Validation of Two Commercial Multiplex Real-Time PCR Assays for Detection of SARS-CoV-2 in Stool Donors for Fecal Microbiota Transplantation
Abstract
:1. Introduction
2. Materials and Methods
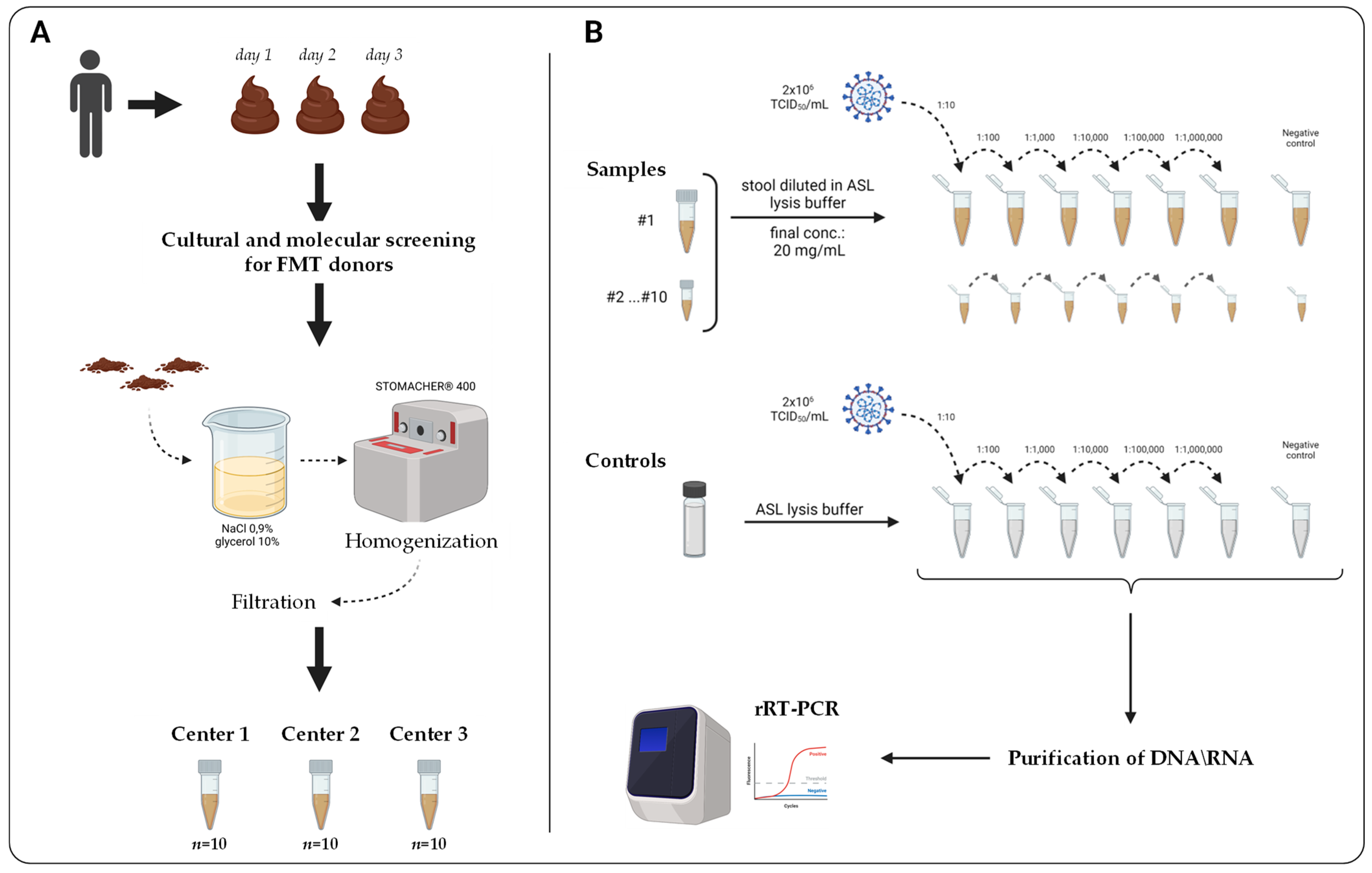
2.1. Study Design
2.2. Stool Specimens and Donors’ Clinical Characteristics
2.3. Cell Culture and Propagation of SARS-CoV-2
2.4. Preparation of Stool Samples Spiked with SARS-CoV-2 RNA
2.5. RNA Extraction and rRT-PCR Testing
2.6. Analytical Evaluations
2.7. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. China Novel Coronavirus Investigating and Research Team. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Brooks, E.F.; Bhatt, A.S. The Gut Microbiome: A Missing Link in Understanding the Gastrointestinal Manifestations of COVID-19? Mol. Case Stud. 2021, 7, a006031. [Google Scholar] [CrossRef] [PubMed]
- Mao, R.; Qiu, Y.; He, J.-S.; Tan, J.-Y.; Li, X.-H.; Liang, J.; Shen, J.; Zhu, L.-R.; Chen, Y.; Iacucci, M.; et al. Manifestations and prognosis of gastrointestinal and liver involvement in patients with COVID-19: A systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 2020, 5, 667–678. [Google Scholar] [CrossRef]
- CDC Symptoms of Coronavirus. Available online: https://www.cdc.gov/coronavirus/2019-ncov/symptoms-testing/symptoms.html (accessed on 16 December 2021).
- Jeong, H.W.; Kim, S.-M.; Kim, H.-S.; Kim, Y.-I.; Kim, J.H.; Cho, J.Y.; Kim, S.-H.; Kang, H.; Kim, S.-G.; Park, S.-J.; et al. Viable SARS-CoV-2 in various specimens from COVID-19 patients. Clin. Microbiol. Infect. 2020, 26, 1520–1524. [Google Scholar] [CrossRef] [PubMed]
- Wölfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Müller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C.; et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020, 581, 465–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.; Guo, C.; Tang, L.; Hong, Z.; Zhou, J.; Dong, X.; Yin, H.; Xiao, Q.; Tang, Y.; Qu, X.; et al. Prolonged Presence of SARS-CoV-2 Viral RNA in Faecal Samples. Lancet Gastroenterol. Hepatol. 2020, 5, 434–435. [Google Scholar] [CrossRef]
