Bacillus subtilis RBT-7/32 and Bacillus licheniformis RBT-11/17 as New Promising Strains for Use in Probiotic Feed Additives
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microorganisms
2.2. Media Composition for the Strain Maintenance and Morphological and Cultural Studies
2.3. Strain Isolation and Identification
2.4. Cultivation of Bacillus Strains on a Liquid Nutrient Medium
- Medium 1: soybean meal, 20.0; NaNO3, 3.0; K2HPO4, 1.0; MgSO4, 0.2; NaCl, 3.0;
- Medium 2: corn meal, 25.0; NaNO3, 3.0; K2HPO4, 1.0; MgSO4, 0.2; NaCl, 3.0;
- Medium 3: pea meal, 25.0; NaNO3, 3.0; K2HPO4, 1.0; MgSO4, 0.2; NaCl, 3.0;
- All three media were prepared using a tap water (pH 6.8–7.0).
2.5. Obtaining of a Dry Biomass of Bacillus Strains
2.6. UV Mutagenesis of Bacillus Strains
2.7. Antagonistic Activity Assay
2.8. Evaluation of the Bacterial Growth Dynamics
2.9. Cellulolytic Activity Assay
2.10. Amylolytic Activity Assay
2.11. Evaluation of Spore Resistance to Divverent pH Levels
2.12. Determination of the Viability of Bacillus Cells after Exposure to Organic Acid Solutions
2.13. Determination of Spore Resistance to High Temperatures
2.14. Antibiotic Sensitivity Assay
2.15. Statistical Data Treatment
3. Results
3.1. Strain Isolation and Identification
3.2. Improvement of the Target Activity of B. subtilis and B. licheniformis
3.3. Growth of B. subtilis RBT-7/32 and B. licheniformis RBT-11/17 Strains on Different Substrates
3.4. Evaluation of the Spore Germination of Mutant Strains at Different pH Levels
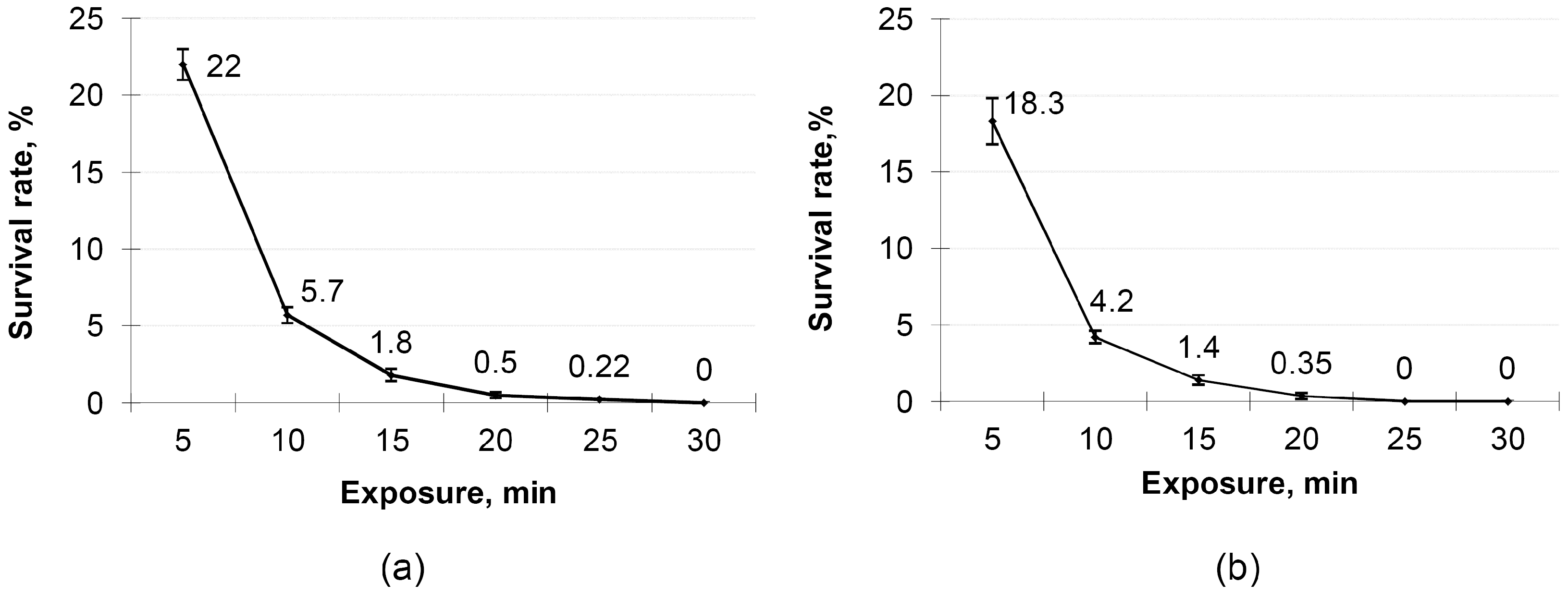
3.5. Evaluation of the Heat Tolerance of Mutant B. subtilis and B. licheniformis Strains
3.6. Antibiotic Sensitivity of Mutant B. subtilis RBT-7/32 and B. licheniformis RBT-11/17 Strains
3.7. Survivability of Mutant B. subtilis RBT-7/32 and B. licheniformis RBT-11/17 Strains in Organic Acid Solutions
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Adedokun, S.A.; Olojede, O.C. Optimizing gastrointestinal integrity in poultry: The role of nutrients and feed additives. Front. Vet. Sci. 2019, 31, 348. [Google Scholar] [CrossRef] [PubMed]
- Alagawany, M.; Elnesr, S.S.; Farag, M.R.; Abd El-Hack, M.E.; Barkat, R.A.; Gabr, A.A.; Foda, M.A.; Noreldin, A.E.; Khafaga, A.F.; El-Sabrout, K.; et al. Potential role of important nutraceuticals in poultry performance and health—A comprehensive review. Res. Vet. Sci. 2021, 137, 9–29. [Google Scholar] [CrossRef] [PubMed]
- El-Sabrout, K.; Khalifah, A.; Mishra, B. Application of botanical products as nutraceutical feed additives for improving poultry health and production. Vet. World 2023, 16, 369–379. [Google Scholar] [CrossRef] [PubMed]
- O’Hara, E.; Neves, A.L.; Song, Y.; Guan, L.L. The role of the gut microbiome in cattle production and health: Driver or passenger? Ann. Rev. Anim. Biosci. 2020, 8, 199–220. [Google Scholar] [CrossRef]
