Immaturin-Nuclease as a Model System for a Gene-Programmed Sexual Development and Rejuvenescence in Paramecium Life History
Abstract
1. Introduction
2. Materials and Methods
2.1. Stocks and Cell Culture
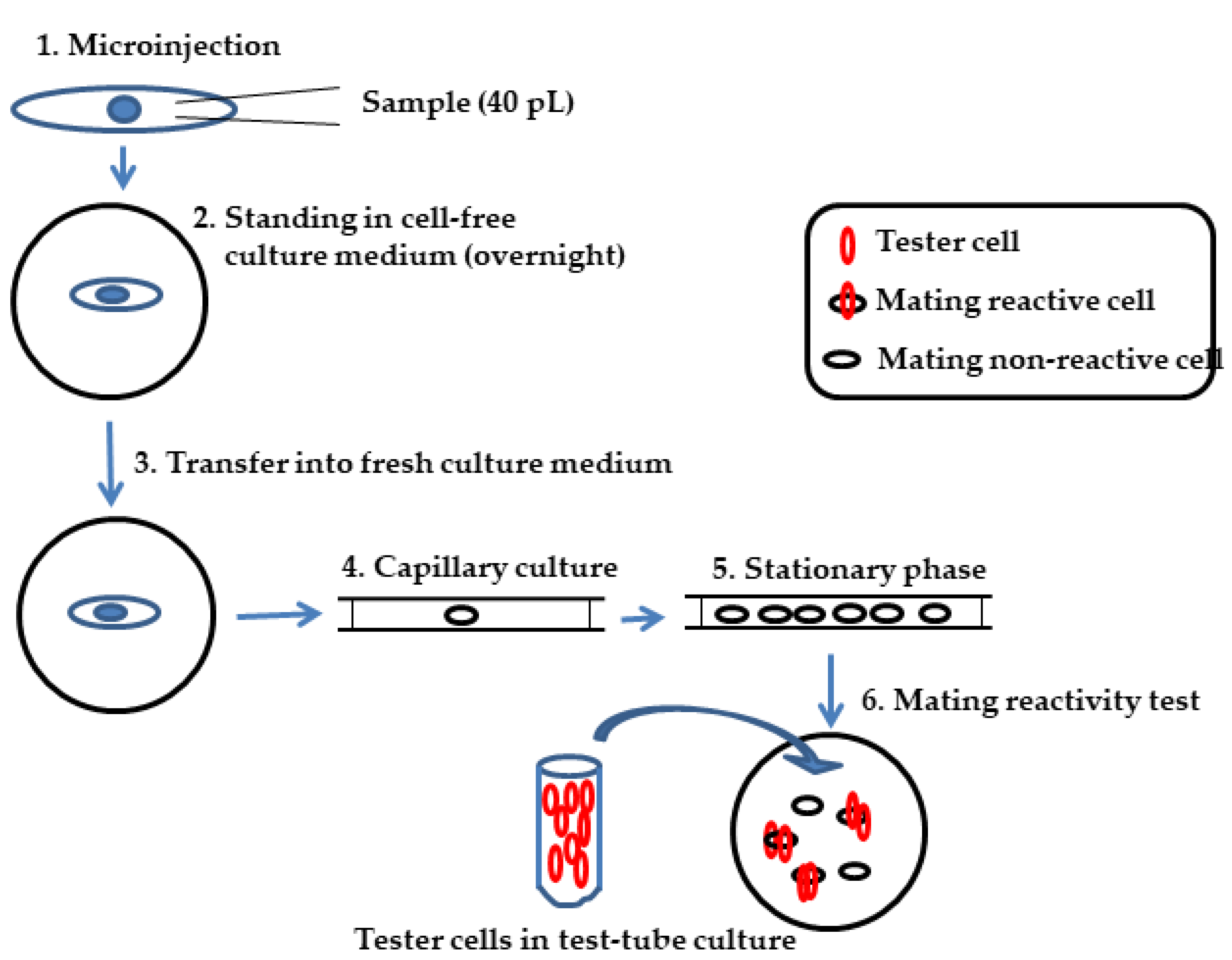
2.2. Microinjection
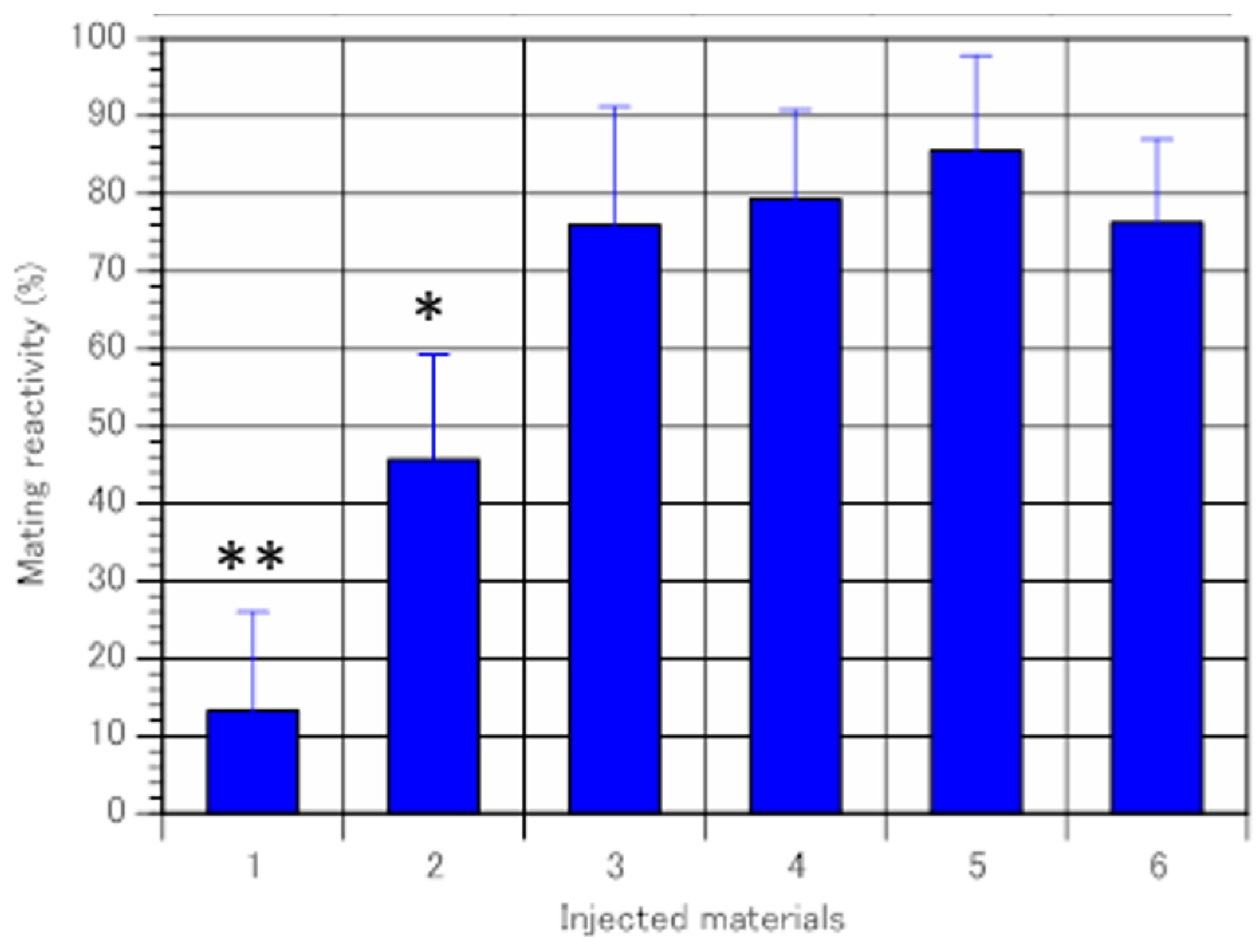
2.3. Immaturin Assay
2.4. Immaturin Inhibition by Anti-Immaturin Antibody
2.5. Preparation of Recombinant Immaturin and Its Effects
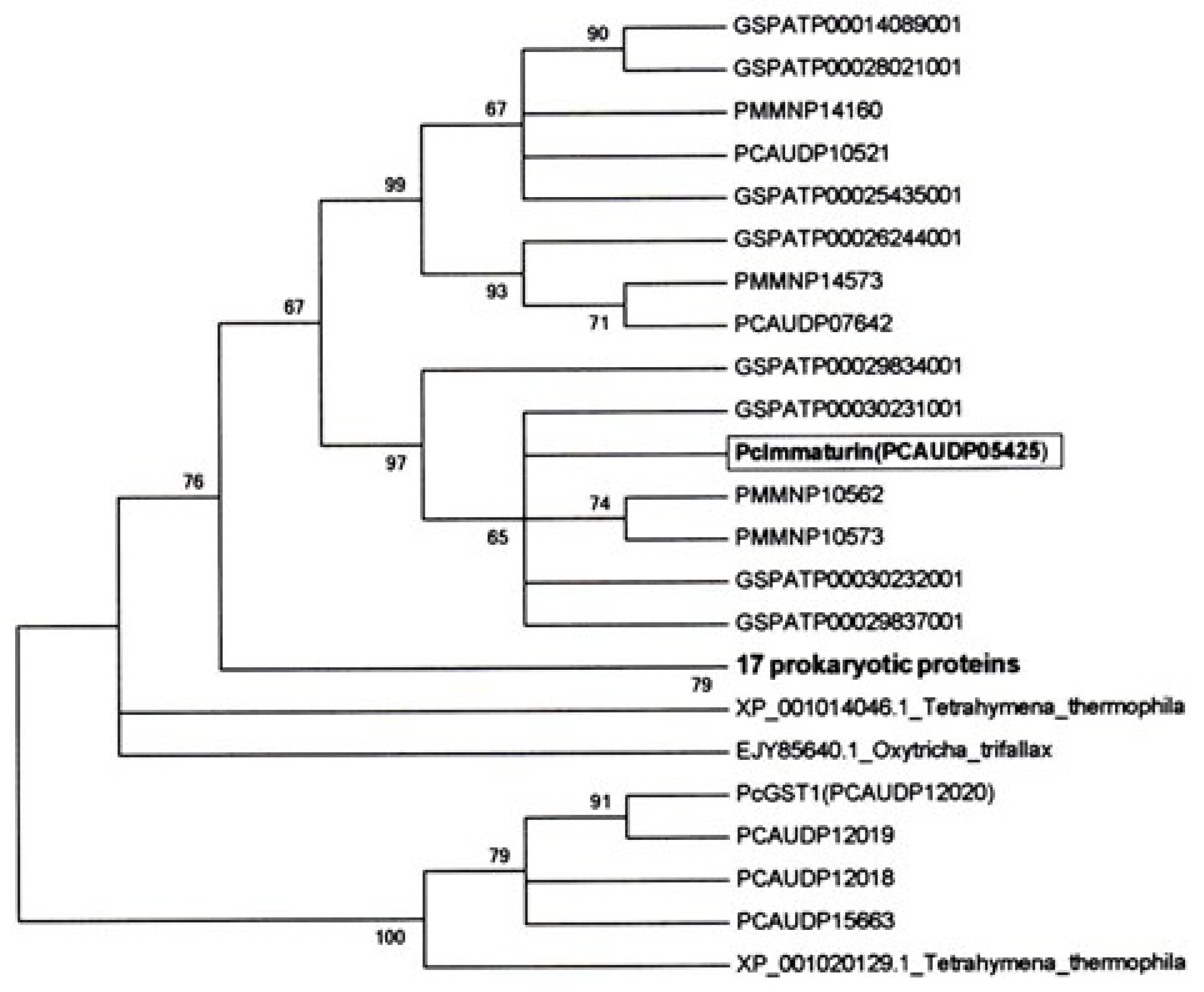
2.6. The Maximum Likelihood (ML) Consensus Tree of Immaturin
2.7. Immaturin Imaging in Intracellular Localizations: Direct Immunofluorescent Staining
2.8. Indirect Immunofluorescent Staining
2.9. Assay for Immaturin-Nuclease Activity
2.10. GST Assay
2.11. Paramecium Expression Vector and Transformation
3. Results
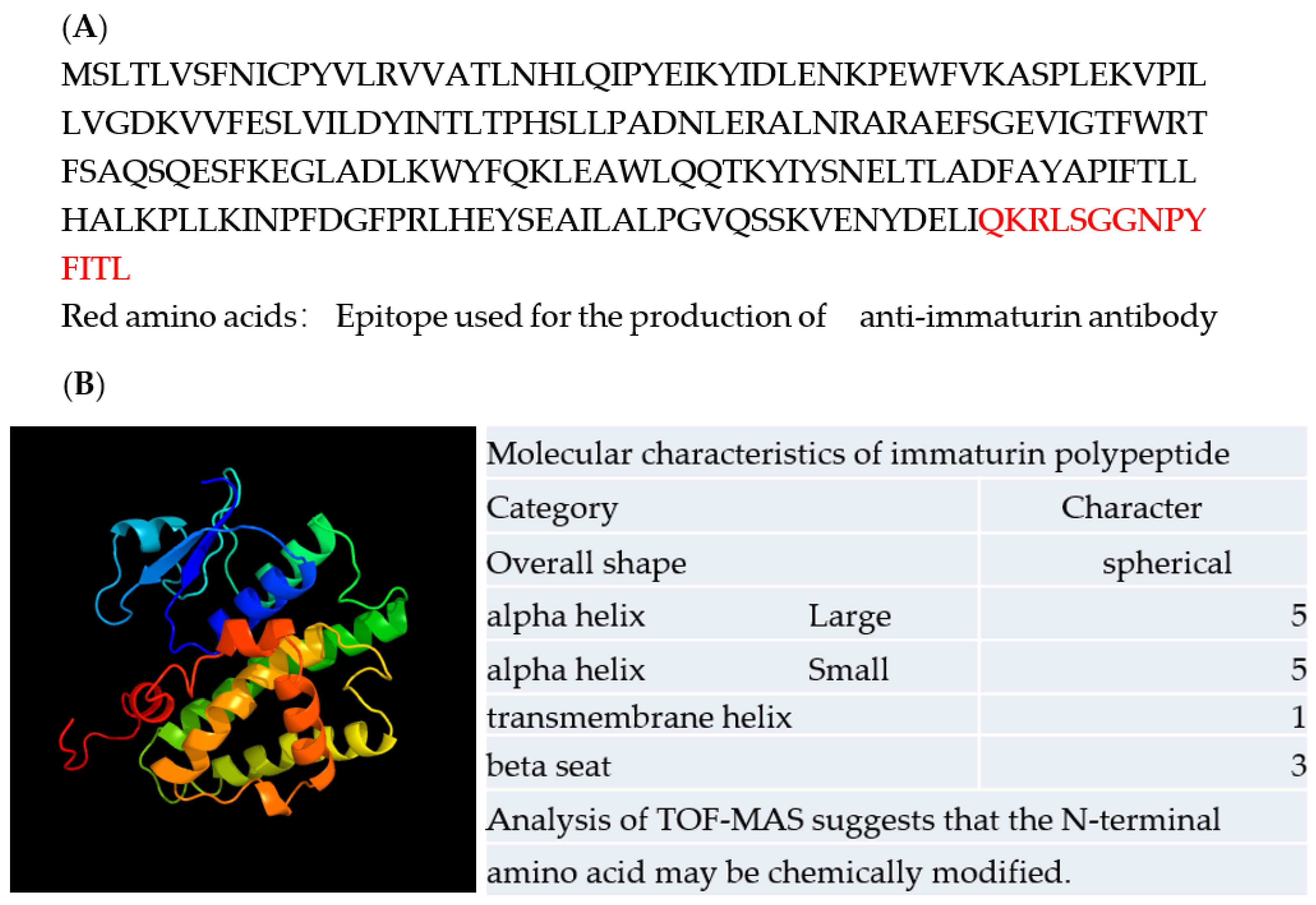
3.1. Identification of the Immaturin Gene and Phylogenetic Analysis of the Original Gene
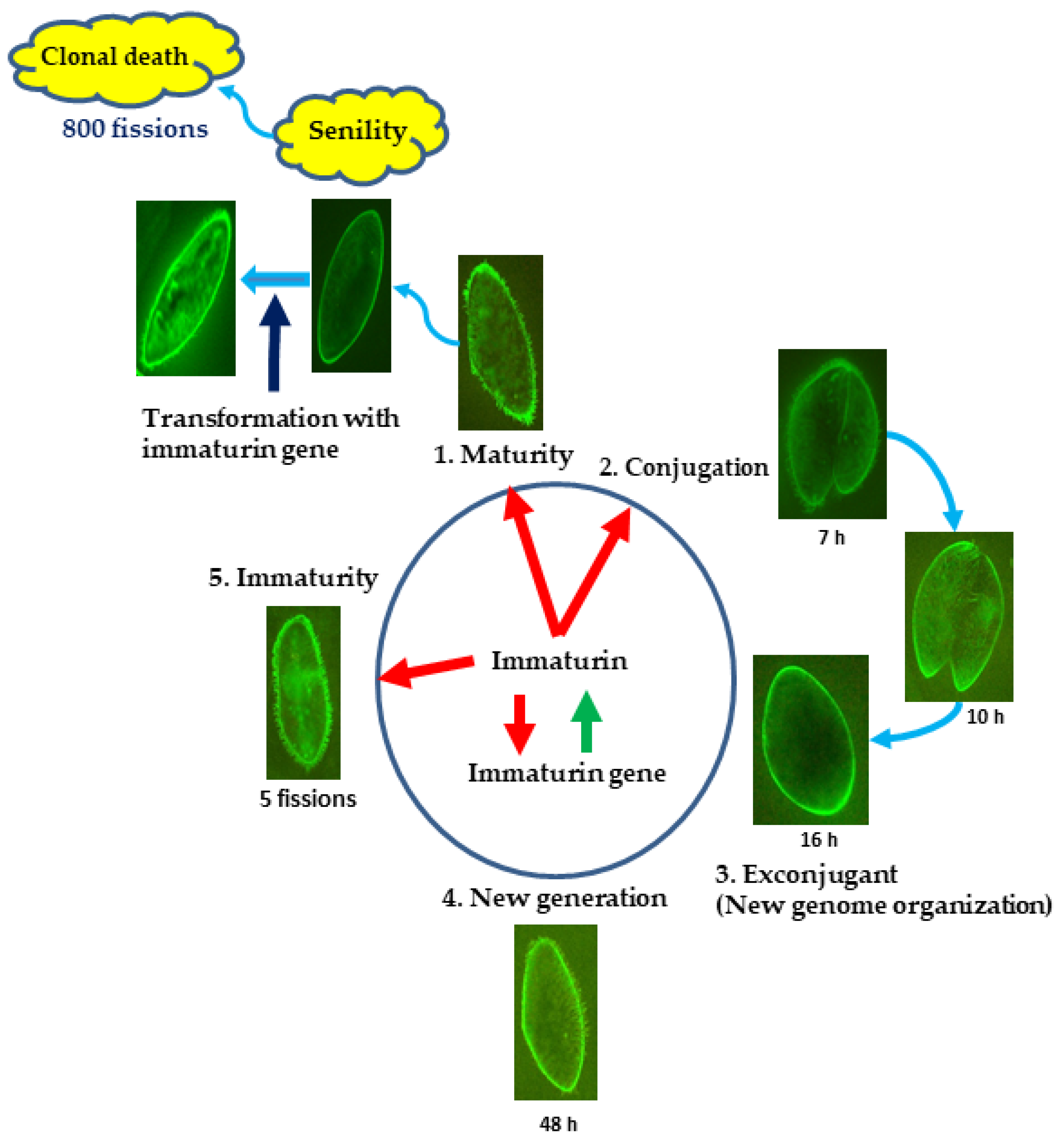
3.2. Immaturin Localization in Subcellular Compartments
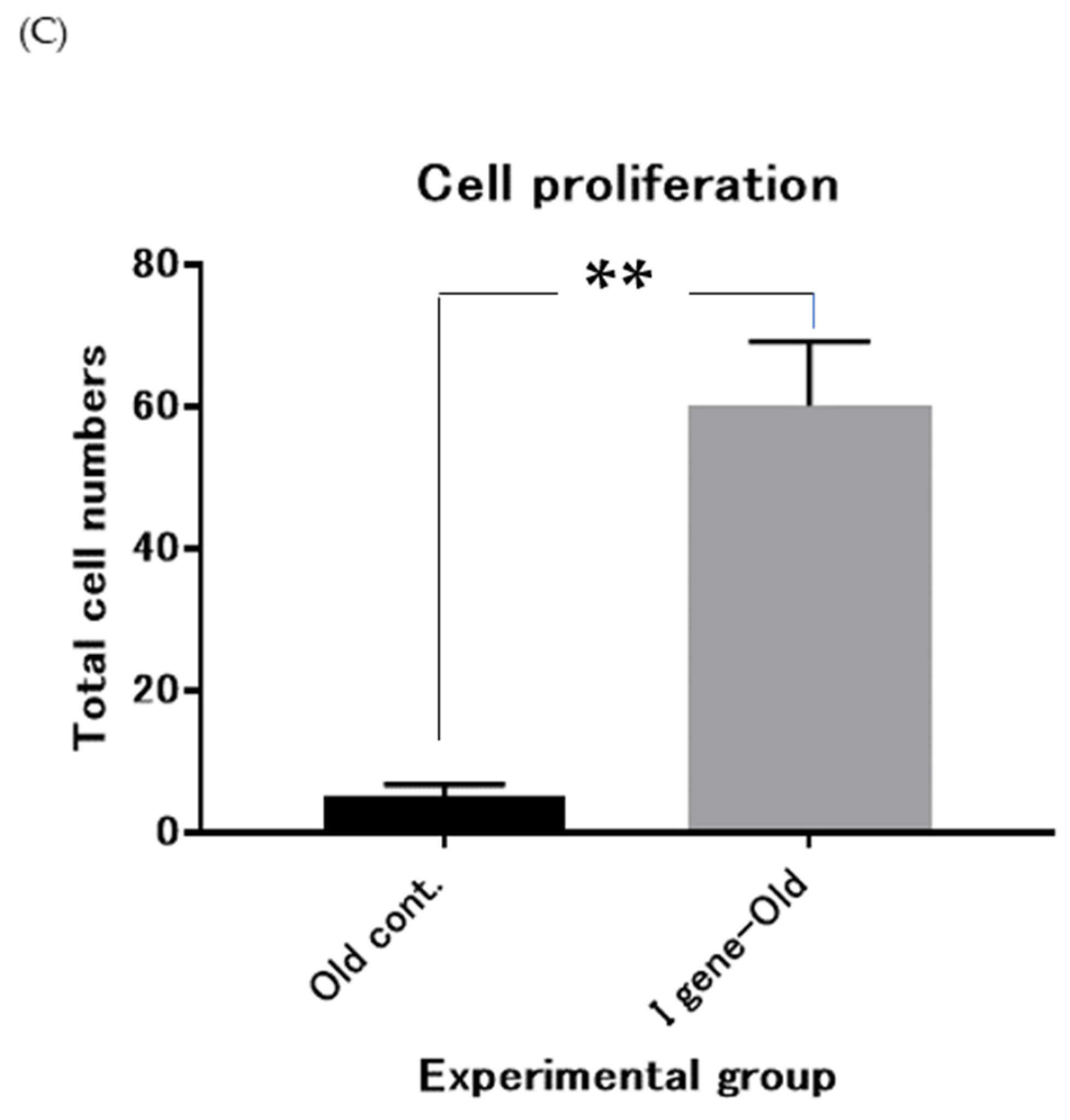
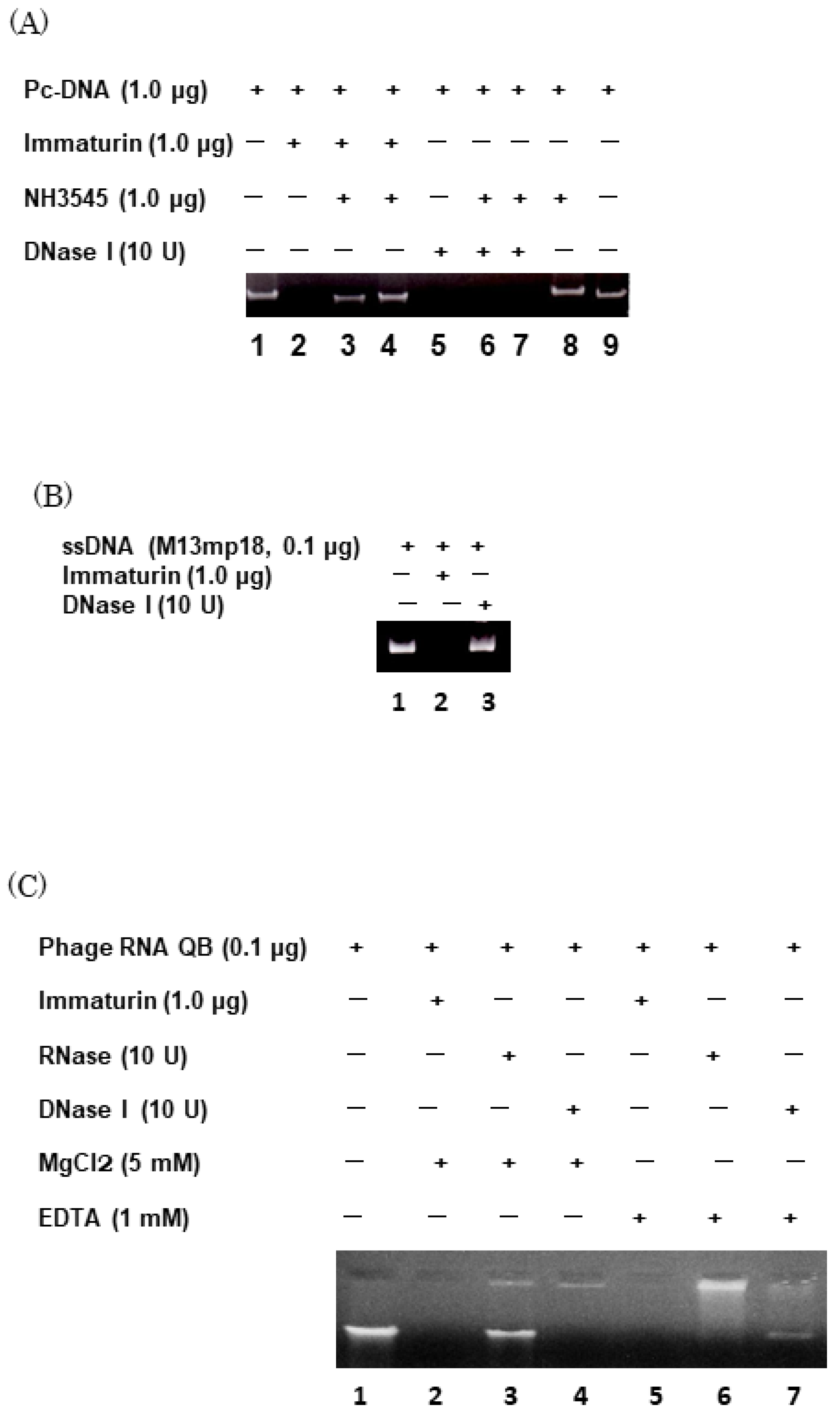
3.3. Immaturin Digests DNA and Retroviral Genomic RNA
