Novel Unspecific Peroxygenase from Truncatella angustata Catalyzes the Synthesis of Bioactive Lipid Mediators
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Enzymes
2.3. Enzyme Assays
2.4. Enzymatic Conversion of Arachidonic Acid
2.5. Analytical Methods
2.5.1. High Performance Liquid Chromatography/ELSD
2.5.2. High Resolution Mass Spectrometry
3. Results
3.1. Small-Scale Conversion of AA by Different UPOs
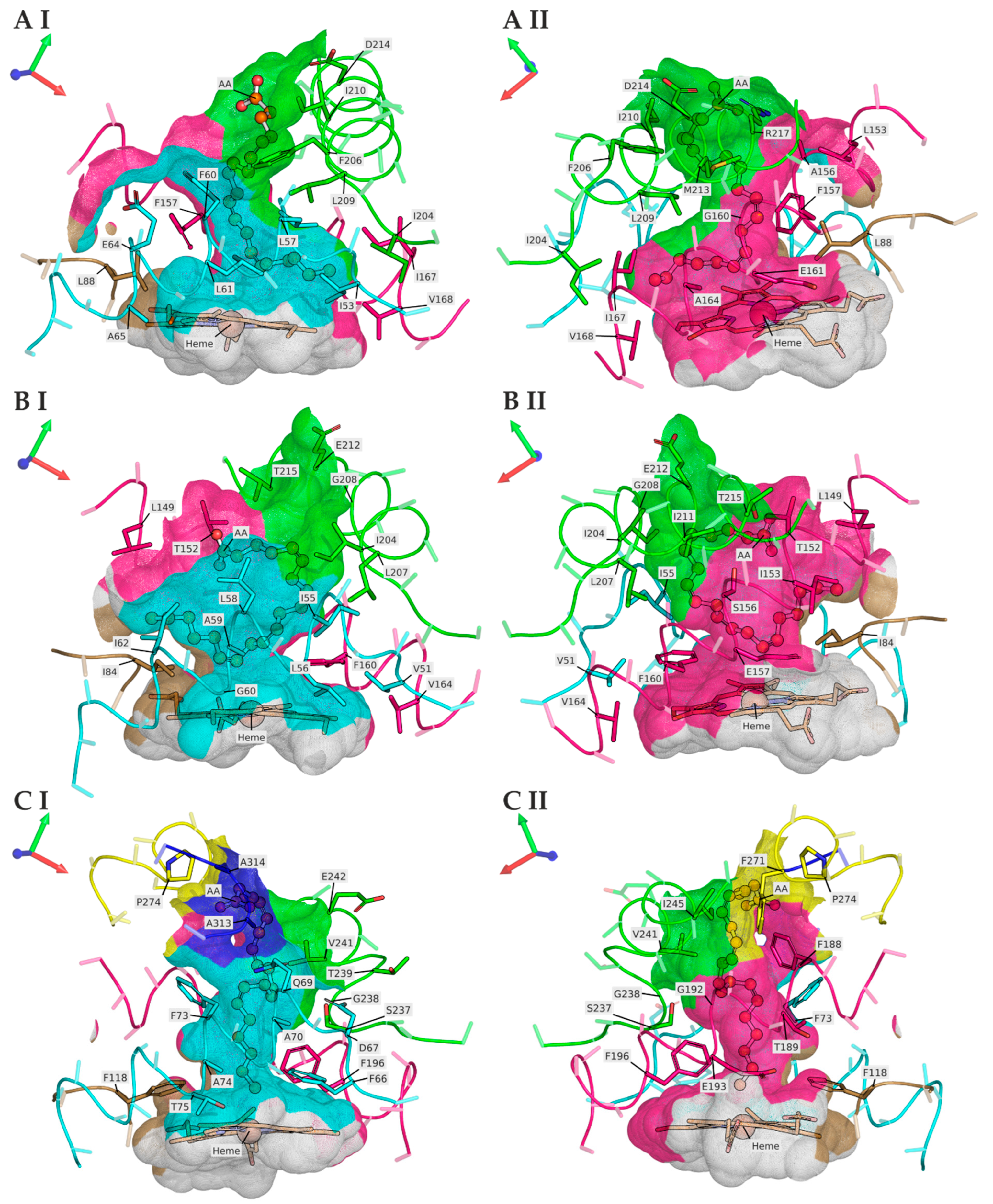
3.2. TanUPO Protein-Structure Simulation and Ligand Docking
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Eckenfels, A.; Vane, J.R. Prostaglandins, oxygen tension and smooth muscle tone. Br. J. Pharmacol. 1972, 45, 451–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Main, I.H.M. The inhibitory actions of prostaglandins on respiratory smooth muscle. Br. J. Pharmacol. Chemother. 1964, 22, 511–519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hertting, G.; Seregi, A. Formation and function of eicosanoids in the central nervous system. Ann. N. Y. Acad. Sci. 1989, 559, 84–99. [Google Scholar] [CrossRef]
- Simmet, T.; Peskar, B.A. Lipoxygenase products of polyunsaturated fatty acid metabolism in the central nervous system: Biosynthesis and putative functions. Pharmacol. Res. 1990, 22, 667–682. [Google Scholar] [CrossRef]
- Feldberg, W.; Saxena, P.N. Fever produced by prostaglandin E1. J. Physiol. 1971, 217, 547–556. [Google Scholar] [CrossRef]
- Serhan, C.N.; Chiang, N.; Dalli, J.; Levy, B.D. Lipid mediators in the resolution of inflammation. Cold Spring Harb. Perspect. Biol. 2015, 7, a016311. [Google Scholar] [CrossRef] [Green Version]
- Borgeat, P.; Naccache, P.H. Biosynthesis and biological activity of leukotriene B4. Clin. Biochem. 1990, 23, 459–468. [Google Scholar] [CrossRef]
- Ott, V.L.; Cambier, J.C.; Kappler, J.; Marrack, P.; Swanson, B.J. Mast cell–dependent migration of effector CD8+ T cells through production of leukotriene B 4. Nat. Immunol. 2003, 4, 974–981. [Google Scholar] [CrossRef]
- Harizi, H.; Gualde, N. The impact of eicosanoids on the crosstalk between innate and adaptive immunity: The key roles of dendritic cells. Tissue Antigens 2005, 65, 507–514. [Google Scholar] [CrossRef] [PubMed]
- Denisenko, Y.K.; Lobanova, E.G.; Novgorodtseva, T.P.; Gvozdenko, T.A.; Nazarenko, A.V. The role of arachidonic acid metabolites (endocannabinoids and eicosanoids) in the immune processes: A review. Int. J. Chem. Biomed. Sci. 2015, 1, 70–78. [Google Scholar]
- Esser-von Bieren, J. Immune-regulation and-functions of eicosanoid lipid mediators. Biol. Chem. 2017, 398, 1177–1191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, T.J. Prostaglandin E2, prostaglandin I2 and the vascular changes of inflammation. Br. J. Pharmacol. 1979, 65, 517–524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drenjančević, I.; Jukić, I.; Mihaljević, Z.; Ćosić, A.; Kibel, A. The metabolites of arachidonic acid in microvascular function. In Microcirculation Revisited-from Molecules to Clinical Practice/Helena Lenasi (ur.); IN TECH doo: Rijeka, Croatia, 2016; pp. 101–133. [Google Scholar]