- Aruwa, C.E.; Pillay, C.; Nyaga, M.M.; Sabiu, S. Poultry gut health–microbiome functions, environmental impacts, microbiome engineering and advancements in characterization technologies. J. Anim. Sci. Biotechnol. 2021, 12, 119. [Google Scholar] [CrossRef]
- Plaizier, J.C.; Danesh Mesgaran, M.; Derakhshani, H.; Golder, H.; Khafipour, E.; Kleen, J.L.; Lean, I.; Loor, J.; Penner, G.; Zebeli, Q. Review: Enhancing gastrointestinal health in dairy cows. Animal 2018, 12, 399–418. [Google Scholar] [CrossRef]
- Pluske, J.R.; Turpin, D.L.; Kim, J.-C. Gastrointestinal tract (gut) health in the young pig. Anim. Nutr. 2017, 4, 187–196. [Google Scholar] [CrossRef]
- Villena, J.; Aso, H.; Rutten, V.P.M.G.; Takahashi, H.; van Eden, W.; Kitazawa, H. Immunobiotics for the bovine host: Their interaction with intestinal epithelial cells and their effect on antiviral immunity. Front. Immunol. 2018, 2, 326. [Google Scholar] [CrossRef]
- Álvarez-Martínez, F.J.; Barrajón-Catalán, E.; Micol, V. Tackling antibiotic resistance with compounds of natural origin: A comprehensive review. Biomedicines 2020, 8, 405. [Google Scholar] [CrossRef]
- Iramiot, J.S.; Kajumbula, H.; Bazira, J.; Kansiime, C.; Asiimwe, B.B. Antimicrobial resistance at the human-animal interface in the pastoralist communities of Kasese District, South Western Uganda. Sci. Rep. 2020, 10, 14737. [Google Scholar] [CrossRef]
- Fathima, S.; Shanmugasundaram, R.; Adams, D.; Selvaraj, R. Gastrointestinal microbiota and their manipulation for improved growth and performance in chickens. Foods 2022, 11, 1401. [Google Scholar] [CrossRef] [PubMed]
- Melara, E.G.; Avellaneda, M.C.; Valdivié, M.; García-Hernández, Y.; Aroche, R.; Martínez, Y. Probiotics: Symbiotic relationship with the animal host. Animals 2022, 12, 719. [Google Scholar] [CrossRef] [PubMed]
- Al-Khalaifah, H.S. Benefits of probiotics and/or prebiotics for antibiotic-reduced poultry. Poult. Sci. 2018, 97, 3807–3815. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Suzuki, Y.; Guan, L.L. Dissect the mode of action of probiotics in affecting host-microbial interactions and immunity in food producing animals. Vet. Immunol. Immunopathol. 2018, 205, 35–48. [Google Scholar] [CrossRef] [PubMed]
- AlGburi, A.; Volski, A.; Cugini, C.; Walsh, E.M.; Chistyakov, V.A.; Mazanko, M.S.; Bren, A.B.; Dicks, L.M.T.; Chikindas, M.L. Safety properties and probiotic potential of Bacillus subtilis KATMIRA1933 and Bacillus amyloliquefaciens B-1895. Adv. Microbiol. 2016, 6, 432–452. [Google Scholar] [CrossRef]
- Ruiz Sella, S.R.B.R.; Bueno, T.; de Oliveira, A.A.B.; Karp, S.G.; Soccol, C.R. Bacillus subtilis natto as a potential probiotic in animal nutrition. Crit. Rev. Biotechnol. 2021, 41, 355–369. [Google Scholar] [CrossRef]
- Hong, H.A.; To, E.; Fakhry, S.; Baccigalupi, L.; Ricca, E.; Cutting, S.M. Defining the natural habitat of Bacillus spore-formers. Res. Microbiol. 2009, 160, 375–379. [Google Scholar] [CrossRef]