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hiwatashi, K. Determination and inheritance of mating type in Paramecium caudatum. Genetics 1968, 58, 373–386. [Google Scholar] [CrossRef] [PubMed]
- Hiwatashi, K. Localization of mating reactivity on the surface of Paramecium caudatum. Sci. Rep. Tohoku Univ. Ser. 1961, 4, 93–99. [Google Scholar]
- Sonneborn, T.M. Breeding systems, reproductive methods and species problems in Protozoa. In The Species Problem; Mayer, E., Ed.; American Association for the Advancement of Science: Washington, DC, USA, 1957; pp. 155–324. [Google Scholar]
- Takagi, Y. Expression of the mating-type trait in the clonal history after conjugation in Paramecium multimicronucleatum and Paramecium caudatum. Japan J. Genet. 1970, 45, 11–21. [Google Scholar] [CrossRef]
- Miwa, I.; Hiwatashi, K. Effect of mitomycin C on the expression of mating ability in Paramecium caudatum. Japan J. Gent. 1970, 45, 269–275. [Google Scholar] [CrossRef]
- Bleyman, L.K. Temporal patterns in the ciliated Protozoa. In Developmental Aspects of the Cell Cycle; Cameron, T.L., Padilla, G.M., Zimmerman, A.Z., Eds.; Academic Press: New York, NY, USA; London, UK, 1971; pp. 67–91. [Google Scholar]