- Moncada, S.; Gryglewski, R.J.; Bunting, S.; Vane, J.R. An enzyme isolated from arteries transforms prostaglandin endoperoxides to an unstable substance that inhibits platelet aggregation. Nature 1976, 263, 663–665. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, F.A.; Ennis, M.D.; Baze, M.E.; Wynalda, M.A.; McGee, J.E.; Liggett, W.F. Inhibition of cyclooxygenase activity and platelet aggregation by epoxyeicosatrienoic acids. Influence of stereochemistry. J. Biol. Chem. 1986, 261, 15334–15338. [Google Scholar] [CrossRef]
- Rich, M.R. Conformational analysis of arachidonic and related fatty acids using molecular dynamics simulations. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 1993, 1178, 87–96. [Google Scholar] [CrossRef]
- Brash, A.R. Arachidonic acid as a bioactive molecule. J. Clin. Investig. 2001, 107, 1339–1345. [Google Scholar] [CrossRef] [Green Version]
- Dennis, E.A.; Cao, J.; Hsu, Y.-H.; Magrioti, V.; Kokotos, G. Phospholipase A2 enzymes: Physical structure, biological function, disease implication, chemical inhibition, and therapeutic intervention. Chem. Rev. 2011, 111, 6130–6185. [Google Scholar] [CrossRef] [Green Version]
- Needleman, P.; Jakschik, B.A.; Morrison, A.R.; Lefkowith, J.B. Arachidonic acid metabolism. Annu. Rev. Biochem. 1986, 55, 69–102. [Google Scholar] [CrossRef]
- Dubois, R.N.; Abramson, S.B.; Crofford, L.; Gupta, R.A.; Simon, L.S.; van de Putte, L.B.A.; Lipsky, P.E. Cyclooxygenase in biology and disease. FASEB J. 1998, 12, 1063–1073. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, T.; Rådmark, O.; Samuelsson, B. Enzyme with dual lipoxygenase activities catalyzes leukotriene A4 synthesis from arachidonic acid. Proc. Natl. Acad. Sci. USA 1984, 81, 689–693. [Google Scholar] [CrossRef] [Green Version]
- Crooks, S.W.; Stockley, R.A. Leukotriene B4. Int. J. Biochem. Cell Biol. 1998, 30, 173–178. [Google Scholar] [CrossRef]
- Ueda, N.; Yokoyama, C.; Yamamoto, S.; Fitzsimmons, B.J.; Rokach, J.; Oates, J.A.; Brash, A.R. Lipoxin synthesis by arachidonate 12-lipoxygenase purified from porcine leukocytes. Biochem. Biophys. Res. Commun. 1987, 149, 1063–1069. [Google Scholar] [CrossRef]
- Edenius, C.; Stenke, L.; Lindgren, J.Å. On the mechanism of transcellular lipoxin formation in human platelets and granulocytes. Eur. J. Biochem. 1991, 199, 401–409. [Google Scholar] [CrossRef] [PubMed]
- Capdevila, J.H.; Falck, J.R.; Harris, R.C. Cytochrome P450 and arachidonic acid bioactivation: Molecular and functional properties of the arachidonate monooxygenase. J. Lipid Res. 2000, 41, 163–181. [Google Scholar] [CrossRef]