- Bernardeau, M.; Lehtinen, M.J.; Forssten, S.D.; Nurminen, P. Importance of the gastrointestinal life cycle of Bacillus for probiotic functionality. J. Food Sci. Technol. 2017, 54, 2570–2584. [Google Scholar] [CrossRef]
- Alagawany, M.; Abd El-Hack, M.E.; Farag, M.R.; Sachan, S.; Karthik, K.; Dhama, K. The use of probiotics as eco-friendly alternatives for antibiotics in poultry nutrition. Environ. Sci. Pollut. Res. 2018, 25, 10611–10618. [Google Scholar] [CrossRef]
- Efremenkova, O.; Gabrielyan, N.; Malanicheva, I.; Demiankova, M.; Efimenko, T.; Rogozhin, E.; Sharapchenko, S.; Krupenio, T.; Davydov, D.; Kornilov, M. Antimicrobial properties of the probiotic strain Bacillus subtilis 534. Int. Arch. Med. Microbiol. 2019, 2, 119. [Google Scholar]
- Tran, C.; Cock, I.E.; Chen, X.; Feng, Y. Antimicrobial bacillus: Metabolites and their mode of action. Antibiotics 2022, 11, 88. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Chen, Y.; Imre, K.; Arslan-Acaroz, D.; Istanbullugil, F.R.; Fang, Y.; Ros, G.; Zhu, K.; Acaroz, U. Mechanisms of probiotic Bacillu against enteric bacterial infections. One Health Adv. 2023, 1, 21. [Google Scholar] [CrossRef]
- Newton, G.G. Antibiotics from a strain of B. subtilis; bacilipin A and B and bacilysin. Br. J. Exp. Pathol. 1949, 30, 306–319. [Google Scholar]
- Kenig, M.; Abraham, E.P. Antimicrobial activities and antagonists of bacilysin and anticapsin. J. Gen. Microbiol. 1976, 94, 37–45. [Google Scholar] [CrossRef]
- Brötz, H.; Bierbaum, G.; Leopold, K.; Reynolds, P.E.; Sahl, H.G. The lantibiotic mersacidin inhibits peptidoglycan synthesis by targeting lipid II. Antimicrob. Agents Chemother. 1998, 42, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Sass, P.; Jansen, A.; Szekat, C.; Sass, V.; Sahl, H.G.; Bierbaum, G. The lantibiotic mersacidin is a strong inducer of the cell wall stress response of Staphylococcus aureus. BMC Microbiol. 2008, 8, 186. [Google Scholar] [CrossRef]
- Rodrigues, B.; Morais, T.P.; Zaini, P.A.; Campos, C.S.; Almeida-Souza, H.O.; Dandekar, A.M.; Nascimento, R.; Goulart, L.R. Antimicrobial activity of epsilon-poly-l-lysine against phytopathogenic bacteria. Sci. Rep. 2020, 10, 11324. [Google Scholar] [CrossRef]
- Das, B.K.; Neha Nidhi, R.G.; Pragyan, R.; Muduli, A.K.; Swain, P.; Mishra, S.S.; Jayasankar, P. Antagonistic activity of cellular components of Bacillus subtilis AN11 against bacterial pathogens. Int. J. Curr. Microbiol. App. Sci. 2014, 3, 795–809. [Google Scholar]
- Wang, T.; Liang, Y.; Wu, M.; Chen, Z.; Lin, J.; Yang, L. Natural products from Bacillus subtilis with antimicrobial properties. Chin. J. Chem. Eng. 2015, 23, 744–754. [Google Scholar] [CrossRef]