- Wang, W.; Xu, Y.; Gao, R.; Lu, R.; Han, K.; Wu, G.; Tan, W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA J. Am. Med. Assoc. 2020, 323, 1843–1844. [Google Scholar] [CrossRef] [Green Version]
- Kitajima, M.; Ahmed, W.; Bibby, K.; Carducci, A.; Gerba, C.P.; Hamilton, K.A.; Haramoto, E.; Rose, J.B. SARS-CoV-2 in wastewater: State of the knowledge and research needs. Sci. Total Environ. 2020, 739, 139076. [Google Scholar] [CrossRef]
- Baunwall, S.M.D.; Terveer, E.M.; Dahlerup, J.F.; Erikstrup, C.; Arkkila, P.; Vehreschild, M.J.; Ianiro, G.; Gasbarrini, A.; Sokol, H.; Kump, P.K.; et al. The use of Faecal Microbiota Transplantation (FMT) in Europe: A Europe-wide survey. Lancet Reg. Health Eur. 2021, 9, 100181. [Google Scholar] [CrossRef]
- Van Prehn, J.; Reigadas, E.; Vogelzang, E.H.; Bouza, E.; Hristea, A.; Guery, B.; Krutova, M.; Norén, T.; Allerberger, F.; Coia, J.E.; et al. European Society of Clinical Microbiology and Infectious Diseases: 2021 update on the treatment guidance document for Clostridioides difficile infection in adults. Clin. Microbiol. Infect. 2021, 27, S1–S21. [Google Scholar] [CrossRef]
- Wilcox, M.H.; McGovern, B.H.; Hecht, G.A. The Efficacy and Safety of Fecal Microbiota Transplant for Recurrent Clostridium Difficile Infection: Current Understanding and Gap Analysis. Open Forum Infect. Dis. 2020, 7, ofaa114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ianiro, G.; Mullish, B.H.; Kelly, C.R.; Kassam, Z.; Kuijper, E.J.; Ng, S.C.; Iqbal, T.H.; Allegretti, J.R.; Bibbò, S.; Sokol, H.; et al. Reorganisation of faecal microbiota transplant services during the COVID-19 pandemic. Gut 2020, 69, 1555–1563. [Google Scholar] [CrossRef] [PubMed]
- Ianiro, G.; Mullish, B.; Kelly, C.R.; Sokol, H.; Kassam, Z.; Ng, S.C.; Fischer, M.; Allegretti, J.R.; Masucci, L.; Zhang, F.; et al. Screening of faecal microbiota transplant donors during the COVID-19 outbreak: Suggestions for urgent updates from an international expert panel. Lancet Gastroenterol. Hepatol. 2020, 5, 430–432. [Google Scholar] [CrossRef]