- Carroll, M.A.; McGiff, J.C. A new class of lipid mediators: Cytochrome P450 arachidonate metabolites. Thorax 2000, 55, S13–S16. [Google Scholar] [CrossRef] [Green Version]
- Capdevila, J.; Chacos, N.; Werringloer, J.; Prough, R.A.; Estabrook, R.W. Liver microsomal cytochrome P-450 and the oxidative metabolism of arachidonic acid. Proc. Natl. Acad. Sci. USA 1981, 78, 5362–5366. [Google Scholar] [CrossRef] [Green Version]
- Morrison, A.R.; Pascoe, N. Metabolism of arachidonate through NADPH-dependent oxygenase of renal cortex. Proc. Natl. Acad. Sci. USA 1981, 78, 7375–7378. [Google Scholar] [CrossRef] [Green Version]
- Munro, A.W.; McLean, K.J.; Grant, J.L.; Makris, T.M. Structure and function of the cytochrome P450 peroxygenase enzymes. Biochem. Soc. Trans. 2018, 46, 183–196. [Google Scholar] [CrossRef] [Green Version]
- Hardwick, J.P. Cytochrome P450 omega hydroxylase (CYP4) function in fatty acid metabolism and metabolic diseases. Biochem. Pharmacol. 2008, 75, 2263–2275. [Google Scholar] [CrossRef]
- Waldman, M.; Peterson, S.J.; Arad, M.; Hochhauser, E. The role of 20-HETE in cardiovascular diseases and its risk factors. Prostaglandins Other Lipid Mediat. 2016, 125, 108–117. [Google Scholar] [CrossRef]
- Rocic, P.; Schwartzman, M.L. 20-HETE in the regulation of vascular and cardiac function. Pharmacol. Ther. 2018, 192, 74–87. [Google Scholar] [CrossRef] [PubMed]
- Shoieb, S.M.; El-Sherbeni, A.A.; El-Kadi, A.O.S. Subterminal hydroxyeicosatetraenoic acids: Crucial lipid mediators in normal physiology and disease states. Chem. Biol. Interact. 2019, 299, 140–150. [Google Scholar] [CrossRef] [PubMed]
- Dakarapu, R.; Errabelli, R.; Manthati, V.L.; Adebesin, A.M.; Barma, D.K.; Barma, D.; Garcia, V.; Zhang, F.; Schwartzman, M.L.; Falck, J.R. 19-Hydroxyeicosatetraenoic acid analogs: Antagonism of 20-hydroxyeicosatetraenoic acid-induced vascular sensitization and hypertension. Bioorg. Med. Chem. Lett. 2019, 29, 126616. [Google Scholar] [CrossRef]
- Shoieb, S.M.; El-Kadi, A.O.S. S-enantiomer of 19-hydroxyeicosatetraenoic acid preferentially protects against angiotensin II-induced cardiac hypertrophy. Drug Metab. Dispos. 2018, 46, 1157–1168. [Google Scholar] [CrossRef] [Green Version]
- Node, K.; Huo, Y.; Ruan, X.; Yang, B.; Spiecker, M.; Ley, K.; Zeldin, D.C.; Liao, J.K. Anti-inflammatory properties of cytochrome P450 epoxygenase-derived eicosanoids. Science 1999, 285, 1276–1279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spector, A.A.; Fang, X.; Snyder, G.D.; Weintraub, N.L. Epoxyeicosatrienoic acids (EETs): Metabolism and biochemical function. Prog. Lipid Res. 2004, 43, 55–90. [Google Scholar] [CrossRef]
- Thomson, S.J.; Askari, A.; Bishop-Bailey, D. Anti-inflammatory effects of epoxyeicosatrienoic acids. Int. J. Vasc. Med. 2012, 2012, 605101. [Google Scholar] [CrossRef] [PubMed]