- Siegel, R.W. Nuclear differentiation and transitional cellular phenotypes in the life cycle of Paramecium. Expl. Cell Res. 1961, 24, 6–20. [Google Scholar] [CrossRef]
- Bleyman, L.K.; Simon, E.M. Genetic control of maturity in Tetrahymena pyriformis. Genet. Res. 1967, 10, 319–321. [Google Scholar] [CrossRef]
- Myohara, K.; Hiwatashi, K. Mutants of sexual maturity in Paramecium caudatum selected by erythromycin resistance. Genetics 1978, 90, 227–241. [Google Scholar] [CrossRef]
- Miwa, I.; Haga, N.; Hiwatashi, K. Immaturity substances: Material basis for immaturity in Paramecium. J. Cell Sci. 1975, 19, 369–378. [Google Scholar] [CrossRef]
- Miwa, I. Specificity of the immaturity substances in Paramecium. J. Cell Sci. 1979, 36, 253–260. [Google Scholar] [CrossRef]
- Miwa, I. Immaturity substances in Paramecium primaurelia and their specificity. J. Cell Sci. 1979, 38, 193–199. [Google Scholar] [CrossRef]
- Haga, N.; Hiwatashi, K. A protein called immaturin controlling sexual immaturity in Paramecium. Nature 1981, 289, 177–179. [Google Scholar] [CrossRef] [PubMed]
