Abstract
The migratory behavior of wild birds contributes to the geographical spread of ticks and their microorganisms. In this study, we aimed to investigate the dispersal and co-occurrence of Francisella and spotted fever group Rickettsia (SFGR) in ticks infesting birds migrating northward in the African-Western Palaearctic region (AWPR). Birds were trapped with mist nests across the Mediterranean basin during the 2014 and 2015 spring migration. In total, 575 ticks were collected from 244 birds. We screened the ticks for the species Francisella tularensis, the genus Francisella, and SFGR by microfluidic real-time PCR. Confirmatory analyses and metagenomic sequencing were performed on tick samples that putatively tested positive for F. tularensis during initial screenings. Hyalomma rufipes was the most common tick species and had a high prevalence of Francisella, including co-occurrence of Francisella and SFGR. Metagenomic analysis of total DNA extracted from two H. rufipes confirmed the presence of Francisella, Rickettsia, and Midichloria. Average nucleotide identity and phylogenetic inference indicated the highest identity of the metagenome-assembled genomes to a Francisella-like endosymbiont (FLE), Rickettsia aeschlimannii, and Midichloria mitochondrii. The results of this study suggest that (i) FLE- and SFGR-containing ticks are dispersed by northbound migratory birds in the AWPR, (ii) H. rufipes likely is not involved in transmission of F. tularensis in the AWPR, and (iii) a dual endosymbiosis of FLEs and Midichloria may support some of the nutritional requirements of H. rufipes.
1. Introduction
Ticks (Acari: Ixodida) transmit pathogens of both human and veterinary importance, such as bacteria in the genera Anaplasma, Borrelia, Coxiella, Francisella, and Rickettsia. They also can be co-infected with different pathogens that potentially can cause co-infections in hosts [1,2]. Additionally, ticks harbor endosymbionts living symbiotically within them. Bacterial endosymbionts of ticks are mostly from the genera Coxiella, Francisella, and Rickettsia; they are closely related to pathogens and may be necessary for the survival of the host [3]. Ticks are strictly hematophagous, meaning their diet consists solely of vertebrate blood, which is nutritionally unbalanced since it contains a high level of proteins but few vitamins [4]. Endosymbiotic bacteria present within tick cells are believed to support the dietary requirements of ticks by providing nutrients that are absent in vertebrate blood [5,6].
The genus Francisella includes both pathogenic and non-pathogenic species, including endosymbionts [7]. Francisella has previously been divided into two major genetic clades [8], but recently four major clades have been recognized (Clade 1–4) [7]. Francisella tularensis and Francisella-like endosymbionts (FLEs) are assigned to Clade 1 [7]. F. tularensis is primarily present in the Northern hemisphere and has a broad host range, including mammals, birds, and arthropods [9]. Furthermore, F. tularensis, the causative agent of tularemia, is regarded as a potential agent of biological warfare [10]. Infection in humans is acquired via direct contact with infected animals, ingestion of contaminated food or water, inhalation of contaminated particles, or bites of blood-feeding arthropods (i.e., ticks, tabanids, and mosquitoes) [9]. Multiple tick species from the genera Amblyomma, Dermacentor, Ixodes, and Haemaphysalis are vectors of F. tularensis [9]. The genomes of FLEs include pseudogenes and inactivated versions of virulence genes of F. tularensis, suggesting they arose from a pathogenic ancestor [5,11]. FLEs replicate intracellularly and can infect the ovaries of female ticks, enabling transovarial transmission (i.e., from the female tick to her offspring) and ensuring the continuation of the symbiotic relationships [12,13]. FLEs are known to have a broad geographical distribution [14,15,16,17,18,19,20,21,22], and are widely distributed across tick taxa, including both soft (Argasidae) and hard (Ixodidae) tick species, such as Ornithodoros moubata, Amblyomma maculatum, Dermacentor andersoni, Dermacentor reticulatus, Dermacentor variabilis, and Hyalomma marginatum [5,15,21,22,23,24,25]. Little is known about FLEs due to culturing difficulties and a limited number of assembled and characterized genomes.
The bacterium Midichloria mitochondrii was first described as an endosymbiont of Ixodes ricinus ticks known to reside primarily in the ovarian primordia or ovaries, enter the mitochondria, and be transmitted by transovarial transmission to the offspring [26,27,28]. The prevalence of M. mitochondrii in female I. ricinus ticks has been reported to be 100% [27]. The bacterium also has been detected in ticks of the genera Hyalomma, Rhipicephalus, Amblyomma, and Haemaphysalis [29]. DNA of M. mitochondrii and of bacteria related to M. mitochondrii has been detected in the salivary glands of I. ricinus ticks [30] and in blood samples from canines [31], respectively, and antibodies against Midichloria have been detected in blood samples collected from canines and humans bitten by ticks [31,32], indicating potential horizontal transmission of the bacterium.
The genus Rickettsia has been divided into four groups [33]. Most tick-borne Rickettsia belong to the spotted fever group (SFG), which includes members that are considered to be emerging human pathogens in Europe (e.g., Rickettsia conorii, Rickettsia massiliae, and Rickettsia aeschlimannii) [34,35]. Rickettsia species in the SFG are transmitted to humans by multiple tick genera, including Rhipicephalus, Ixodes, and Hyalomma [34]. Wild birds are frequently parasitized by Ixodes and Hyalomma ticks, and the migratory behavior of the avian hosts aids in the geographical spread of ticks and their associated microorganisms [36,37,38]. Because they are intracellular tick-borne bacteria, Francisella, Midichloria, and Rickettsia are difficult to culture, and culture-independent generation of genome sequences is of importance for increasing the knowledge and understanding of these bacteria. In this study, we aimed to investigate the dispersal and co-occurrence of Francisella and SFG Rickettsia (SFGR) species in ticks infesting northbound migrating birds in the African-Western Palaearctic region (AWPR), using microfluidic real-time (q) PCR and metagenomics.
2. Materials and Methods
2.1. Trapping of Birds and Collection of Ticks
Birds were trapped using mist nets at bird observatories in Spain (several sites in the provinces of Huelva and Sevilla and the Canary Islands: 37°30′ N, 5°30′ W; 37°33′ N, 6°55′ W; 28°9′ N, 15°25′ W), Italy (Capri: 40°33′ N, 14°15′ E), Greece (Crete and Antikythira; 35°51′ N, 23°18′ E), and Israel (Jerusalem and its vicinity: 31°47′ N, 35°13′ E) during their northbound 2014 and 2015 spring migration. Birds were visually inspected for ticks by blowing apart the feathers. Collected ticks were stored in RNAlaterTM (Invitrogen, ThermoFisher Scientific, Waltham, MA, USA) at −20 °C or at refrigerator temperature during the bird ringing season, a period when birds are trapped, measured, weighed, and ringed (i.e., banded) by ringers/ornithologists at bird observatories. After the ringing season, ticks were stored at −80 °C. Only northward migrating birds were included in the study. See Hoffman et al. 2021 [39] for additional details.
2.2. DNA Extraction
In brief, absolute ethanol (Sigma-Aldrich, Merck, Darmstadt, Germany) and sterile H2O were used for surface sterilization of the ticks before homogenization. Mechanical homogenization was performed using a stainless-steel bead (Qiagen, Hilden, Germany), TRIzolTM (Invitrogen, ThermoFisher Scientific, Waltham, MA, USA), and a TissueLyser II (Qiagen). After homogenization, additional TRIzolTM was added to the homogenate, followed by centrifugation and collection of the supernatant. RNA was isolated and removed using a phase separation technique, in which chloroform (Sigma-Aldrich) was added to the supernatant. Thereafter, DNA was extracted from the organic phase using a back-extraction buffer, inversion, and centrifugation. DNA present in the upper phase was purified using the Nucleospin gDNA Clean up kit (Macherey-Nagel, Bethlehem, PA, USA). The DNA was eluted in DE buffer and stored at −20 °C. For additional details, see Hoffman et al. 2021 [39].