- Shoieb, S.M.; El-Ghiaty, M.A.; El-Kadi, A.O.S. Targeting arachidonic acid–related metabolites in COVID-19 patients: Potential use of drug-loaded nanoparticles. Emergent Mater. 2021, 4, 265–277. [Google Scholar] [CrossRef]
- Hammock, B.D.; Ratcliff, M.; Schooley, D.A. Hydration of an 18O epoxide by a cytosolic epoxide hydrolase from mouse liver. Life Sci. 1980, 27, 1635–1641. [Google Scholar] [CrossRef]
- Motoki, A.; Merkel, M.J.; Packwood, W.H.; Cao, Z.; Liu, L.; Iliff, J.; Alkayed, N.J.; van Winkle, D.M. Soluble epoxide hydrolase inhibition and gene deletion are protective against myocardial ischemia-reperfusion injury in vivo. Am. J. Physiol.-Heart Circ. Physiol. 2008, 295, H2128–H2134. [Google Scholar] [CrossRef] [Green Version]
- VanRollins, M. Synthesis and characterization of cytochrome P-450 epoxygenase metabolites of eicosapentaenoic acid. Lipids 1990, 25, 481–490. [Google Scholar] [CrossRef] [PubMed]
- VanRollins, M.; Kochanek, P.M.; Evans, R.W.; Schiding, J.K.; Nemoto, E.M. Optimization of epoxyeicosatrienoic acid syntheses to test their effects on cerebral blood flow in vivo. Biochim. Biophys. Acta (BBA) Lipids Lipid Metab. 1995, 1256, 263–274. [Google Scholar] [CrossRef]
- Danov, S.M.; Kazantsev, O.A.; Esipovich, A.L.; Belousov, A.S.; Rogozhin, A.E.; Kanakov, E.A. Recent advances in the field of selective epoxidation of vegetable oils and their derivatives: A review and perspective. Catal. Sci. Technol. 2017, 7, 3659–3675. [Google Scholar] [CrossRef]
- Woodman, J.W.; Cinelli, M.A.; Scharmen-Burgdolf, A.; Lee, K.S.S. Enzymatic synthesis of epoxidized metabolites of docosahexaenoic, eicosapentaenoic, and arachidonic acids. J. Vis. Exp. 2019, 148, e59770. [Google Scholar] [CrossRef] [Green Version]
- Hobisch, M.; Holtmann, D.; de Santos, P.G.; Alcalde, M.; Hollmann, F.; Kara, S. Recent developments in the use of peroxygenases–Exploring their high potential in selective oxyfunctionalisations. Biotechnol. Adv. 2021, 51, 107615. [Google Scholar] [CrossRef]
- Hofrichter, M.; Kellner, H.; Pecyna, M.J.; Ullrich, R. Fungal unspecific peroxygenases: Heme-thiolate proteins that combine peroxidase and cytochrome P450 properties. In Monooxygenase, Peroxidase and Peroxygenase Properties and Mechanisms of Cytochrome P450; Springer: Berlin/Heidelberg, Germany, 2015; pp. 341–368. [Google Scholar]
- Hofrichter, M.; Kellner, H.; Herzog, R.; Karich, A.; Liers, C.; Scheibner, K.; Kimani, V.W.; Ullrich, R. Fungal peroxygenases: A phylogenetically old superfamily of heme enzymes with promiscuity for oxygen transfer reactions. In Grand Challenges in Fungal Biotechnology; Springer: Berlin/Heidelberg, Germany, 2020; pp. 369–403. [Google Scholar]
- Kellner, H.; Luis, P.; Pecyna, M.J.; Barbi, F.; Kapturska, D.; Krüger, D.; Zak, D.R.; Marmeisse, R.; Vandenbol, M.; Hofrichter, M. Widespread occurrence of expressed fungal secretory peroxidases in forest soils. PLoS ONE 2014, 9, e95557. [Google Scholar] [CrossRef] [Green Version]