- Ramirez-Olea, H.; Reyes-Ballesteros, B.; Chavez-Santoscoy, R.A. Potential application of the probiotic Bacillus licheniformis as an adjuvant in the treatment of diseases in humans and animals: A systematic review. Front. Microbiol. 2022, 26, 993451. [Google Scholar] [CrossRef]
- Muras, A.; Romero, M.; Mayer, C.; Otero, A. Biotechnological applications of Bacillus licheniformis. Crit. Rev. Biotechnol. 2021, 41, 609–627. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Zhang, Y.; Shi, G.; Li, Y. Recent biotechnological advances and future prospective of Bacillus licheniformis as microbial cell factories. Syst. Microbiol. Biomanuf. 2023, 3, 521–532. [Google Scholar] [CrossRef]
- Camacho, M.I.; García, J.M.; Roget, D.; Ferrer, A.; Wieme, A.D.; Vandamme, P.; Rodríguez, S.; Llauradó, G.; Lescaylle, Y.; Peña, L.; et al. Isolation and identification of a Bacillus sp. from freshwater sediment displaying potent activity against bacteria and phytopathogen fungi. Curr. Microbiol. 2022, 79, 398. [Google Scholar] [CrossRef] [PubMed]
- Vos, P.; Garrity, G.; Jones, D.; Krieg, N.R.; Ludwig, W.; Rainey, F.A.; Schleifer, K.-H.; Whitman, W.B. Bergey’s Manual of Systematic Bacteriology, 2nd ed.; Springer: New York, NY, USA, 2009; Volume 3, p. 1422. [Google Scholar]
- Bulygina, E.S.; Kuznetsov, B.B.; Marusina, A.I.; Kravchenko, I.K.; Bykova, S.A.; Kolganova, T.V.; Galchenko, V.F. Study of nucleotide sequences of nifH genes in methanotrophic bacteria. Microbiology 2002, 71, 425–432. [Google Scholar] [CrossRef]
- Maslennikova, S.N.; Karakotov, S.D. Mixture of Bacterial Strains with Cellulolytic and Fungicidal. Activity. Patent RU № 2752903, 2021. [Google Scholar]
- Donkova, N.V.; Donkov, S.A. The study of antagonistic activity of amylolytic strains of Bacillus subtilis. Mezhduranrodny Vestn. Vet. 2016, 2, 46–50. (In Russian) [Google Scholar]
- Haller, D.; Colbus, H.; Gänzle, M.G.; Scherenbacher, P.; Bode, C.; Hammes, W.P. Metabolic and functional properties of lactic acid bacteria in the gastro-intestinal ecosystem: A comparative in vitro study between bacteria of intestinal and fermented food origin. Syst. Appl. Microbiol. 2001, 24, 218–226. [Google Scholar] [CrossRef]
- EFSA Panel on Additives and Products or Substances Used in Animal Feed (FEEDAP). Guidance on the characterisation of microorganisms used as feed additives or as production organisms. EFSA J. 2018, 16, 5206. [Google Scholar]
- Mingmongkolchai, S.; Panbangred, W. In vitro evaluation of candidate Bacillus spp. for animal feed. J. Gen. Appl. Microbiol. 2017, 63, 147–156. [Google Scholar] [CrossRef]
- Khalid, F.; Khalid, A.; Fu, Y.; Hu, Q.; Zheng, Y.; Khan, S.; Wang, Z. Potential of Bacillus velezensis as a probiotic in animal feed: A review. J. Microbiol. 2021, 59, 627–633. [Google Scholar] [CrossRef]
- Grant, A.; Gay, C.G.; Lillehoj, H.S. Bacillus spp. as direct-fed microbial antibiotic alternatives to enhance growth, immunity, and gut health in poultry. Avian Pathol. 2018, 47, 339–351. [Google Scholar] [CrossRef]
