, H.M.; Yomtovian, R.A. Relationship between Bacterial Load, Species Virulence, and Transfusion Reaction with Transfusion of Bacterially Contaminated Platelets. Clin. Infect. Dis. 2008, 46, 1214–1220. [Google Scholar] [CrossRef] [PubMed]
- Lorusso, A.; Croxon, H.; Faherty-O’Donnell, S.; Field, S.; Fitzpatrick, Á.; Farrelly, A.; Hervig, T.; Waters, A. The impact of donor biological variation on the quality and function of cold-stored platelets. Vox Sang. 2023, 118, 730–737. [Google Scholar] [CrossRef]
- Heltberg, O.; Skov, F.; Gerner-Smidt, P.; Kolmos, H.J.; Dybkjaer, E.; Gutschik, E.; Jerne, D.; Jepsen, O.; Weischer, M.; Frederiksen, W.; et al. Nosocomial epidemic of Serratia marcescens septicemia ascribed to contaminated blood transfusion bags. Transfusion 1993, 33, 221–227. [Google Scholar] [CrossRef]
- Hogman, C.F.; Fritz, H.; Sandberg, L. Posttransfusion Serratia marcescens septicemia. Transfusion 1993, 33, 189–191. [Google Scholar] [CrossRef]
- Loza-Correa, M.; Kou, Y.; Taha, M.; Kalab, M.; Ronholm, J.; Schlievert, P.M.; Cahill, M.P.; Skeate, R.; Cserti-Gazdewich, C.; Ramirez-Arcos, S. Septic transfusion case caused by a platelet pool with visible clotting due to contamination with Staphylococcus aureus. Transfusion 2017, 57, 1299–1303. [Google Scholar] [CrossRef]
- Chi, S.I.; Yousuf, B.; Paredes, C.; Bearne, J.; McDonald, C.; Ramirez-Arcos, S. Proof of concept for detection of staphylococcal enterotoxins in platelet concentrates as a novel safety mitigation strategy. Vox Sang. 2023, 118, 543–550. [Google Scholar] [CrossRef]
- Tan, C.C.S.; Ko, K.K.K.; Chen, H.; Liu, J.; Loh, M.; Chia, M.; Nagarajan, N.; SG10K_Health Consortium. No evidence for a common blood microbiome based on a population study of 9,770 healthy humans. Nat. Microbiol. 2023, 8, 973–985. [Google Scholar] [CrossRef]
- Kumaran, D.; Kalab, M.; Rood, I.G.H.; de Korte, D.; Ramirez-Arcos, S. Adhesion of anaerobic bacteria to platelet containers. Vox Sang. 2014, 107, 188–191. [Google Scholar] [CrossRef] [PubMed]
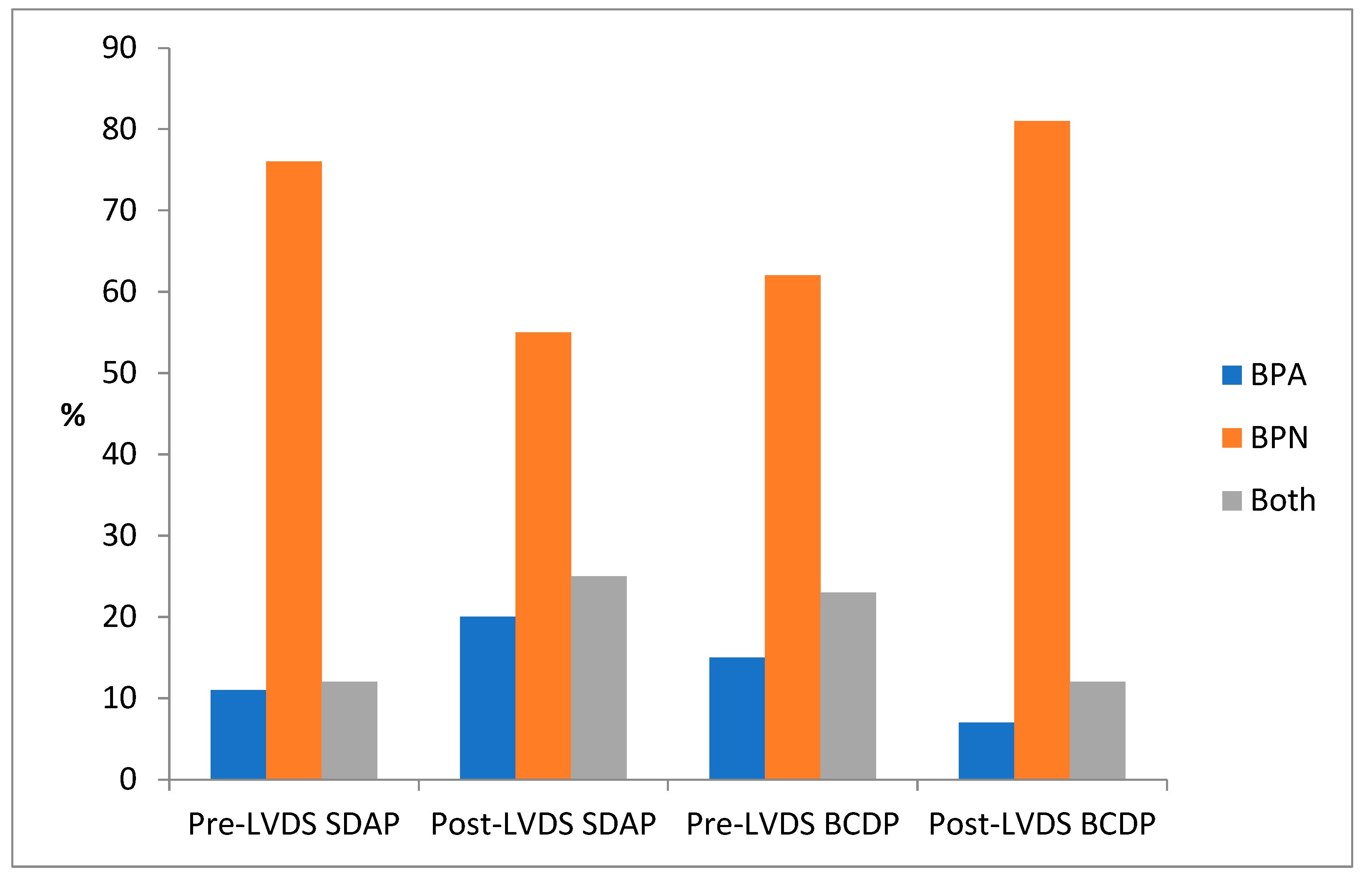
| Primary Culture Time (h) | Volume (mL) | 1st Culture Component(s) Sampled | 2nd Culture Component(s) Sampled | Post-Sample Hold (h) | Length of Screening (d) | ||
|---|---|---|---|---|---|---|---|
| Pre-LVDS (2010–2020) | SDAP | 12–24 (Day-1) | 16 * | Pre-spilt (mother bag) | Each split | No formal quarantine | 5 (7 **) |
| BCDP | ≥36 (Day-2) | 16 * | Each pool | Each pool | No formal quarantine | 5 (7 **) | |
| Post-LVDS (2020-Present) | SDAP | ≥36 (Day-2) | 16 * | Pre-spilt (mother bag) | NA | ≥12 | 7 |
| BCDP | ≥36 (Day-2) | 16 * | Each pool | NA | ≥12 | 7 |
