The Use of Artificial Sputum Media to Enhance Investigation and Subsequent Treatment of Cystic Fibrosis Bacterial Infections
Abstract
1. Introduction
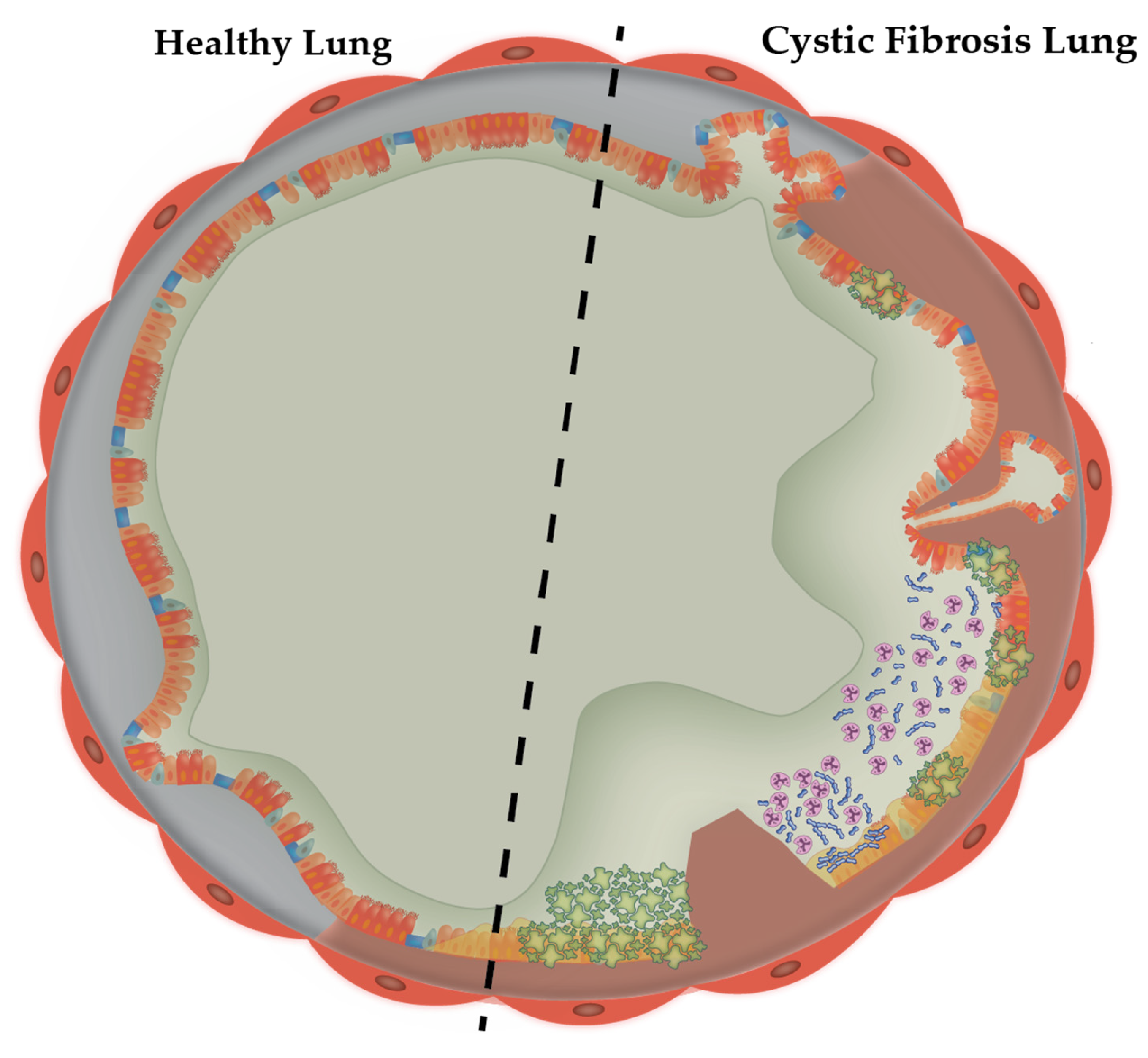
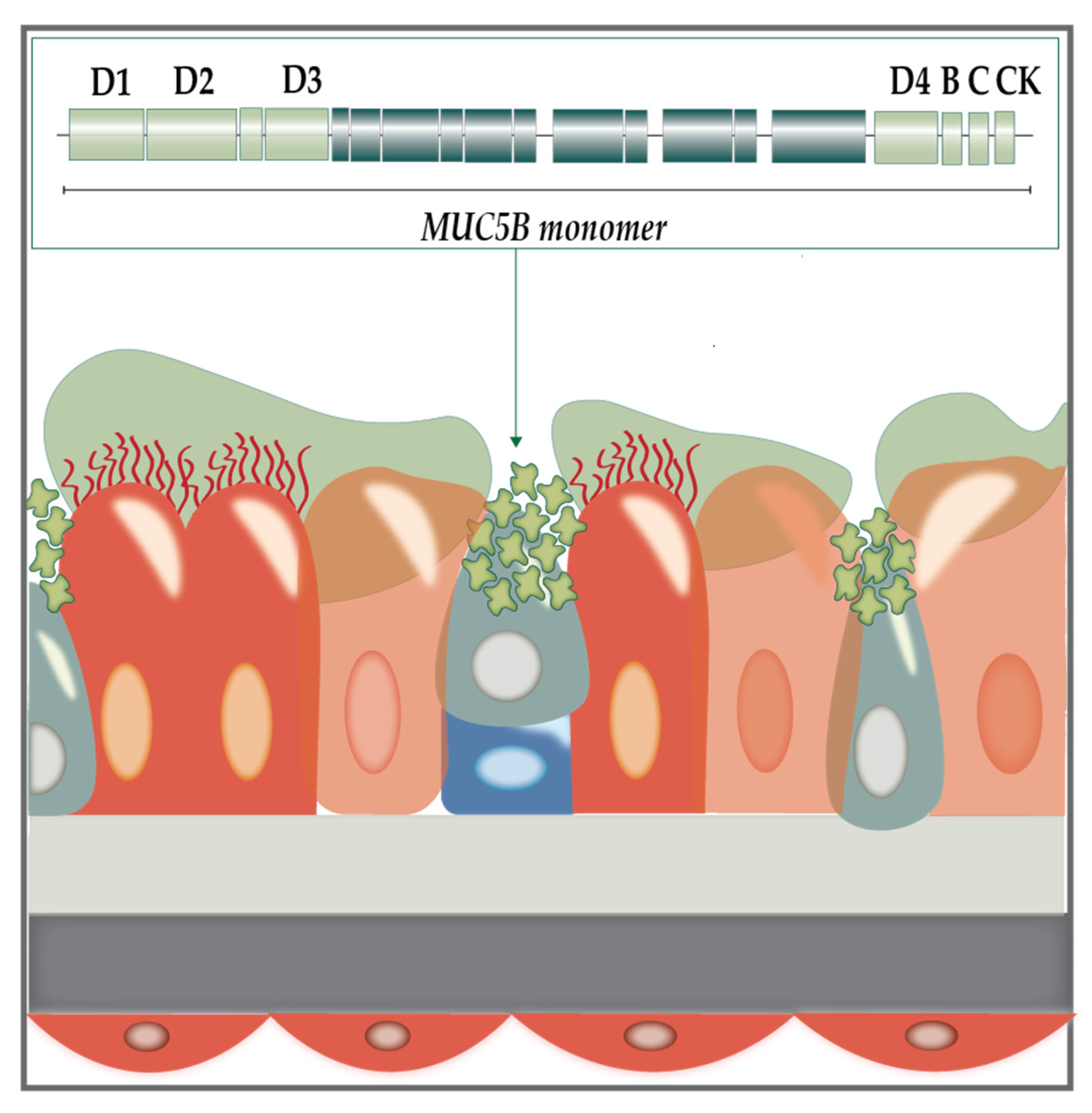
1.1. The Cystic Fibrosis Sputum Microenvironment
1.2. Modelling the CF Lung Mucus Environment
1.3. Artificial Sputum Media: A Lab-Replicable Medium with CF Sputum Roots
1.4. The Significance of Respective Components in Artificial Sputum Media
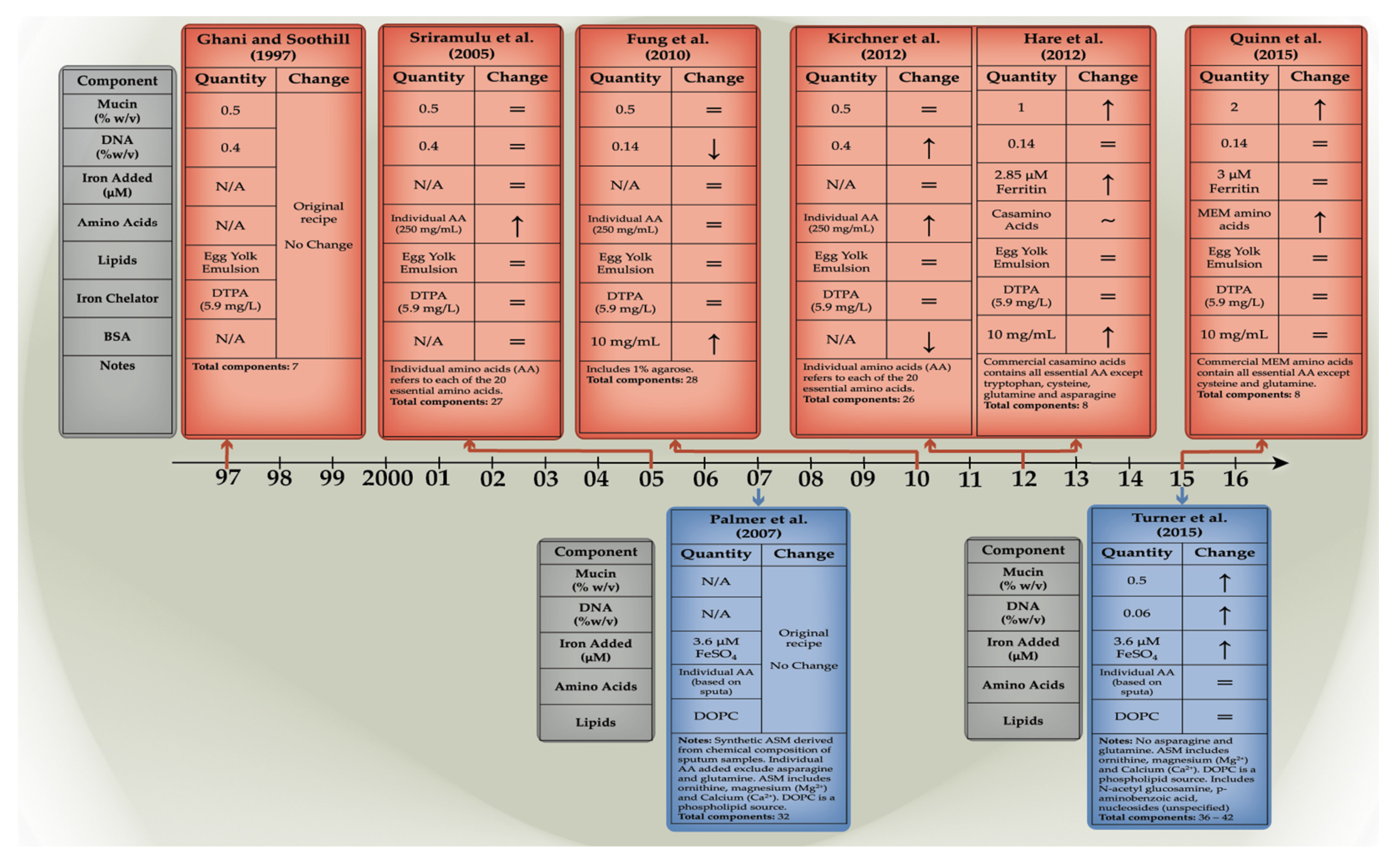
1.5. The Evolution of Artificial Sputum Media
2. Uses of Artificial Sputum Media in the Study of CF Infections
3. Future Directions and Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Hartl, D.; Amaral, M. Cystic fibrosis—From basic science to clinical benefit: A review series. J. Cyst. Fibros 2015, 14, 415–416. [Google Scholar] [CrossRef] [PubMed]
- Riordan, J.R.; Rommens, J.M.; Kerem, B.; Alon, N.; Rozmahel, R.; Grzelczak, Z.; Zielenski, J.; Lok, S.; Plavsic, N.; Chou, J.L.; et al. Identification of the cystic fibrosis gene: Cloning and characterization of complementary DNA. Science 1989, 245, 1066–1073. [Google Scholar] [CrossRef]
- Elborn, J.S. Cystic fibrosis. Lancet 2016, 388, 2519–2531. [Google Scholar] [CrossRef]
- Bhagirath, A.Y.; Li, Y.; Somayajula, D.; Dadashi, M.; Badr, S.; Duan, K. Cystic fibrosis lung environment and Pseudomonas aeruginosa infection. BMC Pulm. Med. 2016, 16, 174. [Google Scholar] [CrossRef] [PubMed]
- Sriramulu, D.D. Amino Acids Enhance Adaptive Behaviour of Pseudomonas aeruginosa in the Cystic Fibrosis Lung Environment. Microbiol. Insights 2010, 3, 17–26. [Google Scholar] [CrossRef]
- Thornton, D.J.; Rousseau, K.; McGuckin, M.A. Structure and function of the polymeric mucins in airways mucus. Annu. Rev. Physiol. 2008, 70, 459–486. [Google Scholar] [CrossRef] [PubMed]