- Łubkowska, B.; Jeżewska-Frąckowiak, J.; Sroczyński, M.; Dzitkowska-Zabielska, M.; Bojarczuk, A.; Skowron, P.M.; Cięszczyk, P. Analysis of industrial Bacillus species as potential probiotics for dietary supplements. Microorganisms 2023, 11, 488. [Google Scholar] [CrossRef] [PubMed]
- Ramlucken, U.; Roets, Y.; Ramchuran, S.O.; Moonsamy, G.; van Rensburg, C.J.; Thantsha, M.S.; Lalloo, R. Isolation, selection and evaluation of Bacillus spp. as potential multi-mode probiotics for poultry. J. Gen. Appl. Microbiol. 2020, 66, 228–238. [Google Scholar] [CrossRef] [PubMed]
- Belyavskaya, V.A.; Lebedev, L.R.; Masycheva, V.I. Biotechnological aspects of designing genetically modified microorganisms. In Biotechnology and Medicine; Zaikov, G.E., Ed.; Nova Science Publishers, Inc.: New York, NY, USA, 2004; pp. 107–119. [Google Scholar]
- Dong, H.; Zhang, D. Current development in genetic engineering strategies of Bacillus species. Microb. Cell Fact. 2014, 13, 63. [Google Scholar] [CrossRef] [PubMed]
- Deckers, M.; Deforce, D.; Fraiture, M.-A.; Roosens, N.H.C. Genetically modified micro-organisms for industrial food enzyme production: An overview. Foods 2020, 9, 326. [Google Scholar] [CrossRef]
- Son, H.I.; Weiss, A.; You, L. Design patterns for engineering genetic stability. Curr. Opin. Biomed. Eng. 2021, 19, 100297. [Google Scholar] [CrossRef]
- Dan, T.; Hu, H.; Tai, J.; He, Y.; He, B. Production and evaluation of a mutant galactose-utilizing strain of Streptococcus thermophilus for application in milk fermentation. LWT 2023, 187, 115284. [Google Scholar] [CrossRef]
- Marcos-Fernández, R.; Blanco-Míguez, A.; Ruiz, L.; Margolles, A.; Ruas-Madiedo, P.; Sánchez, B. Towards the isolation of more robust next generation probiotics: The first aerotolerant Bifidobacterium bifidum strain. Food Res. Int. 2023, 165, 112481. [Google Scholar] [CrossRef]
- Bjerre, K.; Cantor, M.D.; Nørgaard, J.V.; Poulsen, H.D.; Blaabjerg, K.; Canibe, N.; Jensen, B.B.; Stuer-Lauridsen, B.; Nielsen, B.; Derkx, P.M.F. Development of Bacillus subtilis mutants to produce tryptophan in pigs. Biotechnol. Lett. 2017, 39, 289–295. [Google Scholar] [CrossRef]
- Šeme, H.; Bogovič Matijašić, B.; Švigelj, K.; Langerholc, T.; Fujs, Š.; Horvat, J.; Zlatić, E.; Gjuračić, K.; Petković, H.; Štempelj, M.; et al. Generation of Lactobacillus plantarum strains with improved potential to target gastrointestinal disorders related to sugar malabsorption. Food Res. Int. 2017, 94, 45–53. [Google Scholar] [CrossRef]
- Latorre, J.D.; Hernandez-Velasco, X.; Kallapura, G.; Menconi, A.; Pumford, N.R.; Morgan, M.J.; Layton, S.L.; Bielke, L.R.; Hargis, B.M.; Téllez, G. Evaluation of germination, distribution, and persistence of Bacillus subtilis spores through the gastrointestinal tract of chickens. Poult. Sci. 2014, 93, 1793–1800. [Google Scholar] [CrossRef]