- Veitch, N.C. Horseradish peroxidase: A modern view of a classic enzyme. Phytochemistry 2004, 65, 249–259. [Google Scholar] [CrossRef]
- Matsunaga, I.; Sumimoto, T.; Ayata, M.; Ogura, H. Functional modulation of a peroxygenase cytochrome P450: Novel insight into the mechanisms of peroxygenase and peroxidase enzymes. FEBS Lett. 2002, 528, 90–94. [Google Scholar] [CrossRef] [Green Version]
- Prasad, S.; Mitra, S. Substrate modulates compound I formation in peroxide shunt pathway of Pseudomonas putida cytochrome P450cam. Biochem. Biophys. Res. Commun. 2004, 314, 610–614. [Google Scholar] [CrossRef]
- Hrycay, E.G.; Bandiera, S.M. Monooxygenase, peroxidase and peroxygenase properties and reaction mechanisms of cytochrome P450 enzymes. In Monooxygenase, Peroxidase and Peroxygenase Properties and Mechanisms of Cytochrome P450; Springer: Berlin/Heidelberg, Germany, 2015; pp. 1–61. [Google Scholar]
- Aranda, C.; Olmedo, A.; Kiebist, J.; Scheibner, K.; Del Río, J.C.; Martínez, A.T.; Gutiérrez Suárez, A. Selective epoxidation of fatty acids and fatty acid methyl esters by fungal peroxygenases. ChemCatChem 2018, 10, 3964–3968. [Google Scholar] [CrossRef] [Green Version]
- Municoy, M.; González-Benjumea, A.; Carro, J.; Aranda, C.; Linde, D.; Renau-Mínguez, C.; Ullrich, R.; Hofrichter, M.; Guallar, V.; Gutiérrez, A. Fatty-Acid Oxygenation by Fungal Peroxygenases: From Computational Simulations to Preparative Regio-and Stereoselective Epoxidation. ACS Catal. 2020, 10, 13584–13595. [Google Scholar] [CrossRef]
- González-Benjumea, A.; Marques, G.; Herold-Majumdar, O.M.; Kiebist, J.; Scheibner, K.; Del Río, J.C.; Martínez, A.T.; Gutiérrez, A. High epoxidation yields of vegetable oil hydrolyzates and methyl esters by selected fungal peroxygenases. Front. Bioeng. Biotechnol. 2021, 8, 1470. [Google Scholar] [CrossRef] [PubMed]
- Ullrich, R.; Nüske, J.; Scheibner, K.; Spantzel, J.; Hofrichter, M. Novel haloperoxidase from the agaric basidiomycete Agrocybe aegerita oxidizes aryl alcohols and aldehydes. Appl. Environ. Microbiol. 2004, 70, 4575–4581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anh, D.H.; Ullrich, R.; Benndorf, D.; Svatos, A.; Muck, A.; Hofrichter, M. The coprophilous mushroom Coprinus radians secretes a haloperoxidase that catalyzes aromatic peroxygenation. Appl. Environ. Microbiol. 2007, 73, 5477–5485. [Google Scholar] [CrossRef] [Green Version]
- Gröbe, G.; Ullrich, R.; Pecyna, M.J.; Kapturska, D.; Friedrich, S.; Hofrichter, M.; Scheibner, K. High-yield production of aromatic peroxygenase by the agaric fungus Marasmius rotula. AMB Express 2011, 1, 31. [Google Scholar] [CrossRef] [Green Version]
- Kiebist, J.; Schmidtke, K.-U.; Zimmermann, J.; Kellner, H.; Jehmlich, N.; Ullrich, R.; Zänder, D.; Hofrichter, M.; Scheibner, K. A peroxygenase from Chaetomium globosum catalyzes the selective oxygenation of testosterone. ChemBioChem 2017, 18, 563. [Google Scholar] [CrossRef] [Green Version]