- Thornton, D.J.; Sheehan, J.K. From mucins to mucus: Toward a more coherent understanding of this essential barrier. Proc. Am. Thorac. Soc. 2004, 1, 54–61. [Google Scholar] [CrossRef]
- Fahy, J.V.; Dickey, B.F. Airway mucus function and dysfunction. N. Engl. J. Med. 2010, 363, 2233–2247. [Google Scholar] [CrossRef]
- Hattrup, C.L.; Gendler, S.J. Structure and function of the cell surface (tethered) mucins. Annu. Rev. Physiol. 2008, 70, 431–457. [Google Scholar] [CrossRef]
- Rose, M.C.; Voynow, J.A. Respiratory tract mucin genes and mucin glycoproteins in health and disease. Physiol. Rev. 2006, 86, 245–278. [Google Scholar] [CrossRef]
- Rubin, B.K. Cystic Fibrosis 2017-The Year in Review. Respir. Care 2018, 63, 238–241. [Google Scholar] [CrossRef] [PubMed]
- Hoiby, N.; Ciofu, O.; Johansen, H.K.; Song, Z.J.; Moser, C.; Jensen, P.O.; Molin, S.; Givskov, M.; Tolker-Nielsen, T.; Bjarnsholt, T. The clinical impact of bacterial biofilms. Int. J. Oral Sci. 2011, 3, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Fleming, D.; Rumbaugh, K. The Consequences of Biofilm Dispersal on the Host. Sci. Rep. 2018, 8, 10738. [Google Scholar] [CrossRef] [PubMed]
- Kolpen, M.; Kragh, K.N.; Enciso, J.B.; Faurholt-Jepsen, D.; Lindegaard, B.; Egelund, G.B.; Jensen, A.V.; Ravn, P.; Mathiesen, I.H.M.; Gheorge, A.G.; et al. Bacterial biofilms predominate in both acute and chronic human lung infections. Thorax 2022. [Google Scholar] [CrossRef]
- Palmer, K.L.; Aye, L.M.; Whiteley, M. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J. Bacteriol. 2007, 189, 8079–8087. [Google Scholar] [CrossRef] [PubMed]
- Zemanick, E.T.; Hoffman, L.R. Cystic Fibrosis: Microbiology and Host Response. Pediatr. Clin. N. Am. 2016, 63, 617–636. [Google Scholar] [CrossRef] [PubMed]
- Kirchner, S.; Fothergill, J.L.; Wright, E.A.; James, C.E.; Mowat, E.; Winstanley, C. Use of artificial sputum medium to test antibiotic efficacy against Pseudomonas aeruginosa in conditions more relevant to the cystic fibrosis lung. J. Vis. Exp. 2012, 64, e3857. [Google Scholar] [CrossRef]