2.3. Molecular Screening and Confirmation Analyses
2.3.1. Francisella
Tick extracts were screened for the presence of DNA from the genus Francisella and the species F. tularensis specifically by microfluidic qPCR (BioMarkTM Dynamic Arrays, Fluidigm, CA, USA) and with FopA (genus-specific) and Tul4 (species-specific) primers and probes (Table 1), at the Animal Health Laboratory (Paris, France) according to Michelet et al. [40]. Samples with a cycle threshold (Ct) value higher than 30 were considered negative [40]. To confirm the initial putative findings of F. tularensis (Tul4+ samples), subsequent qPCRs were performed using the Francisella qPCR assays in Table 1 [40,41,42,43]. In brief, the DNA was pre-amplified (due to the limited amount of DNA) using the RepliG midi kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. The PCRs (25 µL) consisted of 2X PerfeCTa qPCR ToughMix (VWR, Radnor, PA, USA), 0.5 µM (final concentration) of each primer, 0.1 µM (final concentration) of each probe, and 1 µL template. The temperature profile was as follows: 95 °C for 10 min, followed by a two-step cycle of 15 s at 95 °C and 60 s at 60 °C. Positive and negative controls were included. The tick DNA samples were tested for PCR inhibitor with an additional qPCR using seal herpesvirus type 1 [44].

Table 1.
Primers and probes used for real-time PCR assays targeting tick taxa, Francisella, and Rickettsia.
2.3.2. Spotted Fever Group Rickettsia
Ticks also were screened for SFGR DNA by microfluidic qPCR, using primers and probes targeting the gltA gene, according to Michelet et al. [40]. Samples with a Ct-value higher than 30 were considered negative [40]. Confirmation analyses were executed on a small set of ticks (n = 38) with Ct-valuesgltA ranging from 5.6 to 29.6, using primers targeting the 17 kDa gene by Carl et al. [45] (Table 1). This was done because the confirmation PCR did not include a pre-amplification step, as was the case with the microfluidic qPCR, making it a less sensitive method. In brief, the reaction comprised of 1X Phusion Green HF buffer (ThermoFisher Scientific, Waltham, MA, USA), 200 µM dNTP (Invitrogen, Thermo Fisher Scientific), 0.25 µM (final concentration) of each primer (Invitrogen), 0.02 U/µL Phusion HotStart II DNA polymerase (ThermoFisher Scientific), 8.4 µL sterile H2O, and 5 µL template. The reaction profile was as follows: 98 °C for 30 s followed by 35 cycles of 30 s at 98 °C, 30 s at 62 °C, and 30 s at 72 °C, and a final extension at 72 °C for 10 min.
2.4. Tick Taxon
Tick taxa and Hyalomma species were determined by PCR, using primers by Beati and Keirans [43] (Table 1). See Hoffman et al. 2021 [39] for further details. Morphological determination was not performed since identification of species level of immature ticks belonging to the tick complex H. marginatum, including the species H. rufipes and H. marginatum, is difficult and not recommended [46]. Furthermore, life stage determination was not performed for the infesting ticks. However, the majority of the avian-associated ticks were likely immatures [36,47].
2.5. Characterization
2.5.1. Sanger Sequencing
12S rDNA and 17 kDa amplicons were treated with illustra ExoProStarTM 1-step kit (Cytiva, Marlborough, MA, USA), according to the instructions by the manufacturer, prior Sanger sequencing at Macrogen (Amsterdam, the Netherlands).
2.5.2. Spotted Fever Group Rickettsia
The CLC Main Workbench 7 by Qiagen (Aarhus, Denmark) was used for assembling partial 17 kDa sequences, which were compared to sequences deposited in the GenBank database [48] using the nucleotide Basic Local Alignment Search Tool (BLASTN) (v.2.10.0) [49].
2.5.3. Metagenomic Sequencing
Two DNA samples putatively positive for F. tularensis by microfluidic qPCR were whole genome amplified (RepliG midi kit, Qiagen, Hilden, Germany) before being subjected to further characterization using two sequencing technologies. Non-enriched samples were prepared using TruSeq PCR-free library kits (Illumina, San Diego, CA, USA) and sequenced as 2 × 150 base pairs (bp) in one lane on an S4 flow cell in a NovaSeq 6000 sequencing instrument (Illumina) at the SNP&SEQ platform at NGI Uppsala (Sweden). Enriched samples were prepared using Nextera library kits (Illumina) and sequenced as 2 × 150 bp on a 300-cycle sequencing flow cell using a NextSeq instrument (Illumina) at NAU (Northern Arizona University). Non-enriched samples were also prepared using LSK-109 library kits (Oxford Nanopore Technologies, Oxford, UK) and sequenced in MinION R9.4.1 flow cells using a MinION sequencing device (Oxford Nanopore Technologies).
2.5.4. Enrichment
Due to few metagenomic reads mapping to Francisella, RNA baiting was performed at NAU to enrich Francisella DNA present in the tick extracts. Briefly, pre-amplified tick DNA extracts were uniquely indexed in ~300 bp sequencing libraries and exposed to RNA hybridization baits (probes) of 120 bp (Agilent Technologies Inc., Santa Clara, CA, USA). The RNA baits were designed against a Francisella pan-genome defined by 498 Francisella genomes examined in [7]. Sequences <120 bp were removed, and regions with a homology of ≥80% with non-Francisella bacteria and ribosomal RNA genes were excluded, yielding 188,430 unique probe signatures, which included 2X tiling to ensure 50% sequence overlap to optimize capture. To further refine optimal capture, manufacturing of replicate copies of ~20,000 probes comprised of high (≥50%) or low (≤22%) GC content was performed. Bait capture was executed twice for increased purification of Francisella sequences from background tick DNA.
2.5.5. Taxonomic Classification of Sequence Reads
Sequenced tick samples were characterized with a custom-made database containing bacteria, eukaryotes, and viruses, using Kraken 2 [50]. The database was created using FlexTaxD [51] with bacteria based on the taxonomy from the Genome Taxonomy Database (GTDB) together with eukaryotes and viruses from the National Center for Biotechnology Information (NCBI). The post-processing tool StringMeUp (v.0.1.4) (https://github.com/danisven/StringMeUp, accessed on 15 May 2021) was used for adjusting the results for different confidence scores.
2.5.6. Genome Assembly
Enriched Samples
Illumina reads from enriched tick samples were analyzed using a pipeline controlled by Snakemake (v.6.2.1) [52]. Initially, the data were pre-processed by using BBMap (v.38.90) (https://sourceforge.net/projects/bbmap/, accessed on 15 May 2021) to map reads to the collection of 498 Francisella genomes [7] that were used in the design of hybridization baits and only keeping mapped reads followed by digital normalization step using bbnorm in BBMap with the settings k = 31 and kmer coverage 100. The remaining reads were de novo assembled using SPAdes (v.3.15.3) [53]. Post-processing of assemblies was performed by removing contigs <500 bp and keeping contigs matching BLAST results containing the ‘*rancisella’ string (to include all different genera inside the family) through BLAST-based filtering. The following blastn settings were used: culling_limit = 5, evalue = 1e-25. Finally, two rounds of Pilon (v.1.24) [54] polishing finalized the assembled sequences before calculating summary statistics using assembly-stats (v.1.0.1) (https://github.com/sanger-pathogens/assembly-stats, accessed on 15 May 2021). The quality of metagenome-assembled genomes (MAGs) was evaluated using checkM (v.1.1.3) [55] and BUSCO (v.5.1.3) [56].
Non-Enriched Samples
Illumina reads from non-enriched tick samples were analyzed using the same workflow as enriched samples but with the modification of keeping contigs that matched ‘Rickettsia’ and ‘Midichloria’. Nanopore reads were assembled using Flye (v.2.8.3) [57] and polished using Medaka (v.1.3.0) (https://github.com/nanoporetech/medaka, accessed on 15 May 2021) followed by BLAST-based filtering, keeping contigs matching tick mitochondrial genomes.
2.5.7. Tick Species Confirmation
Species confirmation of the metagenomic characterized ticks was performed using assembled mitochondrial genomes and the animal identification engine provided by BOLD [58], in which the mitochondrial cytochrome oxidase subunit 1 (COI) gene was used.
2.5.8. Phylogenetic Analyses
Tick Phylogenies
Assembly of 12S rDNA sequences was performed in the CLC Main Workbench 7 (Qiagen, Aarhus, Denmark). Partial 12S rDNA sequences were aligned using the MAFFT algorithm and compared to sequences available in GenBank [48] using BLASTN (v.2.10.0) [49] and to sequences from morphologically determined reference specimens of multiple species of Hyalomma. Maximum likelihood 12S rDNA phylogenies were built in MEGA7 [59], and tick sequences were grouped based on their position in the 12S rDNA phylogenies. See Hoffman et al. 2021 [39] for details.