- Xiao, F.; Tang, M.; Zheng, X.; Liu, Y.; Li, X.; Shan, H. Evidence for Gastrointestinal Infection of SARS-CoV-2. Gastroenterology 2020, 158, 1831–1833.e3. [Google Scholar] [CrossRef] [PubMed]
- Szymczak, W.A.; Goldstein, D.Y.; Orner, E.P.; Fecher, R.A.; Yokoda, R.; Skalina, K.A.; Narlieva, M.; Gendlina, I.; Fox, A.S. Utility of Stool PCR for the Diagnosis of COVID-19: Comparison of Two Commercial Platforms. J. Clin. Microbiol. 2020, 58, e01369-20. [Google Scholar] [CrossRef]
- Cao, R.; Bao, L.; Pan, M.; Zhang, C.; Liao, H.; Liu, L.; Li, Y.; Li, M. Detection of SARS-CoV-2 in fecal samples with different pretreatment methods and PCR kits. BMC Microbiol. 2021, 21, 56. [Google Scholar] [CrossRef]
- Schrader, C.; Schielke, A.; Ellerbroek, L.; Johne, R. PCR Inhibitors—Occurrence, Properties and Removal. J. Appl. Microbiol. 2012, 113, 1014–1026. [Google Scholar] [CrossRef]
- Cammarota, G.; Ianiro, G.; Tilg, H.; Rajilic-Stojanovic, M.; Kump, P.; Satokari, R.; Sokol, H.; Arkkila, P.; Pintus, C.; Hart, A.; et al. European consensus conference on faecal microbiota transplantation in clinical practice. Gut 2017, 66, 569–580. [Google Scholar] [CrossRef]
- Pastorino, B.; Touret, F.; Gilles, M.; De Lamballerie, X.; Charrel, R.N. Heat Inactivation of Different Types of SARS-CoV-2 Samples: What Protocols for Biosafety, Molecular Detection and Serological Diagnostics? Viruses 2020, 12, 735. [Google Scholar] [CrossRef]
- Kralik, P.; Ricchi, M. A Basic Guide to Real Time PCR in Microbial Diagnostics: Definitions, Parameters, and Everything. Front. Microbiol. 2017, 8, 108. [Google Scholar] [CrossRef] [Green Version]
- Smith, M. Validating Real-Time Polymerase Chain Reaction (PCR) Assays. Encycl. Virol. 2020, 5, 35–44. [Google Scholar] [CrossRef]
- Muhammad, A.; Ameer, H.; Haider, S.A.; Ali, I. Detection of SARS-CoV-2 Using Real-Time Polymerase Chain Reaction in Different Clinical Specimens: A Critical Review. Allergol. Immunopathol. 2021, 49, 159–164. [Google Scholar] [CrossRef] [PubMed]