- Haley, C.L.; Colmer-Hamood, J.A.; Hamood, A.N. Characterization of biofilm-like structures formed by Pseudomonas aeruginosa in a synthetic mucus medium. BMC Microbiol. 2012, 12, 181. [Google Scholar] [CrossRef]
- Smith, D.J.; Lamont, I.L.; Anderson, G.J.; Reid, D.W. Targeting iron uptake to control Pseudomonas aeruginosa infections in cystic fibrosis. Eur. Respir. J. 2013, 42, 1723–1736. [Google Scholar] [CrossRef]
- Kumar, B.; Cardona, S.T. Synthetic Cystic Fibrosis Sputum Medium Regulates Flagellar Biosynthesis through the flhF Gene in Burkholderia cenocepacia. Front. Cell Infect. Microbiol. 2016, 6, 65. [Google Scholar] [CrossRef]
- Yang, Y.; Pollard, A.M.; Höfler, C.; Poschet, G.; Wirtz, M.; Hell, R.; Sourjik, V. Relation between chemotaxis and consumption of amino acids in bacteria. Mol. Microbiol. 2015, 96, 1272–1282. [Google Scholar] [CrossRef] [PubMed]
- Sriramulu, D.D.; Lunsdorf, H.; Lam, J.S.; Romling, U. Microcolony formation: A novel biofilm model of Pseudomonas aeruginosa for the cystic fibrosis lung. J. Med. Microbiol. 2005, 54, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Ramon-Perez, M.L.; Diaz-Cedillo, F.; Ibarra, J.A.; Torales-Cardena, A.; Rodriguez-Martinez, S.; Jan-Roblero, J.; Cancino-Diaz, M.E.; Cancino-Diaz, J.C. D-Amino acids inhibit biofilm formation in Staphylococcus epidermidis strains from ocular infections. J. Med. Microbiol. 2014, 63, 1369–1376. [Google Scholar] [CrossRef] [PubMed]
- Tong, Z.; Zhang, L.; Ling, J.; Jian, Y.; Huang, L.; Deng, D. An in vitro study on the effect of free amino acids alone or in combination with nisin on biofilms as well as on planktonic bacteria of Streptococcus mutans. PLoS ONE 2014, 9, e99513. [Google Scholar] [CrossRef]