Whole Genome and Mitochondrion Phylogenies
Publicly available sequences for Francisella, Rickettsia, Midichloria, and tick mitochondria, as determined by the GTDB (bacteria) and NCBI (ticks) taxonomies, were downloaded from NCBI (Table S1 in the Supplementary Materials) using NCBI-genome-download (v.0.3.0) (https://github.com/kblin/ncbi-genome-download, accessed on 15 May 2021). Using the workflow manager Snakemake (v.6.2.1) [52], the genome assemblies and the public genomes were aligned pairwise with prograssiveMauve (v.2015_02_13) to selected reference genomes: F. tularensis tularensis strain SCHUS4 (GCF_000008985.1) for Francisella-positive samples, Rickettsia rickettsii strain Iowa (GCA_000017445.3) for Rickettsia-positive samples, Midichloria mitochondrii strain IricVA (GCA_000219355.1) for Midichloria-positive samples, and Hyalomma asiaticum strain WY042-2 (NC_053941) for tick mitochondrial sequences. The Python script included in CanSNPer (v.1.0.8) [60] was used to set the alignments to the reference coordinates and to merge them into a multi-FASTA file. IQ-TREE (v.2.1.2) [61] with ModelFinder setting (-m TEST) was used to create the four separate phylogenies. The selected best fit models according to Bayesian Information Criterion (BIC) for Francisella was GTR + F + I + G4, for Rickettsia TVM + F + I + G4, for Midichloria TVM + F + I + G4, and for tick mitochondria K3Pu + F + I + G4. The trees were recalculated with the selected models, and support values were calculated with bootstrap –b 100. The trees were visualized using iTOL [62]. The Francisella tree was rooted in Clade 2 according to a previous publication [7], the Midichloria and Rickettsia phylogenies were rooted in Orientia tsutsugamushi (Genome: Orientia tsutsugamushi strain Karp GCF_900327275.1) according to the Encyclopedia of Life [63], and the tick mitochondrion phylogeny was rooted by Rhipicephalus decoloratus (NC_053941) [64].
2.6. Genome Analysis
2.6.1. Average Nucleotide Identity
The similarity between two genomes at the nucleotide-level, average nucleotide identity (ANI), was calculated pairwise for all genomes within each dataset using pyANI (v.0.2.10) with ANIb (BLASTN+) method setting [65].
2.6.2. Biotin Synthesis Pathways
Previous analyses have identified multiple genes involved in the biotin synthesis pathway in the FLE of the tick O. moubata, including bioA, bioB, bioC, bioD, and bioF [11,13]. Homologs to these genes are present in the genome of the FLE of the tick species Argus arboreus (Francisella persica) [66]. To assess the conservation of these same genes in the genomes of the Francisella-like and Midichloria endosymbionts from H. rufipes samples D14IT15.2 and D14IT20, sequencing reads from both the enriched and non-enriched metagenomic data for these samples were mapped to these genes in the F. persica and M. mitochondrii genomes with minimap2 (v.2.22) [67] and the breadth of coverage was calculated with Samtools (v.1.11) [68] at a minimum depth of 3X.
3. Results
3.1. Bird Trapping and Tick Collection
In total, 10,209 birds were trapped and screened for ticks. Of these, 244 (2.4%) birds were found to be infested by ticks (n = 575) (Table S2 in the Supplementary Materials). Most of the tick-infested birds were long-distance migrants (98.0%). See Hoffman et al. [39] for additional details and information about the distribution pattern of ticks on the bird species.
3.2. Tick Determination
The collected ticks were assigned to the Ixodidae genera Hyalomma, Ixodes, Amblyomma, and Haemaphysalis, according to their position in phylogenies based on partial 12S rDNA sequences [39]. The assignment was not possible for 11.5% of the ticks due to the absence of PCR amplicons. The most common tick species were H. rufipes and H. marginatum [39]. The two metagenomics characterized ticks were confirmed as H. rufipes based on their similarity to a known COI sequence from H. rufipes and their positioning in the Hyalomma 12S rDNA [39] and mitochondrion (Figure 1) phylogenies.
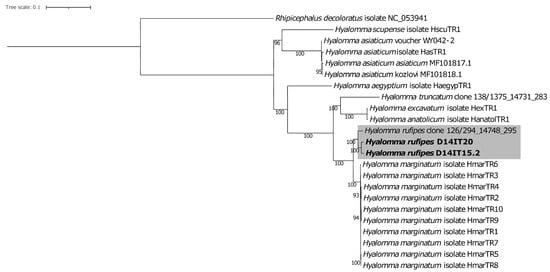
Figure 1.
Maximum likelihood phylogeny of Hyalomma based on mitochondrial genomes. Study genomes (in bold) form a highly supported clade together with the species Hyalomma rufipes (shaded area). Rhipicephalus decoloratus was used to root the tree. Bootstrap values ≥ 75 are presented at the nodes. The scale bar represents the expected number of substitutions per site.
3.3. Detection and Determination
3.3.1. Francisella
Results from the microfluidic qPCR suggested the presence of Francisella spp. in 72.5% (417/575; fopA+) of the total collected ticks, including 77.0% (371/482) of the Hyalomma ticks (H. rufipes: 76.7% (343/447); H. marginatum: 75% (18/24)), 5.9% (1/17) of the Ixodes ticks, 100% (2/2) of the Haemaphysalis ticks, 62.5% (5/8) of the Amblyomma ticks, and 57.6% (38/66) of the undetermined ticks. Furthermore, screening results from the microfluidic qPCR suggested the putative presence of F. tularensis in two H. rufipes ticks (0.3%; 2/575; fopA+ and lpnA/Tul4+) collected from two whinchats (Saxicola rubetra) trapped on the island of Capri (Italy) in 2014, a result that was not confirmed using the F. tularensis specific Tul4 and iQFt1 primers and probes (Table 2).

Table 2.
Confirmation results of the two ticks that tested positive for the putative presence of Francisella tularensis DNA during screening.
3.3.2. Spotted Fever Group Rickettsia
The microfluidic qPCR data suggested the presence of SFGR in 59.1% (340/575, gltA+) of the total collected ticks, including 60.0% (289/482) of Hyalomma ticks (H. rufipes: 61.5% (275/447); H. marginatum: 50.0% (12/24)), 17.6% (3/17) of Ixodes ticks, 100% (2/2) of Haemaphysalis ticks, 50.0% (4/8) of Amblyomma ticks, and 63.6% (42/66) of the undetermined ticks. Presence of SFGR was confirmed in 26 out of 38 analyzed samples by comparison with 17 kDa sequences deposited in GenBank.
3.3.3. Co-Occurrence
Screening data suggested the presence of both Francisella and SFGR spp. in 47.1% (271/575; fopA+ and gltA+) of the total collected ticks, including 48.8% (235/482) of Hyalomma ticks (H. rufipes: 50.6% (226/447); H. marginatum: 29.2% (7/24)), 5.9% (1/17) of Ixodes ticks, 100% (2/2) of Haemaphysalis ticks, 25% (2/8) of Amblyomma ticks, and 47% (31/66) of the undetermined ticks.
3.3.4. Metagenomics
Classification of metagenomic sequencing reads from tick samples that were putatively positive for F. tularensis by microfluidic qPCR confirmed the presence of FLEs and not F. tularensis. However, assembly of complete Francisella genomes was not possible from these data due to low read count and coverage (Table 3). Two rounds of Francisella enrichment were therefore performed, resulting in >95% Francisella DNA after enrichment. The assembly of Francisella DNA present in the sequenced enriched sample D14IT15.2 was in total 1,413,985 bp divided into 510 contigs with N50 = 10,189 and N50n = 36. The assembly of Francisella DNA present in the sequenced enriched sample D14IT20 was, in total, 1,415,455 bp divided into 506 contigs with N50 = 11,392 and N50n = 35.

Table 3.
Relative abundance of bacterial reads of selected species in metagenomic sequence data according to Kraken 2 results for the two ticks testing putatively positive for presence of Francisella tularensis by microfluidic real-time PCR. Species determination according to the Genome Taxonomy Database.
The levels of Rickettsia and Midichloria DNA present in the metagenomic data were relatively high compared to that of Francisella DNA. The assembly of Rickettsia DNA present in the sequenced non-enriched sample D14IT15.2 was in total 1,317,746 bp divided into 20 contigs with N50 = 225,990 and N50n = 3. The assembly of Rickettsia DNA present in the sequenced non-enriched sample D14IT20 was in total 1,318,206 bp divided into 20 contigs with N50 = 225,819 and N50n = 3. The assembly of Midichloria DNA present in the sequenced non-enriched sample D14IT15.2 was in total 1,069,525 bp divided into 105 contigs with N50 = 17,240 and N50n = 23. The assembly of Midichloria DNA present in the sequenced non-enriched sample D14IT20 was in total 952,239 bp divided into 151 contigs with N50 = 9933 and N50n = 34.