- Haga, N.; Karino, S. Microinjection of immaturin rejuvenates sexual activity of old Paramecium. J. Cell Sci. 1986, 86, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Haga, N. Elucidation of nucleus-cytoplasm interaction: Change in the ability of the nucleus to express sexuality according to clonal age in Paramecium. J. Cell Sci. 1995, 108, 3671–3676. [Google Scholar] [CrossRef] [PubMed]
- Yanagi, A. Positional control of the fates of nuclei produced after meiosis in Paramecium caudatum: Analysis by nuclear transplantation. Dev. Biol. 1987, 122, 535–539. [Google Scholar] [CrossRef]
- Koizumi, S. Microinjection and transfer of cytoplasm in Paramecium. Exp. Cell Res. 1974, 88, 74–78. [Google Scholar] [CrossRef] [PubMed]
- Haga, N.; Forte, M.; Saimi, Y.; Kung, C. Microinjection of cytoplasm as a test of complementation in Paramecium. J. Cell Biol. 1982, 82, 559–564. [Google Scholar] [CrossRef]
- Takenaka, Y.; Yanagi, A.; Masuda, H.; Mitsui, Y.; Mizuno, H.; Haga, N. Direct observation of histone H2B-YFP fusion proteins and transport of their mRNA between conjugating Paramecia. Gene 2007, 395, 108–115. [Google Scholar] [CrossRef]