- Warraich, A.A.; Mohammed, A.R.; Perrie, Y.; Hussain, M.; Gibson, H.; Rahman, A. Evaluation of anti-biofilm activity of acidic amino acids and synergy with ciprofloxacin on Staphylococcus aureus biofilms. Sci. Rep. 2020, 10, 9021. [Google Scholar] [CrossRef]
- Cole, A.M.; Waring, A.J. The role of defensins in lung biology and therapy. Am. J. Respir. Med. 2002, 1, 249–259. [Google Scholar] [CrossRef]
- Butt, A.T.; Thomas, M.S. Iron Acquisition Mechanisms and Their Role in the Virulence of Burkholderia Species. Front. Cell Infect. Microbiol. 2017, 7, 460. [Google Scholar] [CrossRef]
- Yang, L.; Barken, K.B.; Skindersoe, M.E.; Christensen, A.B.; Givskov, M.; Tolker-Nielsen, T. Effects of iron on DNA release and biofilm development by Pseudomonas aeruginosa. Microbiology 2007, 153, 1318–1328. [Google Scholar] [CrossRef]
- Neve, R.L.; Carrillo, B.D.; Phelan, V.V. Commercial porcine gastric mucin contributes to variation in production of small molecule virulence factors by Pseudomonas aeruginosa when cultured in different formulations of artificial sputum medium. bioRxiv 2021. [Google Scholar] [CrossRef]
- Sahu, S.; Lynn, W.S. Lipid composition of sputum from patients with asthma and patients with cystic fibrosis. Inflammation 1978, 3, 27–36. [Google Scholar] [CrossRef]
- Harmon, G.S.; Dumlao, D.S.; Ng, D.T.; Barrett, K.E.; Dennis, E.A.; Dong, H.; Glass, C.K. Pharmacological correction of a defect in PPAR-gamma signaling ameliorates disease severity in Cftr-deficient mice. Nat. Med. 2010, 16, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Fung, C.; Naughton, S.; Turnbull, L.; Tingpej, P.; Rose, B.; Arthur, J.; Hu, H.; Harmer, C.; Harbour, C.; Hassett, D.J.; et al. Gene expression of Pseudomonas aeruginosa in a mucin-containing synthetic growth medium mimicking cystic fibrosis lung sputum. J. Med. Microbiol. 2010, 59, 1089–1100. [Google Scholar] [CrossRef] [PubMed]