3.3.5. Phylogenetic Inference of Metagenome-Assembled Genomes
Phylogenetic inference of the MAGs of Francisella revealed that D14IT15.2 and D14IT20 belonged to Clade 1 of Francisella [7], are separated from F. tularensis (FSC200, SCHUS4), and are members of a subclade within the FLE group (GTDB cluster: Francisella sp002095075) (Figure 2, shaded area). The FLE group consists of both cluster F. persica and Francisella sp002095075. Characterization of metagenome-assembled Midichloria and Rickettsia genomes revealed resemblance to M. mitochondrii and R. aeschlimannii, respectively, with the latter within the clade of Rickettsia rhipicephali, according to the taxonomy used by GTDB (Figures S1 and S2 in the Supplementary Materials).
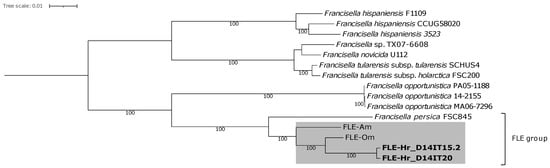
Figure 2.
Whole-genome maximum likelihood phylogeny of Clade 1 Francisella. The phylogeny contains 13 representative Francisella genomes, and the shaded area indicates the subclade within the FLE group (GTDB cluster: Francisella sp002095075) in which the Francisella metagenome-assembled genomes (in bold) generated in this study are assigned. The tree was rooted in Clade 2 of Francisella (not shown). Bootstrap values ≥ 75 are presented at the nodes. The scale bar represents the expected number of substitutions per site. FLE—Francisella-like endosymbiont; FLE-Om (host: Ornithodoros moubata); FLE-Am (host: Amblyomma maculatum); FLE-Hr (detected in Hyalomma rufipes); sp.—species; subsp.—subspecies.
3.4. Genome Analyses
3.4.1. Average Nucleotide Identity
The highest ANI values observed were between the generated MAGs and the genomes of FLE-Om, R. rhipicephali (R. aeschlimannii), M. mitochondrii, and H. rufipes, respectively (Table 4, Figures S3–S6 in the Supplementary Materials). In GTDB R. aeschlimannii belongs to the species R. rhipicephali while it is a recognised species in NCBI. The sequence identity between the two genomes of: (i) FLE-Hr (FLE_D14IT15.2 and FLE_D14IT20) was 99.7–99.8%, (ii) Midichloria-Hr (MID_D14IT15.2 and MID_D14IT20) 99.9–100%, (iii) Rickettsia-Hr (RICK_D14IT15.2 and RICK_D14IT20) 100%, and (iv) Hyalomma (HYA_D14IT15.2 and HYA_D14IT20) 99.3%.

Table 4.
Highest average nucleotide identity for bacterial (n = 6) and tick (n = 2) metagenome-assembled genomes generated in this study detected in two avian-associated Hyalomma rufipes ticks (D14IT15.2 and D14IT20) that tested positive for Francisella tularensis and spotted fever group Rickettsia during screening.
3.4.2. Biotin Gene Conservation
Based upon the mapping of reads to the corresponding F. persica coding DNA sequences (CDSs) encoding homologs to bioA, bioB, bioC, bioD, and bioF, at least bioA appears to be missing in the FLE MAGs generated from the enrichment of samples D14IT15.2 and D14IT20, indicating the biotin pathway is not intact in these FLEs (Table 5). We note that mapping of FLE reads from the non-enriched metagenomic sequencing data to these same CDSs was very limited, owing to the low proportion of FLE reads in these data (Table 3). In contrast, there was significant mapping of reads from the non-enriched metagenomic data to all of the examined biotin genes in the M. mitochondrii genome, indicating that this pathway is likely intact in the Midichloria endosymbiont of H. rufipes. There was a limited mapping of reads from the enriched metagenomic data to the biotin genes in M. mitochondrii, which is not unexpected given the high proportion of Francisella DNA in these samples following enrichment.

Table 5.
Coverage breadth (%) of sequencing reads of samples D14IT15.2 and D14IT20 mapped to genes involved in the biotin synthesis in the genome of Francisella persica and Midichloria mitochondrii, indicating an intact biotin synthesis pathway of Midichloria and a disrupted biotin synthesis pathway of Francisella-like endosymbionts detected in the tick species Hyalomma rufipes.
4. Discussion
In this study, screening of ticks infesting birds migrating in a northward route from wintering areas in Africa revealed a high prevalence of Francisella in the tick species H. rufipes (fopA+: 76.7%), a known vector of SFGR and Crimean-Congo hemorrhagic fever virus [69], which suggest that migratory birds in the AWPR may contribute to northward dispersal of Francisella-infected ticks. The high prevalence of Francisella likely represents a high prevalence of FLEs, as FLEs have previously been detected in multiple species of Hyalomma ticks and at a high prevalence [17,20,70]. The two Francisella MAGs generated from two H. rufipes (Hr) were found to be members of a subclade within the FLE group (GTDB cluster: Francisella sp002095075) in Clade 1 of Francisella (Figure 2), which includes the FLE species FLE-Om (present in the soft tick O. moubata) and FLE-Am (present in the hard tick A. maculatum). The FLE-Hr had the highest identity to FLE-Om (ANIb: 96.7–97.0%). Several ticks tested positive for both Francisella and SFGR spp. (fopA+/gltA+: 47.1%), suggesting that presence of Francisella did not prevent the occurrence of SFGR spp.; a similar observation has been reported by Scoles [19]. ANI and phylogenetic inference indicated the highest similarity of the Rickettsia MAGs detected in H. rufipes (Rickettsia-Hr) with R. aeschlimannii (ANIb: 98.8–99.9%), a SFGR (i) reported to cause human infections [71,72], (ii) associated with Hyalomma ticks, including H. rufipes and H. marginatum [34,73,74,75,76,77], (iii) previously identified together with FLEs in H. marginatum [23], and (iv) detected at similar prevalences as in this study in ticks of the H. marginatum species complex infesting northbound migratory birds [36,78]. Data from the two metagenomic sequenced H. rufipes ticks indicated co-occurrence also with a species of Midichloria. ANI and phylogenetic inference of the Midichloria MAGs detected in H. rufipes (Midichloria-Hr) indicated a close relationship to M. mitochondrii (ANIb: 91.5–92.3%). Midichloria sp. bacteria have previously been detected in H. marginatum species complex ticks infesting northbound trans-Saharan spring migrating birds trapped in Italy [47]. That study found a high prevalence of Midichloria DNA in the investigated Hyalomma ticks (>90%) and in a considerable fraction of the blood samples from the avian hosts (>40%) and suggested that the presence of Midichloria DNA in the blood was associated with lower fat reserves in the tick-infested birds [47].
Ticks may depend on FLEs because they provide nutrients that are absent in the tick diet, such as B vitamins (folate/folic acid (B9), riboflavin (B2), and biotin (B7)) and co-factors, and thereby improve the fitness of the tick [5,11]. It has been suggested that FLEs serve as alternative obligate symbionts in some species of ticks [5,79], whereas Coxiella-like endosymbionts (CLEs) are considered to be obligate symbionts (i.e., present in most specimens) in most tick species [6,80]. Ticks may escape a negative symbiosis by replacing an old symbiont with a new bacterium [81]. FLEs may have replaced CLEs in several tick lineages, including O. moubata and A. maculatum [5,13,79]. M. mitochondrii has also been suggested to be a nutritional endosymbiont since it encodes genes for the production of several co-factors and B vitamin biotin [82]. Buysse et al. [23] showed that the genomes of FLEs detected in H. marginatum included functional biosynthesis pathways for folate and riboflavin but were deprived of a functional biosynthesis pathway for biotin. The authors suggested that this was compensated for by the co-symbiosis with Midichloria bacteria also present in H. marginatum, since their genomes included an intact biotin biosynthesis operon [23]. The Midichloria detected in H. marginatum had a partial riboflavin biosynthesis pathway, indicating that co-occurrence of FLEs and Midichloria may be essential for complete nutritional symbiosis in H. marginatum [23]. We observed similar patterns for H. rufipes: a disrupted biotin biosynthesis pathway in its FLE but an apparently intact biotin biosynthesis pathway within its Midichloria endosymbiont (Table 5), suggesting the co-occurrence of these bacteria also may be essential for nutritional symbiosis in H. rufipes. As noted by Buysse et al. [23], a similar dual symbiosis may be present in several other Hyalomma tick species (Hyalomma aegyptium, Hyalomma anatolicum, Hyalomma dromedarii, Hyalomma excavatum, Hyalomma impeltatum, Hyalomma lusitanicum, and Hyalomma truncatum), as they found evidence for the presence of both FLEs and Midichloria within them. The apparent exception within this tick genus to date appears to be Hyalomma asiaticum, which does not harbor any Midichloria but instead has an FLE with an intact biotin pathway [83].