| Pre-LVDS 2010–2020 | Total Platelets Screened | Total Positive Platelet Rate (%) 95% CI Incidence | Total SDAP Screened (Day-1) | Total SDAP Rate (%) 95% CI Incidence | Total SDAP Screened on Day-4 (Extended SDAP) | Total Extended SDAP Rate (%) 95% CI Incidence | Total BCDP Screened (Day-2) | Total BCDP Rate (%) 95% CI Incidence | Total BCDP Screened on Day-4 (Extended BCDP) | Total Extended BCDP Rate (%) 95% CI Incidence |
|---|---|---|---|---|---|---|---|---|---|---|
| Number of tests | 287,698 | 110,497 | 89,426 * | 64,732 | 23,043 * | |||||
| Confirmed | 101 | 0.035 [0.029–0.043] (1:2848) | 29 | 0.026 [0.018–0.038] (1:3810) | 8 a | 0.009 [0.004–0.018] (1:11,178) | 57 | 0.088 [0.067–0.114] (1:1136) | 7 | 0.030 [0.012–0.063] (1:3292) |
| Indeterminate Positive | 77 | 0.027 [0.021–0.033] (1:3736) | 43 | 0.039 [0.028–0.052] (1:2570) | 7 | 0.008 [0.003–0.016] (1:12,775) | 24 | 0.037 [0.024–0.055] (1:2697) | 3 | 0.013 [0.003–0.038] (1:7681) |
| Combined rate | 178 | 0.062 [0.053–0.072] (1:1616) | 72 | 0.065 [0.051–0.082] (1:1535) | 15 | 0.017 [0.009–0.028] (1:5962) | 81 | 0.125 [0.099–0.156] (1:799) | 10 | 0.043 [0.021–0.080] (1:2304) |
| False Positive | 202 | 0.070 [0.061–0.081] (1:1424) | 95 | 0.086 [0.070–0.105] (1:1163) | 29 | 0.032 [0.022–0.047] (1:3083) | 67 | 0.104 [0.080–0.131] (1:966) | 11 | 0.048 [0.024–0.085] (1:2095) |
| Pre-LVDS | Post-LVDS | |||
|---|---|---|---|---|
| SDAP | BCDP | SDAP | BCDP | |
| All components tested | 20 | 9 | 3 | 3 |
| Some components tested (not all available) | 7 | 18 | 0 | 8 |
| Components not available to test | 23 | 0 | 3 | 0 |
| Post-LVDS 2020–2023 | Total Platelets Screened | Total Positive Platelet Rate (%) 95% CI Incidence | Total SDAP Screened (Day-2) | Total SDAP Rate (%) 95% CI Incidence | Total BCDP Screened (Day-2) | Total BCDP Rate (%) 95% CI Incidence |
|---|---|---|---|---|---|---|
| Number of tests | 44,642 | 23,538 | 20,989 | |||
| Confirmed | 40 | 0.090 [0.064–0.122] (1:1116) | 11 | 0.047 [0.023–0.084] (1:2140) | 29 | 0.138 [0.093–0.198] (1:724) |
| Indeterminate Positive | 17 | 0.038 [0.022–0.061] (1:2626) | 6 | 0.025 [0.009–0.055] (1:3923) | 11 | 0.052 [0.026–0.094] (1:1908) |
| Combined rate | 57 | 0.128 [0.97–0.165] (1:783) | 17 | 0.072 [0.042–0.116] (1:1385) | 40 | 0.191 [0.136–0.259] (1:525) |
| False Positive | 3 | 0.007 [0.001–0.020] (1:14,881) | 3 | 0.013 [0.003–0.037] (1:7846) | 0 | 0.000 [0.000–0.018] (<1:20,989) |
| Group | Organism | SDAP Primary Culture | BCDP Primary Culture | Mean TTD (h) | SDAP Extension Day-4 | BCDP Extension Day-4 | Infused N = 31 (%) |
|---|---|---|---|---|---|---|---|
| Cutibacterium | Cutibacterium acnes/ S. epidermidis | - | 1 | 93.1 | - | - | 0 (0) |