- Arnaiz, O.; Sperling, L. ParameciumDB in 2011: New tools and new data for functional and comparative genomics of the model ciliate Paramecium tetraurelia. Nucleic Acids Res. 2011, 39, 632–636. [Google Scholar] [CrossRef]
- Takenaka, Y.; Haga, N.; Inoue, I.; Nakano, T.; Ikeda, M.; Katayama, S.; Awata, T. Identification of two nickel ion-induced genes, NCI16 and PcGST1, in Paramecium caudatum. Eukaryot. Cell 2014, 13, 1181–1190. [Google Scholar] [CrossRef][Green Version]
- Udomsinprasert, R.; Pongjaroenkit, S.; Wongsantichon, J.; Oakley, A.J.; Prapanthadara, L.A.; Wilce, M.C.; Ketterman, A.J. Identification, characterization and structure of a new Delta class glutathione transferase isoenzyme. Biochem. J. 2005, 388, 763–771. [Google Scholar] [CrossRef]
- Sheehan, D.; Meade, G.; Foley, V.M.; Dowd, C.A. Structure, function and evolution of glutathione transferases: Implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem. J. 2001, 360, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Albcati, N.; Federici, L.; Masulli, M.; Di Ilio, C. Glutathione transferases in bacteria. FEBS J. 2009, 276, 58–75. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, H.J.; Babbitt, P.C. Glutathione transferases are structural and functional outliers in the thioredoxin fold. Biochemistry 2009, 48, 11108–11116. [Google Scholar] [CrossRef] [PubMed]
- Leaver, M.J.; George, S.G. A piscine glutathione-S-transferase which efficiently conjugates the end-products of lipid peroxidation. Mar. Environ. Res. 1998, 46, 71–74. [Google Scholar] [CrossRef]
- Litwack, G.; Ketterer, B.; Arias, I.M. Ligandin: A hepatic protein which binds steroids, bilirubin, carcinogens, and a number of exogenous organic anions. Nature 1971, 234, 466–467. [Google Scholar] [CrossRef]
- Samejima, K.; Earnshaw, W.C. Trashing the genome: The role of nuclease during apoptosis. Nat. Rev. Mol. Cell Biol. 2005, 6, 677–688. [Google Scholar] [CrossRef]
- Zhang, J.; Xu, M. Apoptotic DNA fragmentation and tissue homeostasis. Trend Cell Biol. 2002, 12, 84–89. [Google Scholar] [CrossRef]
- Neph, S.; Vierstra, J.; Stergachis, A.; Reynolds, A.; Haugen, E.; Vernot, B.; Thurman, R.; John, S.; Sandstrom, R.; Johnson, A.; et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature 2012, 489, 83–90. [Google Scholar] [CrossRef]