Molecular species determination of members of the H. marginatum species complex—currently consisting of five species, including H. marginatum and H. rufipes [84]—can be difficult due to the inclusion in public databases of sequences obtained from incorrectly identified specimens [46]. Complete tick mitochondrial MAGs were therefore constructed to verify the initial 12S rDNA-based speciation of the metagenomically characterized ticks. The two characterized tick specimens were found to group in the H. rufipes clade also in the mitochondrion phylogeny, verifying the initial 12S rDNA speciation results.
5. Conclusions
Understanding the biology, ecology, and evolution of tick endosymbionts is important, as they may share a close evolutionary relationship with pathogenic bacteria and also may influence the fitness [4] and even the behavior of the tick host [85]. The results of this study demonstrate that FLEs are present in many H. rufipes ticks, and migratory birds in the AWPR contribute to the northward geographical spread of FLE-containing ticks. The absence of F. tularensis in the investigated ticks does not provide evidence supporting that immature life stages of H. rufipes contribute to the transmission of F. tularensis in the study region. Furthermore, the results suggest that migratory birds also contribute to northward geographical spread in the AWPR of H. rufipes ticks containing SFGR spp., including R. aeschlimannii and Midichloria bacteria, and that a dual endosymbiosis (co-symbiosis) of FLEs and Midichloria may support the nutritional requirements of the medically important tick vector H. rufipes. We acknowledge that the majority of the results of this study are based on unconfirmed screening data and that the reported detection results are therefore conservative estimates of the prevalence. Future studies should therefore focus on verifying the Francisella and SFGR prevalence in H. rufipes as well as investigate the Midichloria prevalence in H. rufipes and the impact that FLEs and Midichloria may have on H. rufipes, including their interaction with bacterial pathogens, such as SFGR.
Supplementary Materials
Supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms10071393/s1, Table S1. Publicly available genomes for Francisella, Rickettsia, Midichloria, and tick mitochondria downloaded from GTDB and NCBI. Table S2. Microfluidic real-time PCR data for ticks collected from bird species trapped in the Mediterranean basin during the spring migration of 2014 and 2015. The ticks were screened for Francisella and spotted fever group Rickettsia species using primers targeting the fopA and gltA genes. Figure S1. Whole genome maximum likelihood phylogeny. Highlighted area indicates Midichloria clade. Study genomes are in bold. Orientia tsutsugamushi strain Karp was used to root the tree. Bootstrap values ≥ 75 are presented at the nodes. The scale bar represents the expected number of substitutions per site. Figure S2. Whole genome maximum likelihood phylogeny of Rickettsia. Highlighted area indicates clade for Rickettsia aeschlimannii. Study genomes are in bold. Orientia tsutsugamushi was used to root the tree. Bootstrap values ≥ 75 are presented at the nodes. The scale bar represents the expected number of substitutions per site. Figure S3. Heatmap of the average nucleotide identity (ANI), demonstrating nucleotide-level genomic similarity between Francisella genomes. The pairwise comparison of 19 Francisella genomes was computed by BLAST, using the pyANI software. Study genomes are in bold. FLE, Francisella-like endosymbiont. Figure S4. Heatmap of the average nucleotide identity (ANI), demonstrating nucleotide-level genomic similarity between Rickettsia genomes. The pairwise comparison of 138 Rickettsia genomes was computed by BLAST, using the pyANI software. Study genomes are in bold. For NCBI organism names, see Table S1. Figure S5. Heatmap of the average nucleotide identity (ANI), demonstrating nucleotide-level genomic similarity between bacterial genomes. The pairwise comparison of 23 genomes was computed by BLAST, using the pyANI software. Study genomes are in bold. Figure S6. Heatmap of the average nucleotide identity (ANI), demonstrating nucleotide-level genomic similarity between Hyalomma mitochondrial genomes. The pairwise comparison of 23 Hyalomma mitochondrial genomes was computed by BLAST, using the pyANI software. Study genomes are in bold.
Author Contributions
T.H. organized the project, analyzed and interpreted data, and wrote the original draft of the manuscript. A.S. performed the bioinformatic analyses and supervised the sequencing. C.Ö. analyzed the whole genome phylogenies and the ANI. L.K. performed molecular confirmation analyses and Nanopore sequencing. R.F.M., J.W.S., D.B. and D.M.W. performed the enrichment, Illumina sequencing of enriched samples, and the biotin gene conservation analysis. L.G.C. assisted in the molecular determination of tick taxa and confirmation analyses. P.W. performed molecular confirmation analyses. C.B., J.F., A.O., Y.K., A.O. and D.P. conducted bird trappings and tick collections. S.M. performed the microfluidic qPCR analyses. J.H.-O.P. supervised and assisted in the molecular analyses. T.G.T.J., P.-E.L., M.F., T.F. and K.N. supervised. Å.L. funded and supervised the project. B.O. funded, organized, and supervised the project. The manuscript has been read and commented on by all authors. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by ALF grants from the Uppsala County Council, the Ax:son Johnson Foundation, the European Union’s Horizon 2020 research innovation program (Grant no: 874735) (VEO), SciLifeLab Pandemic Laboratory Preparedness (LPP1-007), and NGI/Uppmax. S.M.’s research was supported by the French Agency for Food, Environmental, and Occupational Health and Safety (ANSES) (ANSES-2016). J.H.-O.P. is funded by the Swedish research council FORMAS (Grant no: 2015-710) and VR (Grant no: 2020-02593). Furthermore, this work was supported by the Swedish Civil Contingencies Agency (Grant no: TA 014-2010-01) and the US Department of Homeland Security’s Science and Technology Directorate (Award no: HSHQDC-17-C-B0021) pursuant to the agreement between the Kingdom of Sweden and the US government on Cooperation in Science and Technology for Homeland Security Matters.
Institutional Review Board Statement
Collection of ticks was performed by trained and licensed ringers at the bird observatories during the regular bird ringing/banding activities, under the following permits and licenses: 660117 and 180007 issued by the Ministerio de Agricultura, Pesca y Alimentación, and 66042 issued by Consejeria de Agricultura, Ganadería, Pesca y Desarrollo Sostenible, Spain; 59019 issued by L’Instituto Superiore per la Protezione e Ricerca dell’Ambientale (ISPRA), Italy; AΔA:ΒΛ9Σ0-Γ3A, AΔA:Β4ΩΖ0-Ν6Χ, AΔA:ΩHΛΔ465ΦΘH-31Γ, and AΔA: ΩΧΒΠ465ΦΘH-ΒΧΥ issued by the Hellenic Ministry of Environment and Energy, Greece; and A258 issued by the Israel Nature and Parks Authority, Israel.
Informed Consent Statement
Not applicable.
Data Availability Statement
The metagenomic sequence data generated in this study are available under the NCBI BioProjectPRJNA764565.