| Cutibacterium acnes/spp. | 21 | 29 | 92.51 | 1 | 3 | 26 (84) | |
| CNS | Coag Neg Staph | 2 | 1 | 46.96 | 1 | - | 1 (3) |
| S. epidermidis | 1 | 9 | 18.5 | 2 | 3 | 1 (3) | |
| S. capitis | 2 | 2 | 28.92 | - | - | 0 (0) | |
| S. saccharolyticus | 1 | 2 | 61.28 | - | - | 1 (3) | |
| Microaerophilic Staphylococci | - | 1 | 69.4 | - | - | 0 (0) | |
| Gram-positive cocci | Kocuria varians | - | 1 | 59.52 | - | - | 0 (0) |
| Pathogenic Gram-positive rods | Listeria monocytogenes | 1 | - | 18.5 | - | - | 0 (0) |
| Staphylococcus aureus | S. aureus | - | 1 | 12.24 | 3 | 1 | 1 (3) |
| Streptococci and enterococci | S. dysgalactiae | - | 5 | 8.74 | - | - | 1 (3) |
| S. mitis/oralis | 1 | - | 13.4 | - | - | 0 (0) | |
| S. pneumoniae | - | 2 | 16.1 | - | - | 0 (0) | |
| S. infantarius | - | - | - | 1 | - | 0 (0) | |
| Peptostreptococcus micros | - | 1 | 55.9 | - | - | 0 (0) | |
| Gram-negatives | E. coli | - | 1 | 5.3 | - | - | 0 (0) |
| S. marcescens | - | 1 | 7.44 | - | - | 0 (0) |
| All Apheresis Splits Infused (%) | Associated Split(s) Infused (%) | BCDP-only Infused (%) | BCDP and Associated Red Cell Infused (%) | Associated Red Cell-only Infused (%) | ||
|---|---|---|---|---|---|---|
| Pre-LVDS | Apheresis (n = 87) | 24 (28) | 13 (15) | - | - | - |
| BCDP (n = 91) | - | - | 37 (41) | 2 (2) | 4 (4) | |
| Post-LVDS | Apheresis (n = 17) | 3 (18) | 1 (6) | - | - | - |
| BCP (n = 40) | - | - | 10 (25) | 8 (20) | 3 (8) |
| Group | Organism | SDAP Primary Culture | Expired SDAP | BCDP Primary Culture | Expired BCDP | Mean TTD (h) | Infused (%) |
|---|---|---|---|---|---|---|---|
| Skin commensals | C. acnes | 5 | 1 | 17 | 1 | 98.86 | 13 (93) |
| ’Coagulase-negative staphylococci‘ * | S. capitis | 1 | - | 0 | - | 19.92 | 0 (0) |
| S. epidermidis | 1 | - | 1 | - | 19.32 | 0 (0) | |
| S. lugdunensis | - | - | 1 | - | 19.44 | 0 (0) | |
| S. saccharolyticus | - | - | 7 | - | 50.98 | 1 (7) | |
| Mixed skin flora | - | - | 1 | - | 21.12 | 0 (0) | |
| S. aureus | S. aureus | 2 | 1 | 1 | - | 8.64 | 0 (0) |
| Streptococci and Enterococci | S. gallolyticus | 1 | - | - | - | 9.36 | 0 (0) |
| S. dysgalactiae | - | - | 1 | - | 9.12 | 0 (0) | |
| E. casseliflavus | 1 | - | - | - | 4.08 | 0 (0) |
| Expired Platelets Post-LVDS | Total Expired Platelets | Total Expired Platelets Rate (%) 95% CI Incidence | Expired SDAP | Expired SDAP Rate (%) 95% CI Incidence | Expired BCDP | Expired BCDP Rate (%) 95% CI Incidence |
|---|---|---|---|---|---|---|
| Number of Tests | 6809 | 5540 | 1269 | |||
| Confirmed | 3 | 0.044 | 2 | 0.036 | 1 | 0.079 |
| [0.009–0.129] | [0.004–0.130] | [0.002–0.438] | ||||