- Neve, R.L.; Carrillo, B.D.; Phelan, V.V. Impact of Artificial Sputum Medium Formulation on Pseudomonas aeruginosa Secondary Metabolite Production. J. Bacteriol. 2021, 203, e0025021. [Google Scholar] [CrossRef]
- Ghani, M.; Soothill, J.S. Ceftazidime, gentamicin, and rifampicin, in combination, kill biofilms of mucoid Pseudomonas aeruginosa. Can. J. Microbiol. 1997, 43, 999–1004. [Google Scholar] [CrossRef] [PubMed]
- Hare, N.J.; Soe, C.Z.; Rose, B.; Harbour, C.; Codd, R.; Manos, J.; Cordwell, S.J. Proteomics of Pseudomonas aeruginosa Australian epidemic strain 1 (AES-1) cultured under conditions mimicking the cystic fibrosis lung reveals increased iron acquisition via the siderophore pyochelin. J. Proteome Res. 2012, 11, 776–795. [Google Scholar] [CrossRef] [PubMed]
- Quinn, R.A.; Whiteson, K.; Lim, Y.W.; Salamon, P.; Bailey, B.; Mienardi, S.; Sanchez, S.E.; Blake, D.; Conrad, D.; Rohwer, F. A Winogradsky-based culture system shows an association between microbial fermentation and cystic fibrosis exacerbation. ISME J. 2015, 9, 1052. [Google Scholar] [CrossRef] [PubMed]
- Turner, K.H.; Wessel, A.K.; Palmer, G.C.; Murray, J.L.; Whiteley, M. Essential genome of Pseudomonas aeruginosa in cystic fibrosis sputum. Proc. Natl. Acad. Sci. USA 2015, 112, 4110–4115. [Google Scholar] [CrossRef]
- Sagel, S.D.; Kapsner, R.; Osberg, I.; Sontag, M.K.; Accurso, F.J. Airway inflammation in children with cystic fibrosis and healthy children assessed by sputum induction. Am. J. Respir. Crit. Care Med. 2001, 164, 1425–1431. [Google Scholar] [CrossRef]
- Reid, D.W.; Lam, Q.T.; Schneider, H.; Walters, E.H. Airway iron and iron-regulatory cytokines in cystic fibrosis. Eur. Respir. J. 2004, 24, 286–291. [Google Scholar] [CrossRef]
- Stites, S.W.; Plautz, M.W.; Bailey, K.; O’Brien-Ladner, A.R.; Wesselius, L.J. Increased concentrations of iron and isoferritins in the lower respiratory tract of patients with stable cystic fibrosis. Am. J. Respir. Crit. Care Med. 1999, 160, 796–801. [Google Scholar] [CrossRef]