Acknowledgments
We acknowledge the ringers at the bird observatories for trapping the birds and collecting the ticks. The SNP&SEQ Technology Platform at Uppsala University, which is part of the National Genomics Infrastructure, is acknowledged for metagenomic sequencing. This is contribution number 34 from the Antikythira Bird Observatory-Hellenic Ornithological Society.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Moutailler, S.; Valiente Moro, C.; Vaumourin, E.; Michelet, L.; Tran, F.H.; Devillers, E.; Cosson, J.-F.; Gasqui, P.; Van, V.T.; Mavingui, P.; et al. Co-infection of ticks: The rule rather than the exception. PLoS Negl. Trop. Dis. 2016, 10, e0004539. [Google Scholar] [CrossRef] [PubMed]
- Milhanoa, N.; de Carvalhoa, I.L.; Alvesa, A.S.; Arroubeb, S.; Soaresc, J.; Rodriguezc, P.; Carolinod, M.; Núncioa, M.S.; Piesmane, J.; de Sousaa, R. Coinfections of Rickettsia slovaca and Rickettsia helvetica with Borrelia lusitaniae ticks collected in a safari park, Portugal. Ticks Tick Borne Dis. 2010, 1, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, S.I.; Binetruy, F.; Hernandez-Jarguin, A.M.; Duron, O. The tick microbiome: Why non-pathogenic microorganisms matter in tick biology and pathogen transmission. Front. Cell. Infect. Microbiol. 2017, 7, 236. [Google Scholar] [CrossRef]
- Duron, O.; Gottlieb, Y. Convergence of nutritional symbioses in obligate blood feeders. Trends Parasitol. 2020, 36, 816–825. [Google Scholar] [CrossRef] [PubMed]
- Gerhart, J.G.; Moses, A.S.; Raghavan, R. A Francisella-like endosymbiont in the Gulf Coast tick evolved from a mammalian pathogen. Sci. Rep. 2016, 6, 33670. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.A.; Driscoll, T.; Gillespie, J.J.; Raghavan, R. A Coxiella-like endosymbiont is a potential vitamin source for the Lone Star tick. Genome Biol. Evol. 2015, 7, 831–838. [Google Scholar] [CrossRef]
- Öhrman, C.; Sahl, J.W.; Sjödin, A.; Uneklint, I.; Ballard, R.; Karlsson, L.; McDonough, R.F.; Sundell, D.; Soria, K.; Backman, S.; et al. Reorganized genomic taxonomy of Francisellaceae enables design of robust environmental PCR assays for detection of Francisella tularensis. Microorganisms 2021, 9, 146. [Google Scholar] [CrossRef]
- Sjödin, A.; Svensson, K.; Öhrman, C.; Ahlinder, J.; Lindgren, P.; Duodu, S.; Johansson, A.; Colquhoun, D.J.; Larsson, P.; Forsman, M. Genome characterisation of the genus Francisella reveals insight into similar evolutionary paths in pathogens of mammals and fish. BMC Genom. 2012, 13, 268. [Google Scholar] [CrossRef]
- Friend, M. Tularemia; Circular 1297; Reston, V., Ed.; U.S. Geological Survey: Reston, VA, USA, 2006. [Google Scholar]
- Dennis, D.T.; Inglesby, T.V.; Henderson, D.A.; Bartlett, J.G.; Ascher, M.S.; Eitzen, E.; Fine, A.D.; Friedlander, A.M.; Hauer, J.; Layton, M.; et al. Tularemia as a biological weapon: Medical and public health management. JAMA J. Am. Med. Assoc. 2001, 285, 2763–2773. [Google Scholar] [CrossRef] [PubMed]
- Gerhart, J.G.; Auguste Dutcher, H.; Brenner, A.E.; Moses, A.S.; Grubhoffer, L.; Raghavan, R. Multiple acquisitions of pathogen-derived Francisella endosymbionts in soft ticks. Genome Biol. Evol. 2018, 10, 607–615. [Google Scholar] [CrossRef]
- Baldridge, G.D.; Scoles, G.A.; Burkhardt, N.Y.; Schloeder, B.; Kurtti, T.J.; Munderloh, U.G. Transovarial transmission of Francisella-like endosymbionts and Anaplasma phagocytophilum variants in Dermacentor albipictus (Acari: Ixodidae). J. Med. Entomol. 2009, 46, 625–632. [Google Scholar] [CrossRef] [PubMed]
- Duron, O.; Morel, O.; Noêll, V.; Buysse, M.; Binetruy, F.; Lancelot, R.; Loire, E.; Ménard, C.; Bouchez, O.; Vavre, F.; et al. Tick-bacteria mutualism depends on B vitamin synthesis pathways. Curr. Biol. 2018, 28, 1896–1902. [Google Scholar] [CrossRef] [PubMed]
- Machado-Ferreira, E.; Piesman, J.; Zeidner, N.S.; Carlos, A.G.S. Francisella-like endosymbiont DNA and Francisella tularensis virulence-related genes in Brazilian ticks (Acari: Ixodidae). J. Med. Entomol. 2009, 46, 369–374. [Google Scholar] [CrossRef]
- Kaufman, E.L.; Stone, N.E.; Scoles, G.A.; Hepp, C.M.; Busch, J.D.; Wagner, D.M. Range-wide genetic analysis of Dermacentor variabilis and its Francisella-like endosymbionts demonstrates phylogeographic concordance between both taxa. Parasites Vectors 2018, 11, 306. [Google Scholar] [CrossRef]
- Sumrandee, C.; Baimai, V.; Trinachartvanit, W.; Ahantarig, A. Molecular detection of Rickettsia, Anaplasma, Coxiella and Francisella bacteria in ticks collected from Artiodactyla in Thailand. Ticks Tick Borne Dis. 2016, 7, 678–689. [Google Scholar] [CrossRef] [PubMed]
- Szigeti, A.; Kreizinger, Z.; Hornok, S.; Abichu, G.; Gyuranecz, M. Detection of Francisella-like endosymbiont in Hyalomma rufipes from Ethiopia. Ticks Tick Borne Dis. 2014, 5, 818–820. [Google Scholar] [CrossRef] [PubMed]
- Sréter-Lancz, Z.; Széll, Z.; Sréter, T.; Márialigeti, K. Detection of a novel Francisella in Dermacentor reticulatus: A need for careful evaluation of PCR-based identification of Francisella tularensis in Eurasian ticks. Vector Borne Zoonotic Dis. 2009, 9, 123–126. [Google Scholar] [CrossRef]
- Scoles, G.A. Phylogenetic Analysis of the Francisella-like endosymbionts of Dermacentor ticks. J. Med. Entomol. 2004, 41, 277–286. [Google Scholar] [CrossRef]
- Ivanov, I.N.; Mitkova, N.; Reye, A.L.; Hübschen, J.M.; Vatcheva-Dobrevska, R.S.; Dobreva, E.G.; Kantardjiev, T.V.; Muller, C.P. Detection of new Francisella-like tick endosymbionts in Hyalomma spp. and Rhipicephalus spp. (Acari: Ixodidae) from Bulgaria. Appl. Environ. Microbiol. 2011, 77, 5562–5565. [Google Scholar] [CrossRef]
- Sun, L.V.; Scoles, G.A.; Fish, D.; O’Neill, S.L. Francisella-like endosymbionts of ticks. J. Invertebr. Pathol. 2000, 76, 301–303. [Google Scholar] [CrossRef] [PubMed]
- Noda, H.; Munderloh, U.G.; Kurtti, T.J. Endosymbionts of ticks and their relationship to Wolbachia spp. and tick-borne pathogens of humans and animals. Appl. Environ. Microbiol. 1997, 63, 3926–3932. [Google Scholar] [CrossRef] [PubMed]
- Buysse, M.; Floriano, A.M.; Gottlieb, Y.; Nardi, T.; Comandatore, F.; Olivieri, E.; Giannetto, A.; Palomar, A.M.; Makepeace, B.L.; Bazzocchi, C.; et al. A dual endosymbiosis supports nutritional adaptation to hematophagy in the invasive tick Hyalomma marginatum. eLife 2021, 10, e72747. [Google Scholar] [CrossRef] [PubMed]
- Michelet, L.; Bonnet, S.; Madani, N.; Moutailler, S. Discriminating Francisella tularensis and Francisella-like endosymbionts in Dermacentor reticulatus ticks: Evaluation of current molecular techniques. Vet. Microbiol. 2013, 163, 399–403. [Google Scholar] [CrossRef] [PubMed]
- Niebylski, M.L.; Peacock, M.G.; Fischer, E.R.; Porcella, S.F.; Schwan, T.G. Characterization of an endosymbiont infecting wood ticks, Dermacentor andersoni, as a member of the genus Francisella. Appl. Environ. Microbiol. 1997, 63, 3933–3940. [Google Scholar] [CrossRef]