- Koohy, H.; Down, T.A.; Hubbard, T.J. Chromatin accessibility data sets show bias due to sequence specificity of the DNase I enzyme. PLoS ONE 2013, 8, e69853. [Google Scholar] [CrossRef]
- Vaux, D.L.; Flavell, R.A. Apoptosis genes and autoimmunity. Curr. Opin. Immunol. 2000, 12, 719–724. [Google Scholar] [CrossRef]
- Yasutomo, K.; Horiuchi, T.; Kagami, S.; Tsukamoto, H.; Hashimura, C.; Urushihara, M.; Kuroda, Y. Mutation of DNase I in people with systemic Lupus erythematosus. Nat. Genet. 2001, 28, 313–314. [Google Scholar] [CrossRef]
- Rangarajan, E.S.; Shankar, V. Sugar non-specific endonucleases. FEMS Microbiol. Rev. 2001, 25, 583–613. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed]


| Samples | Protein (mg/mL) | GST Activity (Vmax (mU/min)) |
|---|---|---|
| 105,000× g supernatant | 2.30 | 18.34 |
| Sephadex G-50 Immaturin fraction | 0.60 | 0.70 |
| DEAE-Sephadex A25 Immaturin fraction | 0.43 | 0.00 |
| Protein (mg/mL) | Specific Activity | |
|---|---|---|
| (Vmax (mKu/mL)/Protein) | ||
| 105,000× g supernatant | 2.30 | 9.6 |
| Sephadex G-50 Immaturin fraction | 0.60 | 26.7 |
| DEAE-Sephadex A25 Immaturin fraction | 0.43 | 11.6 |