- Van Doorn, A.S.; Meijer, B.; Frampton, C.M.A.; Barclay, M.L.; de Boer, N.K.H. Systematic Review with Meta-Analysis: SARS-CoV-2 Stool Testing and the Potential for Faecal-Oral Transmission. Aliment. Pharmacol. Ther. 2020, 52, 1276–1288. [Google Scholar] [CrossRef] [PubMed]
- Xiao, F.; Sun, J.; Xu, Y.; Li, F.; Huang, X.; Li, H.; Zhao, J.; Huang, J.; Zhao, J. Infectious SARS-CoV-2 in Feces of Patient with Severe COVID-19. Emerg. Infect. Dis. 2020, 26, 1920–1922. [Google Scholar] [CrossRef]
- Guo, M.; Tao, W.; Flavell, R.A.; Zhu, S. Potential Intestinal Infection and Faecal–Oral Transmission of SARS-CoV-2. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 269–283. [Google Scholar] [CrossRef]
- Heller, L.; Mota, C.R.; Greco, D.B. COVID-19 faecal-oral transmission: Are we asking the right questions? Sci. Total Environ. 2020, 729, 138919. [Google Scholar] [CrossRef]
- Hindson, J. COVID-19: Faecal–oral transmission? Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 259. [Google Scholar] [CrossRef]
- FDA Fecal Microbiota for Transplantation: New Safety Information—Regarding Additional Protections for Screening Donors for COVID-19 and Exposure to SARS-CoV-2 and Testing for SARS-CoV-2. Available online: https://www.fda.gov/safety/medical-product-safety-information/fecal-microbiota-transplantation-new-safety-information-regarding-additional-protections-screening (accessed on 16 December 2021).
- Khanna, S.; Tande, A.; Rubin, D.T.; Khoruts, A.; Kahn, S.A.; Pardi, D.S. Fecal Microbiota Transplantation for Recurrent C difficile Infection During the COVID-19 Pandemic. Mayo Clin. Proc. 2021, 96, 1418–1425. [Google Scholar] [CrossRef]
- Parvathy, S.N.; Lenehan, J.G.; Fernandes, R.; Poutanen, S.M.; Hota, S.; Vareki, S.M.; Silverman, M. Enhanced donor screening for faecal microbial transplantation during COVID-19. Gut 2021, 70, 2219–2220. [Google Scholar] [CrossRef]