- Ratledge, C.; Dover, L.G. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 2000, 54, 881–941. [Google Scholar] [CrossRef] [PubMed]
- Sriyosachati, S.; Cox, C.D. Siderophore-mediated iron acquisition from transferrin by Pseudomonas aeruginosa. Infect. Immun. 1986, 52, 885–891. [Google Scholar] [CrossRef]
- Klare, W.; Das, T.; Ibugo, A.; Buckle, E.; Manefield, M.; Manos, J. Glutathione-Disrupted Biofilms of Clinical Pseudomonas aeruginosa Strains Exhibit an Enhanced Antibiotic Effect and a Novel Biofilm Transcriptome. Antimicrob. Agents Chemother. 2016, 60, 4539–4551. [Google Scholar] [CrossRef] [PubMed]
- Lopez, M.J.; Mohiuddin, S.S. Biochemistry, Essential Amino Acids. StatPearls. 2022. Available online: https://www.statpearls.com/ArticleLibrary/viewarticle/36202 (accessed on 16 June 2022).
- La Rosa, R.; Johansen, H.K.; Molin, S. Adapting to the Airways: Metabolic Requirements of Pseudomonas aeruginosa during the Infection of Cystic Fibrosis Patients. Metabolites 2019, 9, 234. [Google Scholar] [CrossRef] [PubMed]
- Goss, C.H.; Burns, J.L. Exacerbations in cystic fibrosis. 1: Epidemiology and pathogenesis. Thorax 2007, 62, 360–367. [Google Scholar] [CrossRef] [PubMed]
- Naughton, S.; Parker, D.; Seemann, T.; Thomas, T.; Turnbull, L.; Rose, B.; Bye, P.; Cordwell, S.; Whitchurch, C.; Manos, J. Pseudomonas aeruginosa AES-1 exhibits increased virulence gene expression during chronic infection of cystic fibrosis lung. PLoS ONE 2011, 6, e24526. [Google Scholar] [CrossRef]
- Leid, J.G.; Willson, C.J.; Shirtliff, M.E.; Hassett, D.J.; Parsek, M.R.; Jeffers, A.K. The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN-gamma-mediated macrophage killing. J. Immunol. 2005, 175, 7512–7518. [Google Scholar] [CrossRef]