- Epis, S.; Mandrioli, M.; Genchi, M.; Montagna, M.; Sacchi, L.; Pistone, D.; Sassera, D. Localization of the bacterial symbiont Candidatus Midichloria mitochondrii within the hard tick Ixodes ricinus by whole-mount FISH staining. Ticks Tick Borne Dis. 2013, 4, 39–45. [Google Scholar] [CrossRef]
- Lo, N.; Beninati, T.; Sassera, D.; Bouman, E.A.; Santagati, S.; Gern, L.; Sambri, V.; Masuzawa, T.; Gray, J.S.; Jaenson, T.G.; et al. Widespread distribution and high prevalence of an alpha-proteobacterial symbiont in the tick Ixodes ricinus. Environ. Microbiol. 2006, 8, 1280–1287. [Google Scholar] [CrossRef] [PubMed]
- Sassera, D.; Beninati, T.; Bandi, C.; Bouman, E.A.P.; Sacchi, L.; Fabbi, M.; Lo, N. ‘Candidatus Midichloria mitochondrii’, an endosymbiont of the tick Ixodes ricinus with a unique intramitochondrial lifestyle. Int. J. Syst. Evol. Microbiol. 2006, 56, 2535–2540. [Google Scholar] [CrossRef]
- Epis, S.; Sassera, D.; Beninati, T.; Lo, N.; Beati, L.; Piesman, J.; Rinaldi, L.; McCoy, K.D.; Torina, A.; Sacchi, L.; et al. Midichloria mitochondrii is widespread in hard ticks (Ixodidae) and resides in the mitochondria of phylogenetically diverse species. Parasitology 2008, 135, 485–494. [Google Scholar] [CrossRef]
- Di Venere, M.; Fumagalli, M.; Cafiso, A.; De Marco, L.; Epis, S.; Plantard, O.; Bardoni, A.; Salvini, R.; Viglio, S.; Bazzocchi, C.; et al. Ixodes ricinus and its endosymbiont Midichloria mitochondrii: A comparative proteomic analysis of salivary glands and ovaries. PLoS ONE 2015, 10, e0138842. [Google Scholar] [CrossRef]
- Bazzocchi, C.; Mariconti, M.; Sassera, D.; Rinaldi, L.; Martin, E.; Cringoli, G.; Urbanelli, S.; Genchi, C.; Bandi, C.; Epis, S. Molecular and serological evidence for the circulation of the tick symbiont Midichloria (Rickettsiales: Midichloriaceae) in different mammalian species. Parasites Vectors 2013, 6, 350. [Google Scholar] [CrossRef]
- Mariconti, M.; Epis, S.; Gaibani, P.; Valle, C.D.; Sassera, D.; Tomao, P.; Fabbi, M.; Castelli, F.; Marone, P.; Sambri, V.; et al. Humans parasitized by the hard tick Ixodes ricinus are seropositive to Midichloria mitochondrii: Is Midichloria a novel pathogen, or just a marker of tick bite? Pathog. Glob. Health 2012, 106, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, J.J.; Beier, M.S.; Rahman, M.S.; Ammerman, N.C.; Shallom, J.M.; Purkayastha, A.; Sobral, B.S.; Azad, A.F. Plasmids and rickettsial evolution: Insight from Rickettsia felis. PLoS ONE 2007, 2, e266. [Google Scholar] [CrossRef] [PubMed]
- Parola, P.; Paddock, C.D.; Socolovschi, C.; Labruna, M.B.; Mediannikov, O.; Kernif, T.; Abdad, M.Y.; Stenos, J.; Bitam, I.; Fournier, P.E.; et al. Update on tick-borne rickettsioses around the world: A geographic approach. Clin. Microbiol. Rev. 2013, 26, 657–702. [Google Scholar] [CrossRef] [PubMed]
- Brouqui, P.; Parola, P.; Fournier, P.E.; Raoult, D. Spotted fever rickettsioses in southern and eastern Europe. FEMS Immunol. Med. Microbiol. 2007, 49, 2–12. [Google Scholar] [CrossRef]
- Wallménius, K.; Barboutis, C.; Fransson, T.; Jaenson, T.G.; Lindgren, P.E.; Nyström, F.; Olsen, B.; Salaneck, E.; Nilsson, K. Spotted fever Rickettsia species in Hyalomma and Ixodes ticks infesting migratory birds in the European Mediterranean area. Parasites Vectors 2014, 7, 318. [Google Scholar] [CrossRef] [PubMed]
- Olsen, B.; Jaenson, T.G.; Bergström, S. Prevalence of Borrelia burgdorferi sensu lato-infected ticks on migrating birds. Appl. Environ. Microbiol. 1995, 61, 3082–3087. [Google Scholar] [CrossRef]
- Hoffman, T.; Wilhelmsson, P.; Barboutis, C.; Fransson, T.; Jaenson, T.G.T.; Lindgren, P.-E.; Von Loewenich, F.D.; Lundkvist, Å.; Olsen, B.; Salaneck, E. A divergent Anaplasma phagocytophilum variant in an Ixodes tick from a migratory bird; Mediterranean basin. Infect. Ecol. Epidemiol. 2020, 10, 1729653. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, T.; Carra, L.G.; Öhagen, P.; Fransson, T.; Barboutis, C.; Piacentini, D.; Figuerola, J.; Kiat, K.; Onrubia, A.; Jaenson, T.G.T.; et al. Association between guilds of birds in the African-Western Palaearctic region and the tick species Hyalomma rufipes, one of the main vectors of Crimean-Congo hemorrhagic fever virus. One Health 2021, 13, 100349. [Google Scholar] [CrossRef] [PubMed]
- Michelet, L.; Delannoy, S.; Devillers, E.; Umhang, G.; Aspan, A.; Juremalm, M.; Chirico, J.; van der Wal, F.J.; Sprong, H.; Boye Pihl, T.P.; et al. High-throughput screening of tick-borne pathogens in Europe. Front. Cell. Infect. Microbiol. 2014, 4, 103. [Google Scholar] [CrossRef] [PubMed]
- Cronhjort, S.; Wilhelmsson, P.; Karlsson, L.; Thelaus, J.; Sjödin, A.; Forsberg, P.; Lindgren, P.-E. The Tick-Borne Diseases STING study: Real-time PCR analysis of three emerging tick-borne pathogens in ticks that have bitten humans in different regions of Sweden and the Åland islands, Finland. Infect. Ecol. Epidemiol. 2019, 9, 1683935. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Thelaus, J.; Andersson, A.; Mathisen, P.; Forslund, A.L.; Noppa, L.; Forsman, M. Influence of nutrient status and grazing pressure on the fate of Francisella tularensis in lake water. FEMS Microbiol. Ecol. 2009, 67, 69–80. [Google Scholar] [CrossRef]
- Beati, L.; Keirans, J.E. Analysis of the systematic relationships among ticks of the genera Rhipicephalus and Boophilus (Acari: Ixodidae) based on mitochondrial 12S ribosomal DNA gene sequences and morphological characters. J. Parasitol. 2001, 87, 32–48. [Google Scholar] [CrossRef]
- Harder, T.C.; Harder, C.M.; Kulonen, H.V.K.; Kennedy-Stoskopf, S.; Liess, B.; Appel, M.J.G.; Osterhaus, A.D.M.E. Characterization of phocid herpesvirus-1 and -2 as putative alpha- and gammaherpesviruses of North American and European pinnipeds. J. Gen. Virol. 1996, 77, 27–35. [Google Scholar] [CrossRef]
- Carl, M.; Tibbs, C.W.; Dobson, M.E.; Paparello, S.; Dasch, G.A. Diagnosis of acute Typhus infection using the polymerase chain reaction. J. Infect. Dis. 1990, 161, 791–793. [Google Scholar] [CrossRef]
- Mihalca, A.D.; Estrada-Peña, A.; Petney, T.N. (Eds.) Ticks of Europe and North Africa. A Guide to Species Identification; Springer: Cham, Switzerland, 2017. [Google Scholar]
- Di Lecce, I.; Bazzocchi, C.; Cecere, J.G.; Epis, S.; Sassera, D.; Villani, B.M.; Bazzi, G.; Negri, A.; Saino, N.; Spina, F.; et al. Patterns of Midichloria infection in avian-borne African ticks and their trans-Saharan migratory hosts. Parasites Vectors 2018, 11, 106. [Google Scholar] [CrossRef]
- NCBI. GenBank. Available online: https://www.ncbi.nlm.nih.gov/genbank/ (accessed on 8 May 2019).
- NCBI. Basic Local Alignment Search Tool (BLAST). Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 8 May 2019).