- Ianiro, G.; Bibbò, S.; Masucci, L.; Quaranta, G.; Porcari, S.; Settanni, C.R.; Lopetuso, L.R.; Fantoni, M.; Sanguinetti, M.; Gasbarrini, A.; et al. Maintaining standard volumes, efficacy and safety, of fecal microbiota transplantation for C. difficile infection during the COVID-19 pandemic: A prospective cohort study. Dig. Liver Dis. 2020, 52, 1390–1395. [Google Scholar] [CrossRef]
- Ng, S.C.; Chan, F.K.L.; Chan, P.K.S. Screening FMT Donors during the COVID-19 Pandemic: A Protocol for Stool SARS-CoV-2 Viral Quantification. Lancet Gastroenterol. Hepatol. 2020, 5, 642–643. [Google Scholar] [CrossRef]
- Jones, D.L.; Baluja, M.Q.; Graham, D.W.; Corbishley, A.; McDonald, J.E.; Malham, S.K.; Hillary, L.; Connor, T.R.; Gaze, W.H.; Moura, I.B.; et al. Shedding of SARS-CoV-2 in feces and urine and its potential role in person-to-person transmission and the environment-based spread of COVID-19. Sci. Total Environ. 2020, 749, 141364. [Google Scholar] [CrossRef] [PubMed]
- Giovacchini, N.; Coppi, M.; Aiezza, N.; Baccani, I.; Malentacchi, F.; Pollini, S.; Antonelli, A.; Rossolini, G.M. Rapid screening for SARS-CoV-2 VOC-Alpha (202012/01, B.1.1.7) using the Allplex™ SARS-CoV-2/FluA/FluB/RSV Assay. Int. J. Infect. Dis. 2021, 113, 207–209. [Google Scholar] [CrossRef] [PubMed]
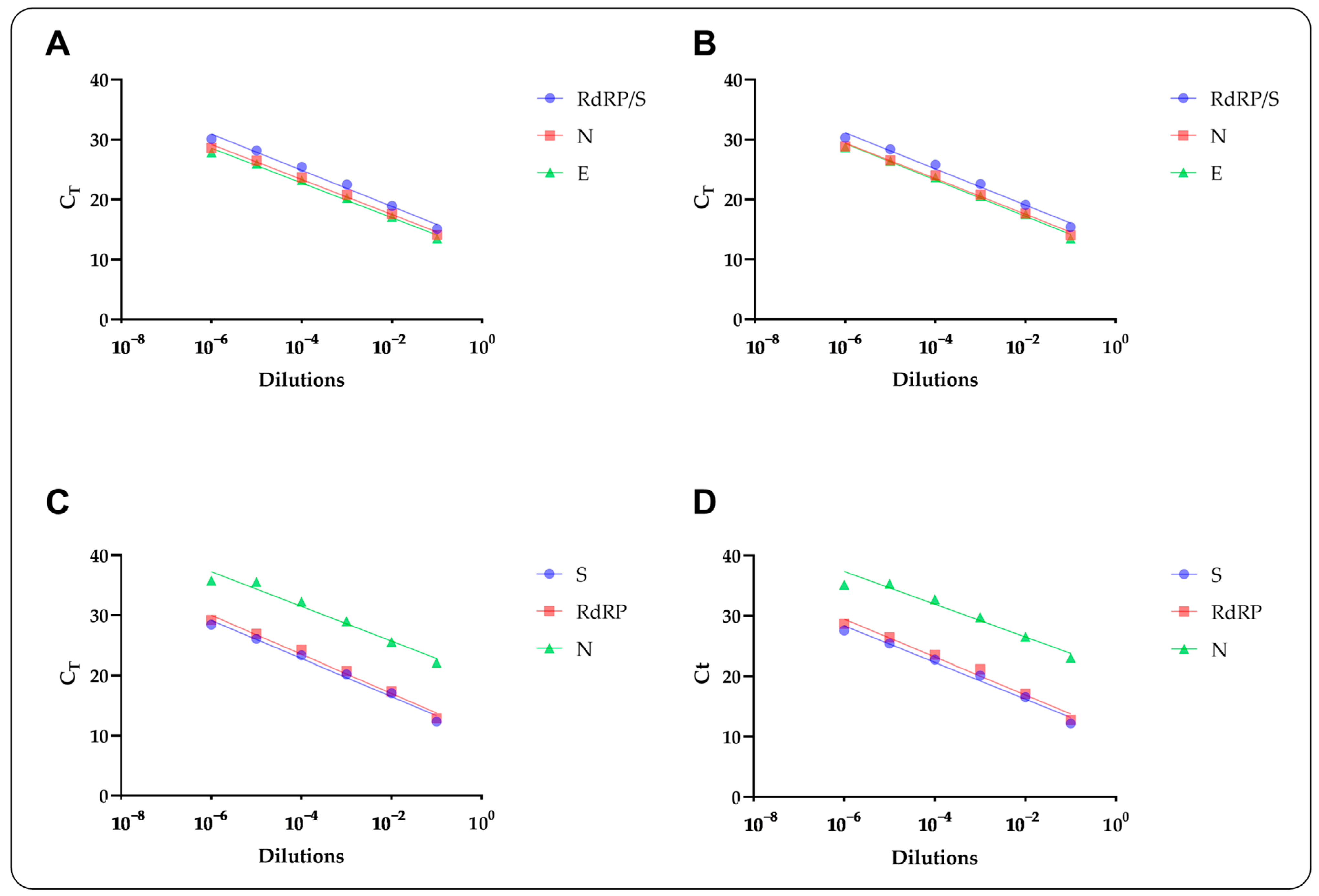
| SC2 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dilution | Viral Titer (TCID50/mL) | RNA cp/µL | RNA cp/mg | RdRP/S (SD) | Percent Positivity (n/N) | CV (%) | CTASL (SD) | CTN (SD) | Percent Positivity (n/N) | CV (%) | CTASL (SD) | CTE (SD) | Percent Positivity (n/N) | CV (%) | CTASL (SD) |
| 10−1 | 2 × 105 | 1.03 × 107 | 5.15 × 108 | 15.12 (1.38) | 100 (30/30) | 9.13 | 15.42 (2.29) | 14.10 (1.29) | 100 (30/30) | 9.15 | 14.04 (2.02) | 13.51 (1.66) | 100 (30/30) | 12.29 | 13.49 (2.68) |
| 10−2 | 2 × 104 | 1.06 × 106 | 5.30 × 107 | 18.94 (1.59) | 100 (30/30) | 8.39 | 19.11 (2.83) | 17.63 (1.27) | 100 (30/30) | 7.20 | 17.69 (1.96) | 17.11 (1.57) | 100 (30/30) | 9.18 | 17.54 (2.33) |
| 10−3 | 2 × 103 | 9.71 × 104 | 4.86 × 106 | 22.51 (1.91) | 100 (30/30) | 8.49 | 22.57 (2.50) | 20.80 (1.16) | 100 (30/30) | 5.58 | 20.82 (2.00) | 20.28 (1.34) | 100 (30/30) | 6.61 | 20.62 (2.11) |