- Banin, E.; Brady, K.M.; Greenberg, E.P. Chelator-induced dispersal and killing of Pseudomonas aeruginosa cells in a biofilm. Appl. Environ. Microbiol. 2006, 72, 2064–2069. [Google Scholar] [CrossRef]
- Kosztolowicz, T.; Metzler, R.; Wa Sik, S.; Arabski, M. Modelling experimentally measured of ciprofloxacin antibiotic diffusion in Pseudomonas aeruginosa biofilm formed in artificial sputum medium. PLoS ONE 2020, 15, e0243003. [Google Scholar] [CrossRef]
- Huang, J.X.; Blaskovich, M.A.; Pelingon, R.; Ramu, S.; Kavanagh, A.; Elliott, A.G.; Butler, M.S.; Montgomery, A.B.; Cooper, M.A. Mucin Binding Reduces Colistin Antimicrobial Activity. Antimicrob. Agents Chemother. 2015, 59, 5925–5931. [Google Scholar] [CrossRef]
- Hall, J.R.; Maloney, S.E.; Jin, H.; Taylor, J.B.; Schoenfisch, M.H. Nitric oxide diffusion through cystic fibrosis-relevant media and lung tissue. RSC Adv. 2019, 9, 40176–40183. [Google Scholar] [CrossRef] [PubMed]
- McMullin, B.B.; Chittock, D.R.; Roscoe, D.L.; Garcha, H.; Wang, L.; Miller, C.C. The antimicrobial effect of nitric oxide on the bacteria that cause nosocomial pneumonia in mechanically ventilated patients in the intensive care unit. Respir. Care 2005, 50, 1451–1456. [Google Scholar] [PubMed]
- Garbe, J.; Wesche, A.; Bunk, B.; Kazmierczak, M.; Selezska, K.; Rohde, C.; Sikorski, J.; Rohde, M.; Jahn, D.; Schobert, M. Characterization of JG024, a Pseudomonas aeruginosa PB1-like broad host range phage under simulated infection conditions. BMC Microbiol. 2010, 10, 301. [Google Scholar] [CrossRef] [PubMed]
- Nafee, N.; Forier, K.; Braeckmans, K.; Schneider, M. Mucus-penetrating solid lipid nanoparticles for the treatment of cystic fibrosis: Proof of concept, challenges and pitfalls. Eur. J. Pharm. Biopharm. 2018, 124, 125–137. [Google Scholar] [CrossRef]