- Ullrich, R.; Poraj-Kobielska, M.; Scholze, S.; Halbout, C.; Sandvoss, M.; Pecyna, M.J.; Scheibner, K.; Hofrichter, M. Side chain removal from corticosteroids by unspecific peroxygenase. J. Inorg. Biochem. 2018, 183, 84–93. [Google Scholar] [CrossRef]
- Geiser, E.; Wiebach, V.; Wierckx, N.; Blank, L.M. Prospecting the biodiversity of the fungal family Ustilaginaceae for the production of value-added chemicals. Fungal Biol. Biotechnol. 2014, 1, 2. [Google Scholar] [CrossRef] [Green Version]
- Grabbe, K.; Haider, K. Die Huminstoffbildung und der Stickstoffumsatz bei der Bereitung des Kultursubstrats und während des Wachstums von Agaricus bisporus. Z. Pflanz. Bodenkd. 1971, 129, 216–226. [Google Scholar] [CrossRef]
- Grabbe, K.; Haider, K. Die Huminstoffbildung und die Stickstoffverteilung bei der Strohrotte in Beziehung zur mikrobiellen Phenolbildung. Z. Pflanz. Bodenkd. 1971, 129, 202–216. [Google Scholar] [CrossRef]
- Ullrich, R.; Dolge, C.; Kluge, M.; Hofrichter, M. Pyridine as novel substrate for regioselective oxygenation with aromatic peroxygenase from Agrocybe aegerita. FEBS Lett. 2008, 582, 4100–4106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campbell, W.B.; Gebremedhin, D.; Pratt, P.F.; Harder, D.R. Identification of epoxyeicosatrienoic acids as endothelium-derived hyperpolarizing factors. Circ. Res. 1996, 78, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Fleming, I. DiscrEET regulators of homeostasis: Epoxyeicosatrienoic acids, cytochrome P450 epoxygenases and vascular inflammation. Trends Pharmacol. Sci. 2007, 28, 448–452. [Google Scholar] [CrossRef] [PubMed]
- Gross, G.J.; Gauthier, K.M.; Moore, J.; Falck, J.R.; Hammock, B.D.; Campbell, W.B.; Nithipatikom, K. Effects of the selective EET antagonist, 14, 15-EEZE, on cardioprotection produced by exogenous or endogenous EETs in the canine heart. Am. J. Physiol.-Heart Circ. Physiol. 2008, 294, H2838–H2844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imig, J.D. Prospective for cytochrome P450 epoxygenase cardiovascular and renal therapeutics. Pharmacol. Ther. 2018, 192, 1–19. [Google Scholar] [CrossRef]
- Imig, J.D.; Jankiewicz, W.K.; Khan, A.H. Epoxy fatty acids: From salt regulation to kidney and cardiovascular therapeutics: 2019 Lewis, K. Dahl memorial lecture. Hypertension 2020, 76, 3–15. [Google Scholar] [CrossRef]
- Imig, J.D.; Hammock, B.D. Soluble epoxide hydrolase as a therapeutic target for cardiovascular diseases. Nat. Rev. Drug Discov. 2009, 8, 794–805. [Google Scholar] [CrossRef] [Green Version]
- Sudhahar, V.; Shaw, S.; Imig, J.D. Epoxyeicosatrienoic acid analogs and vascular function. Curr. Med. Chem. 2010, 17, 1181–1190. [Google Scholar] [CrossRef]