- Patten, J.D.; Waldroup, P.W. Use of organic acids in broiler diets. Poult. Sci. 1998, 67, 1178–1182. [Google Scholar] [CrossRef] [PubMed]
- Yaqoob, M.U.; Wang, G.; Wang, M. An updated review on probiotics as an alternative of antibiotics in poultry—A review. Anim. Biosci. 2022, 35, 1109–1120. [Google Scholar] [CrossRef] [PubMed]
- Al Hakeem, W.G.; Fathima, S.; Shanmugasundaram, R.; Selvaraj, R.K. Campylobacter jejuni in poultry: Pathogenesis and control strategies. Microorganisms 2022, 10, 2134. [Google Scholar] [CrossRef] [PubMed]
- Dittoe, D.K.; Ricke, S.C.; Kiess, A.S. Organic acids and potential for modifying the avian gastrointestinal tract and reducing pathogens and disease. Front. Vet. Sci. 2018, 5, 216. [Google Scholar] [CrossRef] [PubMed]
- Mortada, M.; Cosby, D.E.; Shanmugasundaram, R.; Selvaraj, R.K. In vivo and in vitro assessment of commercial probiotic and organic acid feed additives in broilers challenged with Campylobacter coli. J. Appl. Poult. Res. 2020, 29, 435–446. [Google Scholar] [CrossRef]
- Gadde, U.D.; Oh, S.; Lillehoj, H.S.; Lillehoj, E.P. Antibiotic growth promoters virginiamycin and bacitracin methylene disalicylate alter the chicken intestinal metabolome. Sci. Rep. 2018, 8, 3592. [Google Scholar] [CrossRef] [PubMed]
- EFSA Panel on Additives and Products or Substances Used in Animal Feed (FEEDAP). Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA J. 2012, 10, 2740. [Google Scholar]
- Hung, C.C.; Chen, C.Y.; Chen, B.J. Colistin and tylosin enhances disaccharidase activities, and improves morphology and permeability of the intestine of broilers. Br. Poult. Sci. 2020, 61, 465–470. [Google Scholar] [CrossRef]
- Vinothini, P.; Ramesh, S.; Sooraj Nair, V.; Preetha, S.P.; Sriram, P. Pharmacokinetics and relative bioavailability of tiamulin in broiler chicken as influenced by different routes of administration. J. Vet. Pharmacol. Ther. 2019, 42, 447–451. [Google Scholar] [CrossRef]
- Grozina, A.A.; Ilina, L.A.; Laptev, G.Y.; Yildirim, E.A.; Ponomareva, E.S.; Filippova, V.A.; Tyurina, D.G.; Fisinin, V.I.; Kochish, I.I.; Griffin, D.K.; et al. Probiotics as an alternative to antibiotics in modulating the intestinal microbiota and performance of broiler chickens. J. Appl. Microbiol. 2023, 134, lxad213. [Google Scholar] [CrossRef]
| Parameter | Strains | |||
|---|---|---|---|---|
| B. subtilis B-7 | B. subtilis RBT-7/32 | B. licheniformis RBT-11 | B. licheniformis RBT-11/17 | |
| Diameter of the E. coli growth inhibition zone, mm | 12.4 ± 0.1 | 22.0 ± 0.2 | 11.3 ± 0.2 | 18.5 ± 0.1 |
| Diameter of the S. aureus growth inhibition zone, mm | 11.7 ± 0.1 | 20.8 ± 0.1 | 10.8 ± 0.1 | 19.4 ± 0.1 |
| Diameter of the MCC hydrolysis zone, mm | 10.6 ± 0.2 | 20.3 ± 0.1 | - | - |
| Diameter of the CMC hydrolysis zone, mm | 11.0 ± 0.1 | 22.7 ± 0.1 | - | - |