| 10−4 | 2 × 102 | 9.97 × 103 | 4.99 × 105 | 25.45 (1.44) | 100 (30/30) | 5.66 | 25.82 (2.33) | 23.72 (1.15) | 100 (30/30) | 4.85 | 24.06 (1.59) | 23.26 (1.28) | 100 (30/30) | 5.50 | 23.73 (1.60) |
| 10−5 | 2 × 101 | 9.27 × 102 | 4.64 × 104 | 28.18 (1.80) | 100 (30/30) | 6.39 | 28.37 (2.59) | 26.53 (1.52) | 100 (30/30) | 5.73 | 26.54 (1.98) | 25.99 (1.42) | 100 (30/30) | 5.46 | 26.45 (2.06) |
| 10−6 | 2 | 1.33 × 102 | 6.65 × 103 | 30.11 (2.58) | 100 (30/30) | 8.57 | 30.33 (3.38) | 28.61 (2.55) | 100 (30/30) | 8.91 | 28.89 (2.64) | 27.87 (2.26) | 100 (30/30) | 8.11 | 28.73 (2.52) |
| SC2FABR | |||||||||||||||
| Dilution | Viral Titer (TCID50/mL) | RNA cp/µL | RNA cp/mg | RdRP/S (SD) | Percent Positivity (n/N) | CV (%) | CTASL (SD) | CTN (SD) | Percent Positivity (n/N) | CV (%) | CTASL (SD) | CTS (SD) | Percent Positivity (n/N) | CV (%) | CTASL (SD) |
| 10−1 | 2 × 105 | 1.03 × 107 | 5.15 × 108 | 12.75 (1.49) | 100 (30/30) | 11.69 | 12.88 (1.28) | 23.01 (1.10) | 100 (30/30) | 4.78 | 22.16 (0.45) | 12.17 (1.01) | 100 (30/30) | 8.30 | 12.34 (0.76) |
| 10−2 | 2 × 104 | 1.06 × 106 | 5.30 × 107 | 17.09 (1.20) | 100 (30/30) | 7.02 | 17.39 (0.63) | 26.47 (1.09) | 100 (30/30) | 4.12 | 25.55 (0.62) | 16.56 (0.93) | 100 (30/30) | 5.62 | 17.07 (0.57) |
| 10−3 | 2 × 103 | 9.71 × 104 | 4.86 × 106 | 20.68 (1.48) | 100 (30/30) | 7.16 | 20.74 (0.58) | 29.76 (1.49) | 100 (30/30) | 5.01 | 29.02 (0.27) | 19.79 (0.78) | 100 (30/30) | 3.94 | 20.20 (0.56) |
| 10−4 | 2 × 102 | 9.97 × 103 | 4.99 × 105 | 23.50 (0.93) | 100 (30/30) | 3.96 | 24.28 (0.56) | 32.69 (1.43) | 100 (30/30) | 4.37 | 32.27 (0.99) | 22.73 (0.86) | 100 (30/30) | 3.78 | 23.35 (0.88) |
| 10−5 | 2 × 101 | 9.27 × 102 | 4.64 × 104 | 26.40 (0.98) | 100 (30/30) | 3.71 | 26.90 (1.28) | 35.21 (1.84) | 80 (24/30) | 5.23 | 35.55 (1.84) | 25.40 (0.96) | 100 (30/30) | 3.78 | 26.08 (1.29) |
| 10−6 | 2 | 1.33 × 102 | 6.65 × 103 | 28.55 (2.23) | 100 (30/30) | 7.81 | 29.20 (2.23) | 35.15 (1.95) | 50 (15/30) | 5.55 | 35.77 (1.53) | 27.52 (2.24) | 100 (30/30) | 8.14 | 28.44 (2.17) |