- Wijers, C.D.; Vagedes, R.; Weingart, C. A novel method for investigating Burkholderia cenocepacia infections in patients with cystic fibrosis and other chronic diseases of the airways. BMC Microbiol. 2016, 16, 200. [Google Scholar] [CrossRef]
- Sajjan, U.; Keshavjee, S.; Forstner, J. Responses of well-differentiated airway epithelial cell cultures from healthy donors and patients with cystic fibrosis to Burkholderia cenocepacia infection. Infect. Immun. 2004, 72, 4188–4199. [Google Scholar] [CrossRef]
- Garcia-Romero, I.; Valvano, M.A. Complete Genome Sequence of Burkholderia cenocepacia K56-2, an Opportunistic Pathogen. Microbiol. Resour. Announc. 2020, 9, e01015-20. [Google Scholar] [CrossRef]
- Aiyer, A.; Visser, S.K.; Bye, P.; Britton, W.J.; Whiteley, G.S.; Glasbey, T.; Kriel, F.H.; Farrell, J.; Das, T.; Manos, J. Effect of N-Acetylcysteine in Combination with Antibiotics on the Biofilms of Three Cystic Fibrosis Pathogens of Emerging Importance. Antibiotics 2021, 10, 1176. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aiyer, A.; Manos, J. The Use of Artificial Sputum Media to Enhance Investigation and Subsequent Treatment of Cystic Fibrosis Bacterial Infections. Microorganisms 2022, 10, 1269. https://doi.org/10.3390/microorganisms10071269
Aiyer A, Manos J. The Use of Artificial Sputum Media to Enhance Investigation and Subsequent Treatment of Cystic Fibrosis Bacterial Infections. Microorganisms. 2022; 10(7):1269. https://doi.org/10.3390/microorganisms10071269
Chicago/Turabian StyleAiyer, Aditi, and Jim Manos. 2022. "The Use of Artificial Sputum Media to Enhance Investigation and Subsequent Treatment of Cystic Fibrosis Bacterial Infections" Microorganisms 10, no. 7: 1269. https://doi.org/10.3390/microorganisms10071269
APA StyleAiyer, A., & Manos, J. (2022). The Use of Artificial Sputum Media to Enhance Investigation and Subsequent Treatment of Cystic Fibrosis Bacterial Infections. Microorganisms, 10(7), 1269. https://doi.org/10.3390/microorganisms10071269