| Diameter of the starch hydrolysis zone, mm | 12.5 ± 0.1 | 21.6 ± 0.1 | 11.8 ± 0.1 | 19.7 ± 0.2 |
| Medium | Optical Density OD600, Rel. Units | |||
|---|---|---|---|---|
| B. subtilis RBT-7 | B. subtilis RBT-7/32 | B. licheniformis RBT-11 | B. licheniformis RBT-11/17 | |
| LB medium | 2.2 ± 0.13 | 3.34 ± 0.21 | 2.0 ± 0.13 | 3.04 ± 0.15 |
| Medium 1 (soybean meal) | 1.6 ± 0.21 | 2.98 ± 0.34 | 1.5 ± 0.15 | 2.86 ± 0.25 |
| Medium 2 (pea meal) | 1.5 ± 0.15 | 2.67 ± 0.22 | 1.3 ± 0.1 | 2.70 ± 0.18 |
| Medium 3 (corn meal) | 1.2 ± 0.12 | 2.36 ± 0.53 | 1.2 ± 0.12 | 2.25 ± 0.40 |
| Conditions (pH Value and Exposure Time) | Spore Concentration, CFU/mL | |||
|---|---|---|---|---|
| B. subtilis RBT-7 | B. subtilis RBT-7/32 | B. licheniformis RBT-11 | B. licheniformis RBT-11/17 | |
| Control (initial point) | 4.0 × 107 | 4.0 × 107 | 4.0 × 107 | 4.0 × 107 |
| pH 5.0, 1 h | 3.0 × 107 | 3.3 × 107 | 3.2 × 107 | 3.0 × 107 |
| pH 3.0, 1.5 h | 2.1 × 107 | 2.2 × 107 | 1.8 × 107 | 2.0 × 107 |
| pH 7.0, 2.5 h | 2.0 × 107 | 2.0 × 107 | 1.7 × 107 | 2.0 × 107 |
| Temperature, °C | Exposure, min | Spore Concentration, CFU/mL | |||
|---|---|---|---|---|---|
| B. subtilis RBT-7 | B. subtilis RBT-7/32 | B. licheniformis RBT-11 | B. licheniformis RBT-11/17 | ||
| - | - | 5.0 × 1011 | 5.2 × 1011 | 4.0 × 1011 | 4.8 × 1011 |
| 80 | 10 | 4.8 × 1011 | 5.2 × 1011 | 3.7 × 1011 | 4.7 × 1011 |
| 15 | 4.0 × 1011 | 5.1 × 1011 | 3.2 × 1011 | 4.7 × 1011 | |
| 20 | 3.4 × 1011 | 4.4 × 1011 | 3.0 × 1011 | 4.2 × 1011 | |
| 100 | 10 | 4.0 × 1011 | 4.8 × 1011 | 3.2 × 1011 | 4.3 × 1011 |
| 15 | 3.6 × 1011 | 4.6 × 1011 | 2.6 × 1011 | 4.0 × 1011 | |
| 20 | 3.2 × 1011 | 4.2 × 1011 | 2.2 × 1011 | 3.7 × 1011 | |
| Antibiotic Group | Antibiotic | Dose, µg | Growth Inhibition Zone, (±0.2 mm) 1 | |||
|---|---|---|---|---|---|---|
| B. subtilis RBT-7 | B. subtilis RBT-7/32 | B. licheniformis RBT-11 | B. licheniformis RBT-11/17 | |||
| Tetracyclines | Doxycycline | 30 | 11 | 10 | 8 | 8 |
| Tetracycline | 30 | 16 | 15 | 15 | 15 | |
| Polypeptides | Bacitracin | 0.04 U | 0 | 0 | 0 | 0 |
| 10 U | 0 | 0 | 0 | 0 | ||
| 100 U | 0 | 0 | 0 | 0 | ||
| Colistin sulfate | 10 | 0 | 0 | 0 | 0 | |
| Nitrofurans | Furazolidone | 300 | 14 | 14 | 15 | 15 |
| Penicillins | Amoxicillin | 25 | 24 | 23 | 24 | 22 |
| Lincosamides | Lincomycin | 15 | 23 | 23 | 21 | 22 |
| Streptogramins | Stafac 110 (Virginiamicin) | 50 | 11 | 10 | 10 | 10 |
| 100 | 0 | 0 | 0 | 0 | ||
| Macrolides | Tylosin | 200 | 0 | 0 | 0 | 0 |
| 20 | 10 | 10 | 10 | 10 | ||
| Tiamulin | 300 | 0 | 0 | 0 | 0 | |
| Erythromycin | 15 | 26 | 25 | 25 | 24 | |
| Aminoglycosides | Gentamycin | 120 | 24 | 22 | 22 | 22 |
| Kanamycin | 30 | 22 | 20 | 22 | 21 | |
| Lactams | Ampicillin | 10 | 12 | 12 | 12 | 12 |
| Benzylpenicillin | 10 U | 0 | 0 | 0 | 0 | |
| Quinolones | Enrofloxacin | 5 | 30 | 30 | 26 | 26 |