| DNase I (E. coli) | 1.00 | 4.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haga, N.; Usui, T.; Takenaka, Y.; Chiba, Y.; Abe, T. Immaturin-Nuclease as a Model System for a Gene-Programmed Sexual Development and Rejuvenescence in Paramecium Life History. Microorganisms 2023, 11, 82. https://doi.org/10.3390/microorganisms11010082
Haga N, Usui T, Takenaka Y, Chiba Y, Abe T. Immaturin-Nuclease as a Model System for a Gene-Programmed Sexual Development and Rejuvenescence in Paramecium Life History. Microorganisms. 2023; 11(1):82. https://doi.org/10.3390/microorganisms11010082
Chicago/Turabian StyleHaga, Nobuyuki, Toshinori Usui, Yasuhiro Takenaka, Yuta Chiba, and Tomoaki Abe. 2023. "Immaturin-Nuclease as a Model System for a Gene-Programmed Sexual Development and Rejuvenescence in Paramecium Life History" Microorganisms 11, no. 1: 82. https://doi.org/10.3390/microorganisms11010082
APA StyleHaga, N., Usui, T., Takenaka, Y., Chiba, Y., & Abe, T. (2023). Immaturin-Nuclease as a Model System for a Gene-Programmed Sexual Development and Rejuvenescence in Paramecium Life History. Microorganisms, 11(1), 82. https://doi.org/10.3390/microorganisms11010082