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef] [PubMed]
- Sundell, D.; Öhrman, C.; Svensson, D.; Karlsson, E.; Brindefalk, B.; Myrtennäs, K.; Ahlinder, J.; Antwerpen, M.H.; Walter, M.C.; Forsman, M.; et al. FlexTaxD: Flexible modification of taxonomy databases for improved sequence classification. Bioinformatics 2021, 37, 3932–3933. [Google Scholar] [CrossRef] [PubMed]
- Köster, J.; Rahmann, S. Snakemake—A scalable bioinformatics workflow engine. Bioinformatics 2012, 28, 2520–2522. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Seppey, M.; Manni, M.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness. Methods Mol. Biol. 2019, 1962, 227–245. [Google Scholar] [CrossRef] [PubMed]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Ratnasingham, S.; Hebert, P.D. BOLD: The Barcode of Life Data System (http://www.barcodinglife.org). Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Lärkeryd, A.; Myrtennäs, K.; Karlsson, E.; Dwibedi, C.K.; Forsman, M.; Larsson, P.; Johansson, A.; Sjödin, A. CanSNPer: A hierarchical genotype classifier of clonal pathogens. Bioinformatics 2014, 30, 1762–1764. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Weinert, L.A.; Werren, J.H.; Aebi, A.; Stone, G.N.; Jiggins, F.M. Evolution and diversity of Rickettsia bacteria. BMC Biol. 2009, 7, 6. [Google Scholar] [CrossRef]
- Ciloglu, A.; Ibis, O.; Yildirim, A.; Aktas, M.; Duzlu, O.; Onder, Z.; Simsek, E.; Yetismis, G.; Ellis, V.A.; Inci, A. Complete mitochondrial genome characterization and phylogenetic analyses of the main vector of Crimean-Congo haemorrhagic fever virus: Hyalomma marginatum Koch, 1844. Ticks Tick Borne Dis. 2021, 12, 101736. [Google Scholar] [CrossRef]
- Pritchard, L.; Glover, R.H.; Humphris, S.; Elphinstone, J.G.; Toth, I.K. Genomics and taxonomy in diagnostics for food security: Soft-rotting enterobacterial plant pathogens. Anal. Methods 2018, 8, 12–24. [Google Scholar] [CrossRef]
- Larson, M.A.; Nalbantoglu, U.; Sayood, K.; Zentz, E.B.; Cer, R.Z.; Iwen, P.C.; Francesconi, S.C.; Bishop-Lilly, K.A.; Mokashi, V.P.; Sjöstedt, A.; et al. Reclassification of Wolbachia persica as Francisella persica comb. nov. and emended description of the family Francisellaceae. Int. J. Syst. Evol. Microbiol. 2016, 66, 1200–1205. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.R.; Bouattour, A.; Camicas, J.-L.; Estrada-Peña, A.; Horak, I.G.; Latif, A.A.; Pegram, R.G.; Preston, P.M. Ticks of Domestic Animals in Africa: A Guide to Identification of Species; Bioscience Reports: Edinburgh, UK; Atalanta, The Netherlands, 2003. [Google Scholar]
- Azagi, T.; Klement, E.; Perlman, G.; Lustig, Y.; Mumcuoglu, K.Y.; Apanaskevich, D.A.; Gottlieb, Y. Francisella-like endosymbionts and Rickettsia species in local and imported Hyalomma ticks. Appl. Environ. Microbiol. 2017, 83, 18. [Google Scholar] [CrossRef] [PubMed]
- Raoult, D.; Fournier, P.E.; Abboud, P.; Caron, F. First documented human Rickettsia aeschlimannii infection. Emerg. Infect. Dis. 2002, 8, 748–749. [Google Scholar] [CrossRef]
- Tosoni, A.; Mirijello, A.; Ciervo, A.; Mancini, F.; Rezza, G.; Damiano, F.; Cauda, R.; Gasbarrini, A.; Addolorato, G.; Internal Medicine Sepsis Study Group. Human Rickettsia aeschlimannii infection: First case with acute hepatitis and review of the literature. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 2630–2633. [Google Scholar]
- Palomar, A.M.; Portillo, A.; Mazuelas, D.; Roncero, L.; Arizaga, J.; Crespo, A.; Gutierrez, O.; Marquez, F.J.; Cuadrado, J.F.; Eiros, J.M.; et al. Molecular analysis of Crimean-Congo hemorrhagic fever virus and Rickettsia in Hyalomma marginatum ticks removed from patients (Spain) and birds (Spain and Morocco), 2009–2015. Ticks Tick Borne Dis. 2016, 7, 983–987. [Google Scholar] [CrossRef]
- Djerbouh, A.; Kernif, T.; Beneldjouzi, A.; Socolovschi, C.; Kechemir, N.; Parola, P.; Raoult, D.; Bitam, I. The first molecular detection of Rickettsia aeschlimannii in the ticks of camels from southern Algeria. Ticks Tick Borne Dis. 2012, 3, 374–376. [Google Scholar] [CrossRef]
- Beati, L.; Meskini, M.; Thiers, B.; Raoult, D. Rickettsia aeschlimannii sp. nov., a new spotted fever group rickettsia associated with Hyalomma marginatum ticks. Int. J. Syst. Bacteriol. 1997, 47, 548–554. [Google Scholar] [CrossRef]
- Fernandez-Soto, P.; Encinas-Grandes, A.; Perez-Sanchez, R. Rickettsia aeschlimannii in Spain: Molecular evidence in Hyalomma marginatum and five other tick species that feed on humans. Emerg. Infect. Dis. 2003, 9, 889–890. [Google Scholar] [CrossRef] [PubMed]
- Rumer, L.; Graser, E.; Hillebrand, T.; Talaska, T.; Dautel, H.; Mediannikov, O.; Roy-Chowdhury, P.; Sheshukova, O.; Mantke, O.D.; Niedrig, M. Rickettsia aeschlimannii in Hyalomma marginatum ticks, Germany. Emerg. Infect. Dis. 2011, 17, 325–326. [Google Scholar] [CrossRef] [PubMed]
- Pascucci, I.; Di Domenico, M.; Dondona, G.C.; Di Gennaro, A.; Polci, A.; Dondona, A.C.; Mancuso, E.; Camma, C.; Savini, G.; Cecere, J.G.; et al. Assessing the role of migratory birds in the introduction of ticks and tick-borne pathogens from African countries: An Italian experience. Ticks Tick Borne Dis. 2019, 10, 101272. [Google Scholar] [CrossRef] [PubMed]
- Duron, O.; Binetruy, F.; Noel, V.; Cremaschi, J.; McCoy, K.D.; Arnathau, C.; Plantard, O.; Goolsby, J.; de Leon, A.A.P.; Heylen, D.J.A.; et al. Evolutionary changes in symbiont community structure in ticks. Mol. Ecol. 2017, 26, 2905–2921. [Google Scholar] [CrossRef] [PubMed]
- Gottlieb, Y.; Lalzar, I.; Klasson, L. Distinctive genome reduction rates revealed by genomic analyses of two Coxiella-like endosymbionts in ticks. Genome Biol. Evol. 2015, 7, 1779–1796. [Google Scholar] [CrossRef]
- Bennett, G.M.; Moran, N.A. Heritable symbiosis: The advantages and perils of an evolutionary rabbit hole. Proc. Natl. Acad. Sci. USA 2015, 112, 10169–10176. [Google Scholar] [CrossRef]
- Sassera, D.; Lo, N.; Epis, S.; D’Auria, G.; Montagna, M.; Comandatore, F.; Horner, D.; Pereto, J.; Luciano, A.M.; Franciosi, F.; et al. Phylogenomic evidence for the presence of a flagellum and cbb(3) oxidase in the free-living mitochondrial ancestor. Mol. Biol. Evol. 2011, 28, 3285–3296. [Google Scholar] [CrossRef]
- Buysse, M.; Duron, O. Evidence that microbes identified as tick-borne pathogens are nutritional endosymbionts. Cell 2021, 184, 2259–2260. [Google Scholar] [CrossRef]
- Apanaskevich, D.; Horak, I. The genus Hyalomma Koch, 1844: V. Re-evaluation of the taxonomic rank of taxa comprising the H. (Euhyalomma) marginatum Koch complex of species (Acari: Ixodidae) with redescription of all parasitic stages and notes on biology. Int. J. Acarol. 2008, 34, 13–42. [Google Scholar] [CrossRef]
- Zhong, Z.; Zhong, T.; Peng, Y.; Zhou, X.; Wang, Z.; Tang, H.; Wang, J. Symbiont-regulated serotonin biosynthesis modulates tick feeding activity. Cell Host Microbe 2021, 29, 1545–1557.e1544. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).