| Amphenicols | Florfenicol | 30 | 26 | 25 | 25 | 25 |
| Polymyxins | Polymyxin | 300 | 0 | 0 | 0 | 0 |
| Nitrofurans | Furazolidone | 300 | 15 | 14 | 15 | 15 |
| Antibiotic Group | Antibiotic | Dose, µg | Growth Inhibition Zone, mm | |
|---|---|---|---|---|
| B. subtilis RBT-7/ B. licheniformis RBT-11 | B. subtilis RBT-7/32 B. licheniformis RBT-11/17 | |||
| Tetracyclines | Doxycycline | 30 | 10 | 9 |
| Polypeptides | Bacitracin | 10 U | 0 | 0 |
| Lactams | Ampicillin | 10 | 12 | 11 |
| Streptogramins | Stafac 110 (Virginiamicin) | 100 | 0 | 0 |
| Macrolides | Tylosin | 200 | 0 | 0 |
| Tiamulin | 300 | 0 | 0 | |
| Lactams | Benzylpenicillin | 10 U | 0 | 0 |
| Polymyxins | Polymyxin | 300 | 0 | 0 |
| Organic Acid | Exposure Time, h | Spore Concentration, CFU/mL | |||
|---|---|---|---|---|---|
| B. subtilis RBT-7 | B. subtilis RBT-7/32 | B. licheniformis RBT-11 | B. licheniformis RBT-11/17 | ||
| Control (water) | 3 | 5.00 × 1011 | 5.10 × 1011 | 5.10 × 1011 | 5.20 × 1011 |
| 24 | 4.80 × 1011 | 5.00 × 1011 | 5.00× 1011 | 5.18 × 1011 | |
| Citric acid | 3 | 5.00 × 1011 | 5.14 × 1011 | 5.20 × 1011 | 5.14 × 1011 |
| 24 | 4.82 × 1011 | 5.11 × 1011 | 4.92 × 1011 | 5.05 × 1011 | |
| Ascorbic acid | 3 | 4.82 × 1011 | 5.23 × 1011 | 4.80 × 1011 | 5.10 × 1011 |
| 24 | 4.80 × 1011 | 5.20 × 1011 | 4.75 × 1011 | 5.00 × 1011 | |
| Acetic acid | 3 | 5.03 × 1011 | 5.03 × 1011 | 5.03 × 1011 | 5.03 × 1011 |
| 24 | 5.00 × 1011 | 5.12 × 1011 | 4.90 × 1011 | 5.12 × 1011 | |
| Formic acid | 3 | 5.10 × 1011 | 5.00 × 1011 | 5.00 × 1011 | 5.17 × 1011 |
| 24 | 5.00 × 1011 | 5.14 × 1011 | 4.90 × 1011 | 5.00 × 1011 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yaderets, V.; Karpova, N.; Glagoleva, E.; Shibaeva, A.; Dzhavakhiya, V. Bacillus subtilis RBT-7/32 and Bacillus licheniformis RBT-11/17 as New Promising Strains for Use in Probiotic Feed Additives. Microorganisms 2023, 11, 2729. https://doi.org/10.3390/microorganisms11112729
Yaderets V, Karpova N, Glagoleva E, Shibaeva A, Dzhavakhiya V. Bacillus subtilis RBT-7/32 and Bacillus licheniformis RBT-11/17 as New Promising Strains for Use in Probiotic Feed Additives. Microorganisms. 2023; 11(11):2729. https://doi.org/10.3390/microorganisms11112729
Chicago/Turabian StyleYaderets, Vera, Nataliya Karpova, Elena Glagoleva, Alexandra Shibaeva, and Vakhtang Dzhavakhiya. 2023. "Bacillus subtilis RBT-7/32 and Bacillus licheniformis RBT-11/17 as New Promising Strains for Use in Probiotic Feed Additives" Microorganisms 11, no. 11: 2729. https://doi.org/10.3390/microorganisms11112729
APA StyleYaderets, V., Karpova, N., Glagoleva, E., Shibaeva, A., & Dzhavakhiya, V. (2023). Bacillus subtilis RBT-7/32 and Bacillus licheniformis RBT-11/17 as New Promising Strains for Use in Probiotic Feed Additives. Microorganisms, 11(11), 2729. https://doi.org/10.3390/microorganisms11112729