| Stool Samples | ASL Controls | ||||
|---|---|---|---|---|---|
| Allplex™ Assay | Target | R2 | ε (%) | R2 | ε (%) |
| SC2 | RdRP/S | 0.88 | 114 | 0.84 | 115 |
| N | 0.91 | 120 | 0.90 | 117 | |
| E | 0.90 | 121 | 0.88 | 114 | |
| SC2FABR | RdRP | 0.91 | 114 | 0.96 | 107 |
| S | 0.89 | 108 | 0.96 | 103 | |
| N | 0.87 | 133 | 0.94 | 122 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Pilato, V.; Morecchiato, F.; Rizzato, C.; Quaranta, G.; Fais, R.; Gandolfo, C.; Antonelli, A.; Cusi, M.G.; Pistello, M.; Rossolini, G.M.; et al. Validation of Two Commercial Multiplex Real-Time PCR Assays for Detection of SARS-CoV-2 in Stool Donors for Fecal Microbiota Transplantation. Microorganisms 2022, 10, 284. https://doi.org/10.3390/microorganisms10020284
Di Pilato V, Morecchiato F, Rizzato C, Quaranta G, Fais R, Gandolfo C, Antonelli A, Cusi MG, Pistello M, Rossolini GM, et al. Validation of Two Commercial Multiplex Real-Time PCR Assays for Detection of SARS-CoV-2 in Stool Donors for Fecal Microbiota Transplantation. Microorganisms. 2022; 10(2):284. https://doi.org/10.3390/microorganisms10020284
Chicago/Turabian StyleDi Pilato, Vincenzo, Fabio Morecchiato, Cosmeri Rizzato, Gianluca Quaranta, Roberta Fais, Claudia Gandolfo, Alberto Antonelli, Maria Grazia Cusi, Mauro Pistello, Gian Maria Rossolini, and et al. 2022. "Validation of Two Commercial Multiplex Real-Time PCR Assays for Detection of SARS-CoV-2 in Stool Donors for Fecal Microbiota Transplantation" Microorganisms 10, no. 2: 284. https://doi.org/10.3390/microorganisms10020284
APA StyleDi Pilato, V., Morecchiato, F., Rizzato, C., Quaranta, G., Fais, R., Gandolfo, C., Antonelli, A., Cusi, M. G., Pistello, M., Rossolini, G. M., Sanguinetti, M., Lupetti, A., & Masucci, L. (2022). Validation of Two Commercial Multiplex Real-Time PCR Assays for Detection of SARS-CoV-2 in Stool Donors for Fecal Microbiota Transplantation. Microorganisms, 10(2), 284. https://doi.org/10.3390/microorganisms10020284