- Hwang, S.H.; Wagner, K.; Xu, J.; Yang, J.; Li, X.; Cao, Z.; Morisseau, C.; Lee, K.S.S.; Hammock, B.D. Chemical synthesis and biological evaluation of ω-hydroxy polyunsaturated fatty acids. Bioorg. Med. Chem. Lett. 2017, 27, 620–625. [Google Scholar] [CrossRef] [Green Version]
- Munro, A.W.; Leys, D.G.; McLean, K.J.; Marshall, K.R.; Ost, T.W.B.; Daff, S.; Miles, C.S.; Chapman, S.K.; Lysek, D.A.; Moser, C.C. P450 BM3: The very model of a modern flavocytochrome. Trends Biochem. Sci. 2002, 27, 250–257. [Google Scholar] [CrossRef]
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022. [Google Scholar] [CrossRef] [PubMed]
- El-Sherbeni, A.A.; Aboutabl, M.E.; Zordoky, B.N.M.; Anwar-Mohamed, A.; El-Kadi, A.O.S. Determination of the dominant arachidonic acid cytochrome p450 monooxygenases in rat heart, lung, kidney, and liver: Protein expression and metabolite kinetics. AAPS J. 2013, 15, 112–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, R.; Kim, V.; Chun, Y.; Kim, D. Structure-Functional Analysis of Human Cytochrome P450 2C8 Using Directed Evolution. Pharmaceutics 2021, 13, 1429. [Google Scholar] [CrossRef] [PubMed]
- Rowlatt, B.; Yorke, J.A.; Strong, A.J.; Whitehouse, C.J.C.; Bell, S.G.; Wong, L.-L. Chain length-dependent cooperativity in fatty acid binding and oxidation by cytochrome P450BM3 (CYP102A1). Protein Cell 2011, 2, 656–671. [Google Scholar] [CrossRef] [Green Version]
- Atkins, W.M. Non-Michaelis-Menten kinetics in cytochrome P450-catalyzed reactions. Annu. Rev. Pharmacol. Toxicol. 2005, 45, 291. [Google Scholar] [CrossRef] [Green Version]
- Zeldin, D.C.; Dubois, R.N.; Falck, J.R.; Capdevila, J.H. Molecular cloning, expression and characterization of an endogenous human cytochrome P450 arachidonic acid epoxygenase isoform. Arch. Biochem. Biophys. 1995, 322, 76–86. [Google Scholar] [CrossRef]
- Rifkind, A.B.; Lee, C.; Chang, T.K.H.; Waxman, D.J. Arachidonic acid metabolism by human cytochrome P450s 2C8, 2C9, 2E1, and 1A2: Regioselective oxygenation and evidence for a role for CYP2C enzymes in arachidonic acid epoxygenation in human liver microsomes. Arch. Biochem. Biophys. 1995, 320, 380–389. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold-Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Steinegger, M.; Söding, J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol. 2017, 35, 1026–1028. [Google Scholar] [CrossRef] [Green Version]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Shindyalov, I.N.; Bourne, P.E. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998, 11, 739–747. [Google Scholar] [CrossRef] [PubMed]
- Gaudreault, F.; Morency, L.-P.; Najmanovich, R.J. NRGsuite: A PyMOL plugin to perform docking simulations in real time using FlexAID. Bioinformatics 2015, 31, 3856–3858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
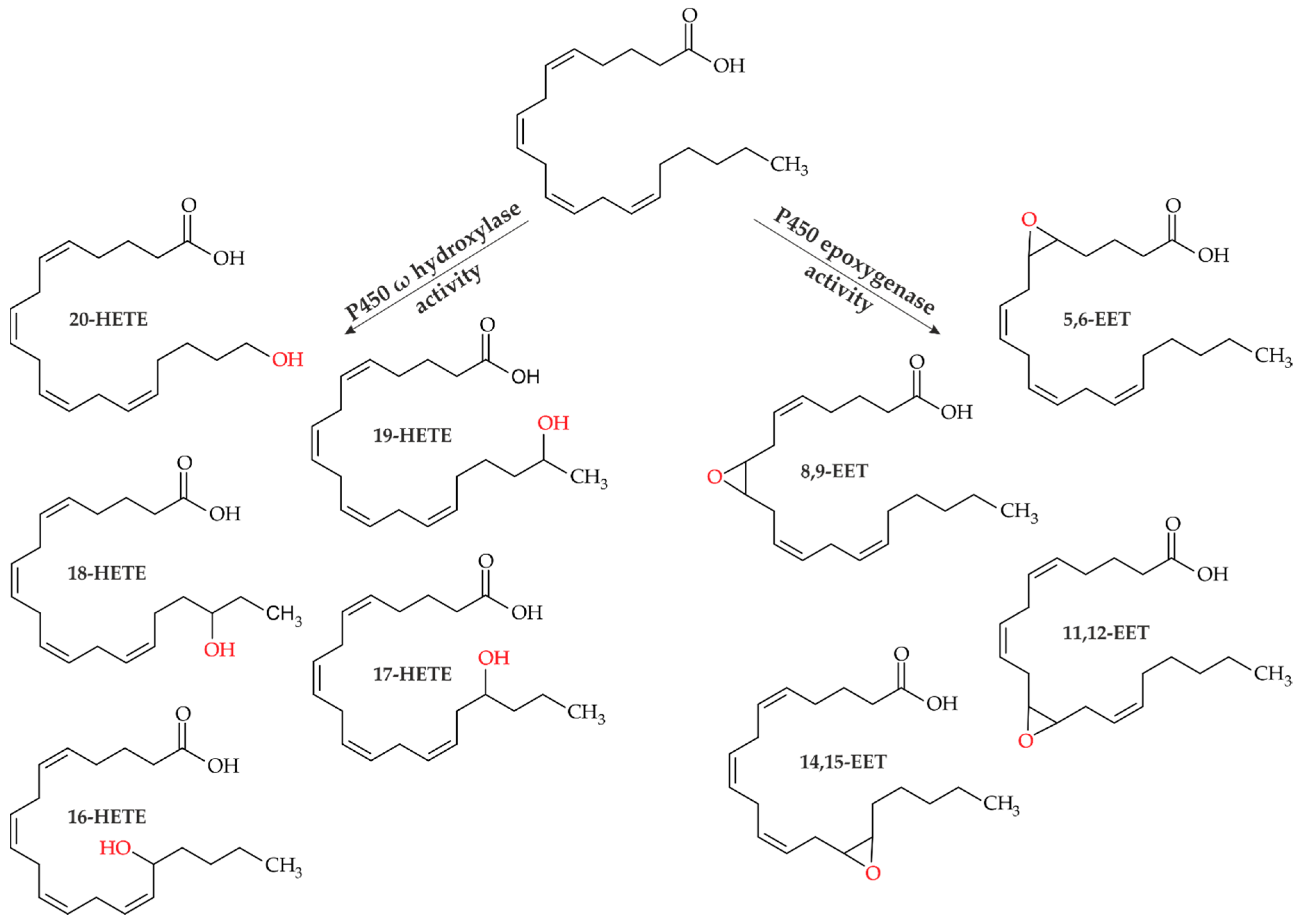
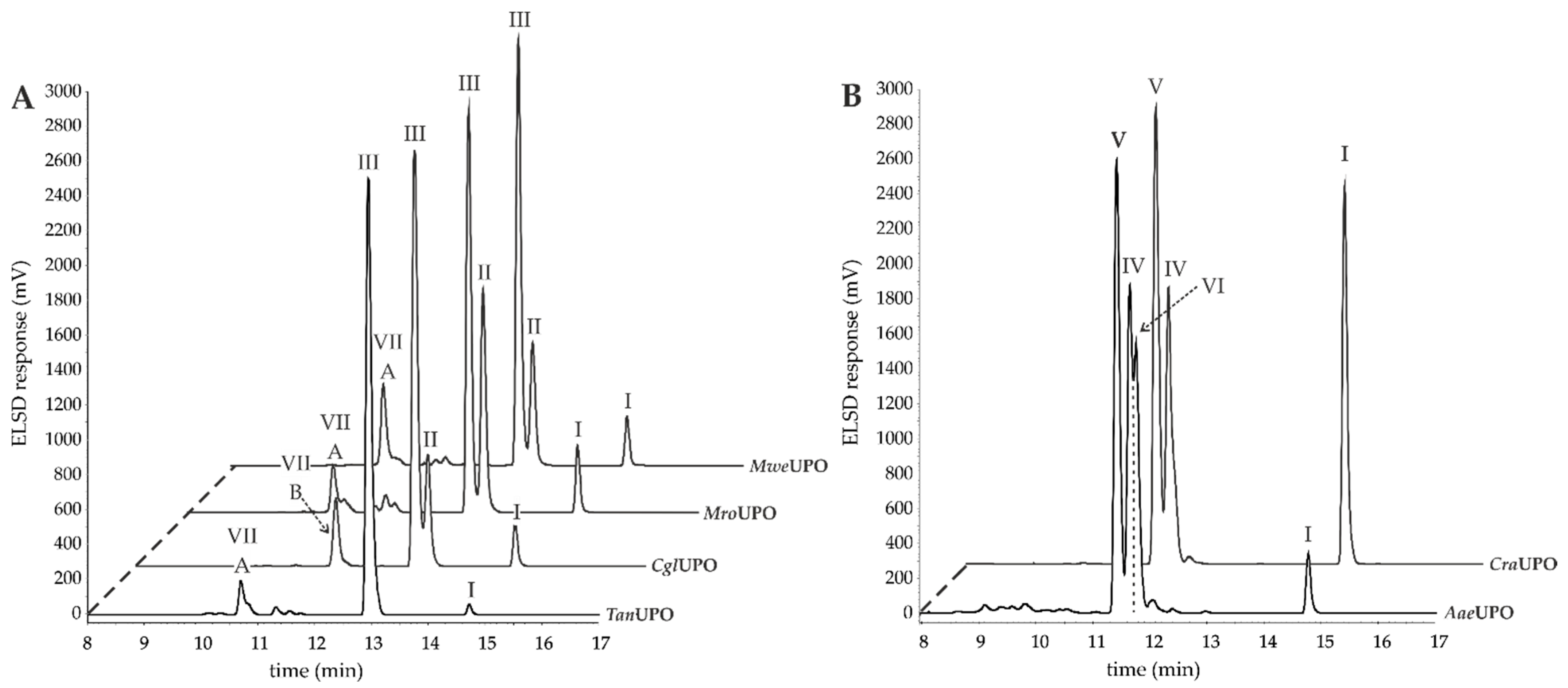