| (1:2270) | (1:2770) | (1:1269) | ||||
| Indeterminate Positive | 1 | 0.015 | 1 | 0.018 | 0 | 0.000 |
| [0.000–0.082] | [0.000–0.101] | [0.000–0.290] | ||||
| (1:6809) | (1:5540) | (<1:1269) | ||||
| Combined rate | 4 | 0.059 | 3 | 0.054 | 1 | 0.079 |
| [0.016–0.150] | [0.011–0.158] | [0.002–0.438] | ||||
| (1:1723) | (1:1847) | (1:1269) | ||||
| False Positive | 3 | 0.044 | 3 | 0.054 | 0 | 0.000 |
| [0.009–0.129] | [0.011–0.158] | [0.000–0.290] | ||||
| (1:2270) | (1:1847) | (<1:1269) |
| Expired Platelets Pre-LVDS | Total Expired Platelets | Total Expired Platelets Rate (%) 95% CI Incidence | Expired SDAP | Expired SDAP Rate (%) 95% CI Incidence | Expired BCDP | Expired BCDP Rate (%) 95% CI Incidence |
|---|---|---|---|---|---|---|
| Number of Tests | 4987 | 3519 | 1468 | |||
| Confirmed | 1 | 0.020 [0.001–0.112] (1:4987) | 0 | 0.000 [0.000–0.105] (<1:3519) | 1 | 0.068 [0.002–0.379] (1:1468) |
| Indeterminate Positive | 4 | 0.080 [0.022–0.205] (1:1247) | 3 | 0.085 [0.018–0.249] (1:1173) | 1 | 0.068 [0.002–0.379] (1:1468) |
| Combined rate | 5 | 0.100 [0.033–0.234] (1:997) | 3 | 0.085 [0.018–0.249] (1:1173) | 2 | 0.136 [0.017–0.491] (1:734) |
| False Positive | 17 | 0.341 [0.199–0.545] (1:293) | 12 | 0.341 [0.176–0.595] (1:293) | 5 | 0.341 [0.111–0.793] (1:294) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
O’Flaherty, N.; Bryce, L.; Nolan, J.; Lambert, M. Changing Strategies for the Detection of Bacteria in Platelet Components in Ireland: From Primary and Secondary Culture (2010–2020) to Large Volume Delayed Sampling (2020–2023). Microorganisms 2023, 11, 2765. https://doi.org/10.3390/microorganisms11112765
O’Flaherty N, Bryce L, Nolan J, Lambert M. Changing Strategies for the Detection of Bacteria in Platelet Components in Ireland: From Primary and Secondary Culture (2010–2020) to Large Volume Delayed Sampling (2020–2023). Microorganisms. 2023; 11(11):2765. https://doi.org/10.3390/microorganisms11112765
Chicago/Turabian StyleO’Flaherty, Niamh, Louise Bryce, James Nolan, and Mark Lambert. 2023. "Changing Strategies for the Detection of Bacteria in Platelet Components in Ireland: From Primary and Secondary Culture (2010–2020) to Large Volume Delayed Sampling (2020–2023)" Microorganisms 11, no. 11: 2765. https://doi.org/10.3390/microorganisms11112765
APA StyleO’Flaherty, N., Bryce, L., Nolan, J., & Lambert, M. (2023). Changing Strategies for the Detection of Bacteria in Platelet Components in Ireland: From Primary and Secondary Culture (2010–2020) to Large Volume Delayed Sampling (2020–2023). Microorganisms, 11(11), 2765. https://doi.org/10.3390/microorganisms11112765