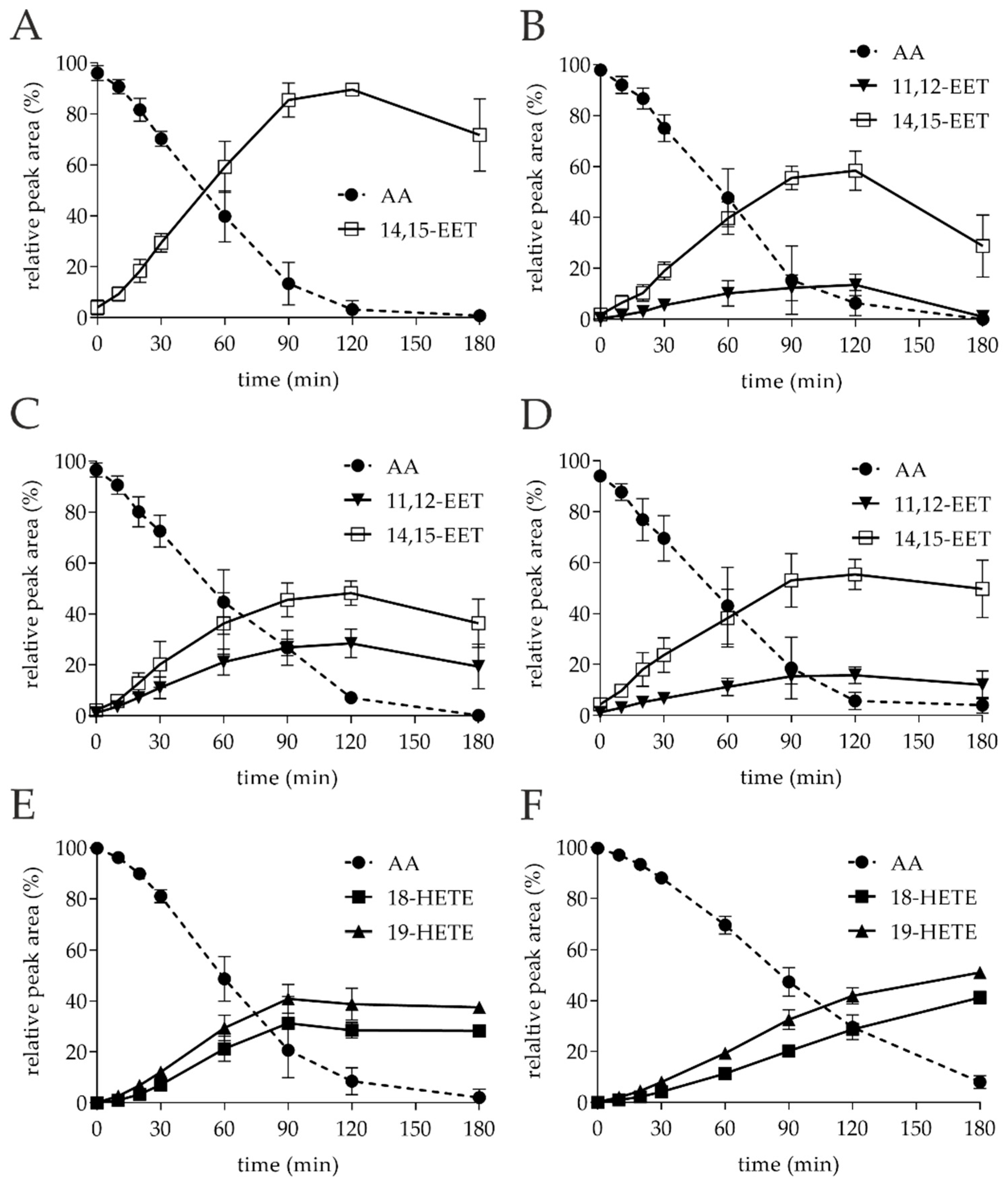
| Entry | Enzyme | AA Conversion Rate (%) 2 | Yield 18-HETE (%) 2 | Yield 19-HETE (%) 2 | Yield 11,12-EET (%) 2 | Yield 14,15-EET (%) 2 |
|---|---|---|---|---|---|---|
| 1 | TanUPO | 95.6 ± 4.4 | - | - | - | 89.2 ± 4.8 |
| 2 | CglUPO | 92.8 ± 6.3 | - | - | 14.5 ± 3.1 | 58.0 ± 7.0 |
| 3 | MroUPO | 91.4 ± 4.3 | - | - | 28.6 ± 4.7 | 48.1 ± 4.2 |
| 4 | MweUPO | 92.5 ± 6.3 | - | - | 15.6 ± 4.1 | 56.3 ± 5.5 |
| 5 | AaeUPO | 89.4 ± 6.2 | 27.0 ± 1.6 | 38.8 ± 4.9 | - | - |
| 6 | CraUPO | 70.8 ± 5.7 | 28.4 ± 2.6 | 42.3 ± 3.6 | - | - |
| Peak | MS1 [M − H]− | MS2 Fragmentation [M − H]− (Relative Abundance %) | Structure |
|---|---|---|---|
| II | 319.2262 theor. for C20H32O3 | 167.1062 (100), 179.1063 (26), 59.0123 (CH3COOH) (24), 257.2265 (-H2O-CO2) (17), 301.2163 (-H2O) (15), 208.1089 (8) | 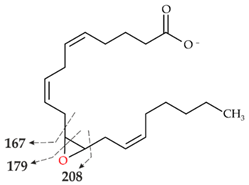 |
| III | 319.2261 theor. for C20H32O3 | 113.0955 (63), 257.2264 (-H2O-CO2) (51), 301.2159 (-H2O) (49), 59.0123 (CH3COOH) (45), 175.1475 (38), 219.1375 (35), 99.0798 (14) | 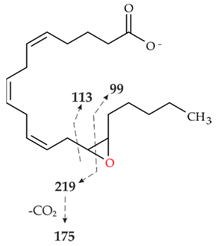 |
| IV | 319.2261 theor. for C20H32O3 | 261.1848 (86), 301.2159 (-H2O)(54), 59.0123 (CH3COOH) (49), 217.1947 (25), 257.2264 (-H2O-CO2)(15) | 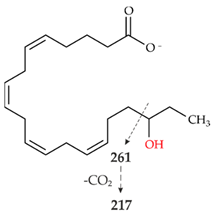 |
| V | 319.2261 theor. for C20H32O3 | 275.2003 (99), 59.0123 (CH3COOH)(74), 301.2159 (-H2O)(50), 231.2104 (20), 257.2264 (-H2O-CO2)(13) | 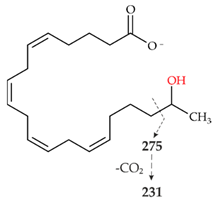 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
König, R.; Kiebist, J.; Kalmbach, J.; Herzog, R.; Schmidtke, K.-U.; Kellner, H.; Ullrich, R.; Jehmlich, N.; Hofrichter, M.; Scheibner, K. Novel Unspecific Peroxygenase from Truncatella angustata Catalyzes the Synthesis of Bioactive Lipid Mediators. Microorganisms 2022, 10, 1267. https://doi.org/10.3390/microorganisms10071267
König R, Kiebist J, Kalmbach J, Herzog R, Schmidtke K-U, Kellner H, Ullrich R, Jehmlich N, Hofrichter M, Scheibner K. Novel Unspecific Peroxygenase from Truncatella angustata Catalyzes the Synthesis of Bioactive Lipid Mediators. Microorganisms. 2022; 10(7):1267. https://doi.org/10.3390/microorganisms10071267
Chicago/Turabian StyleKönig, Rosalie, Jan Kiebist, Johannes Kalmbach, Robert Herzog, Kai-Uwe Schmidtke, Harald Kellner, René Ullrich, Nico Jehmlich, Martin Hofrichter, and Katrin Scheibner. 2022. "Novel Unspecific Peroxygenase from Truncatella angustata Catalyzes the Synthesis of Bioactive Lipid Mediators" Microorganisms 10, no. 7: 1267. https://doi.org/10.3390/microorganisms10071267
APA StyleKönig, R., Kiebist, J., Kalmbach, J., Herzog, R., Schmidtke, K.-U., Kellner, H., Ullrich, R., Jehmlich, N., Hofrichter, M., & Scheibner, K. (2022). Novel Unspecific Peroxygenase from Truncatella angustata Catalyzes the Synthesis of Bioactive Lipid Mediators. Microorganisms, 10(7), 1267. https://doi.org/10.3390/microorganisms10071267







