Fecal Filobasidium Is Associated with Clinical Remission and Endoscopic Response following Fecal Microbiota Transplantation in Mild-to-Moderate Ulcerative Colitis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients and Donors
2.2. Fecal DNA Isolation and Internal Transcribed Spacer 1 (ITS1) Sequencing
2.3. Fungal Strains
2.4. Monocyte-Derived Macrophages Stimulations
2.5. Epithelial Scratch Assays and Stimulation Experiments
2.6. Cytokine Determinations (CBA, ELISA)
2.7. RNA Isolation, First-Strand Synthesis, Qualitative PCR
2.8. Statistical Analysis
2.9. Ethical Considerations
3. Results
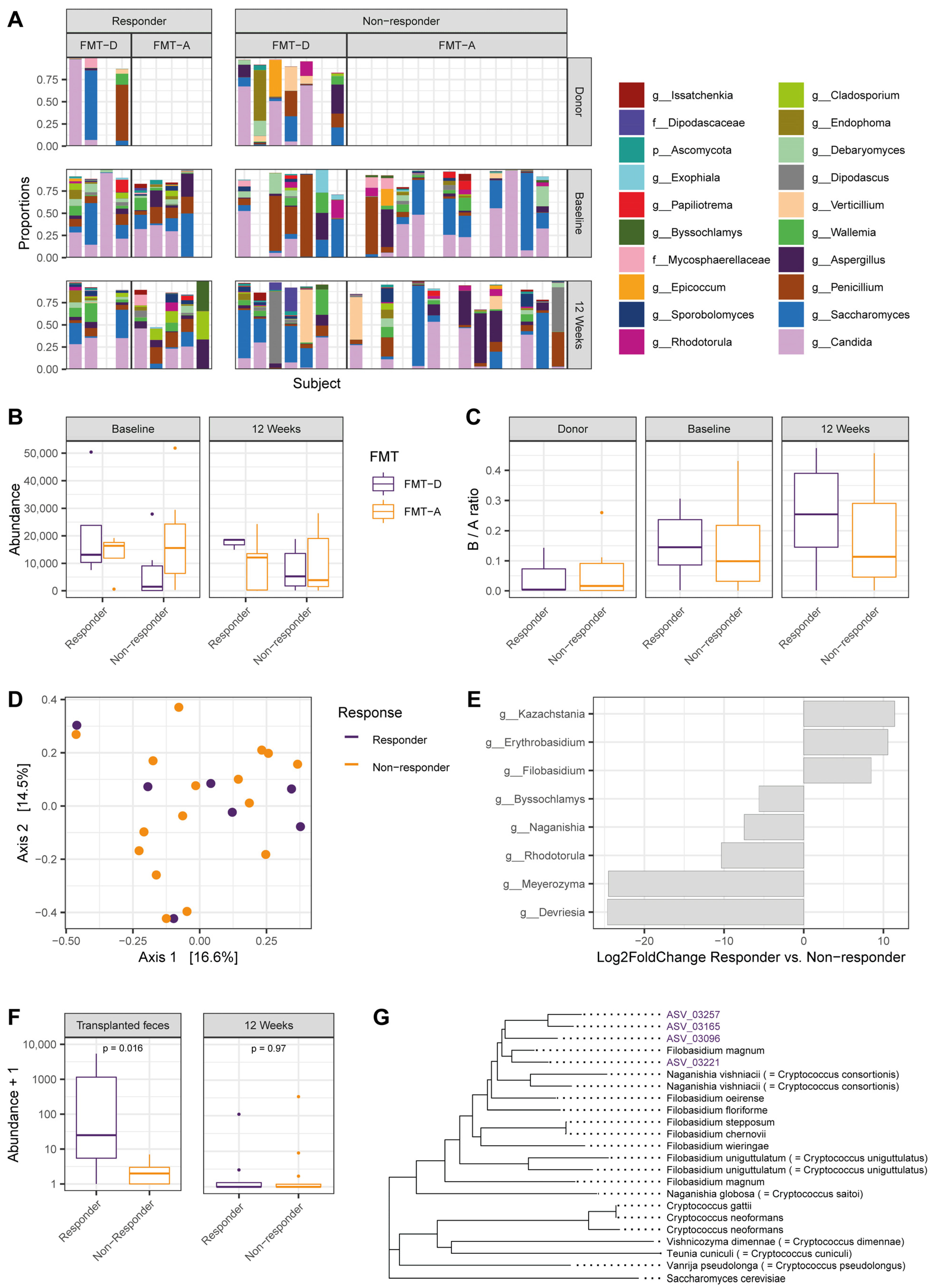
3.1. Elevated Filobasidium spp. Abundance in Transplanted Fecal Material Associates with Positive Response to Fecal Microbiota Transfer
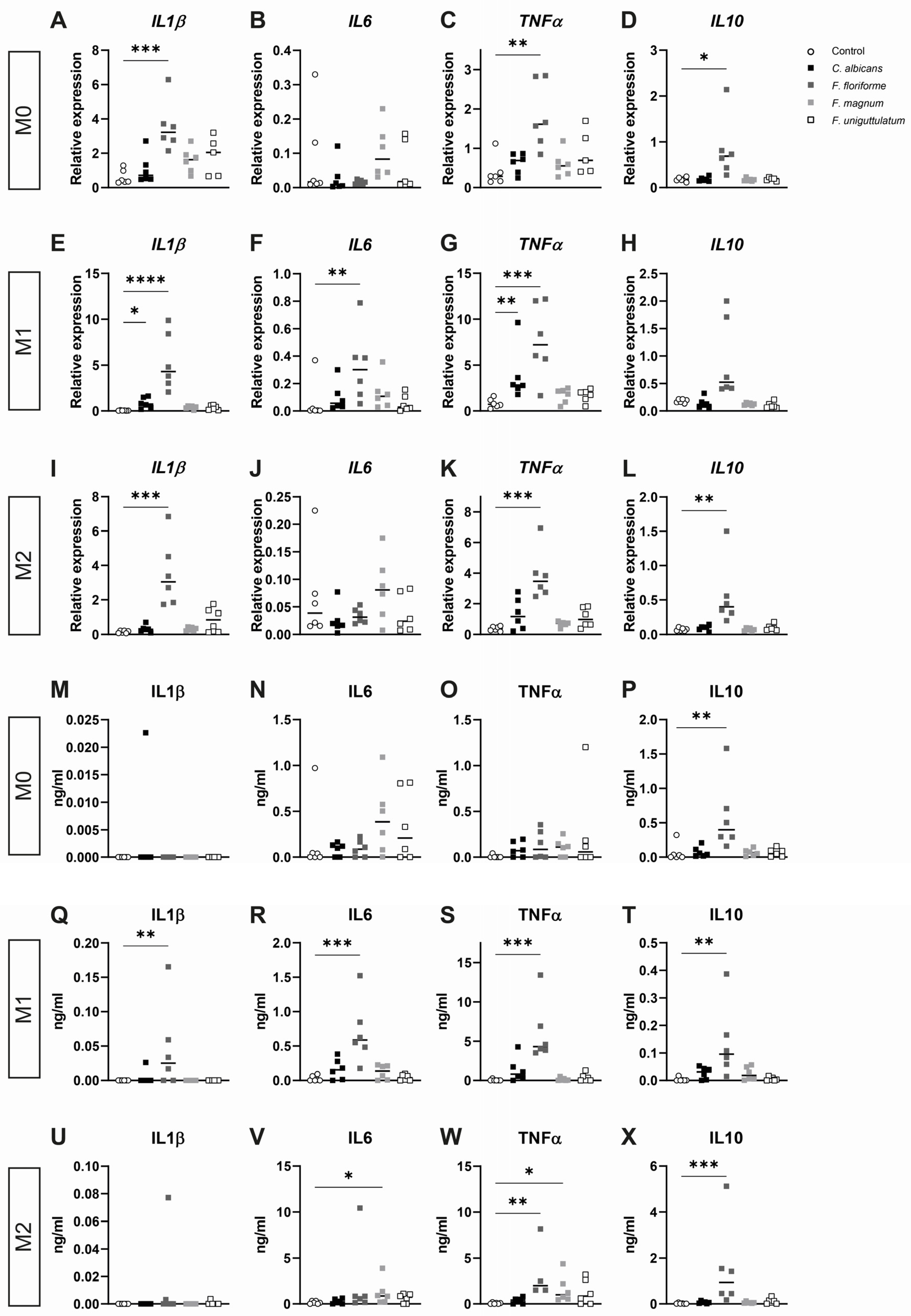
3.2. Filobasidium spp. Stimulation of Monocyte-Derived Macrophages Elicits Cytokine Responses
3.3. Macrophage Cytokine Release after Pre-Treatment with Lipopolysaccharide and Subsequent Stimulation with Filobasidium Species or C. albicans
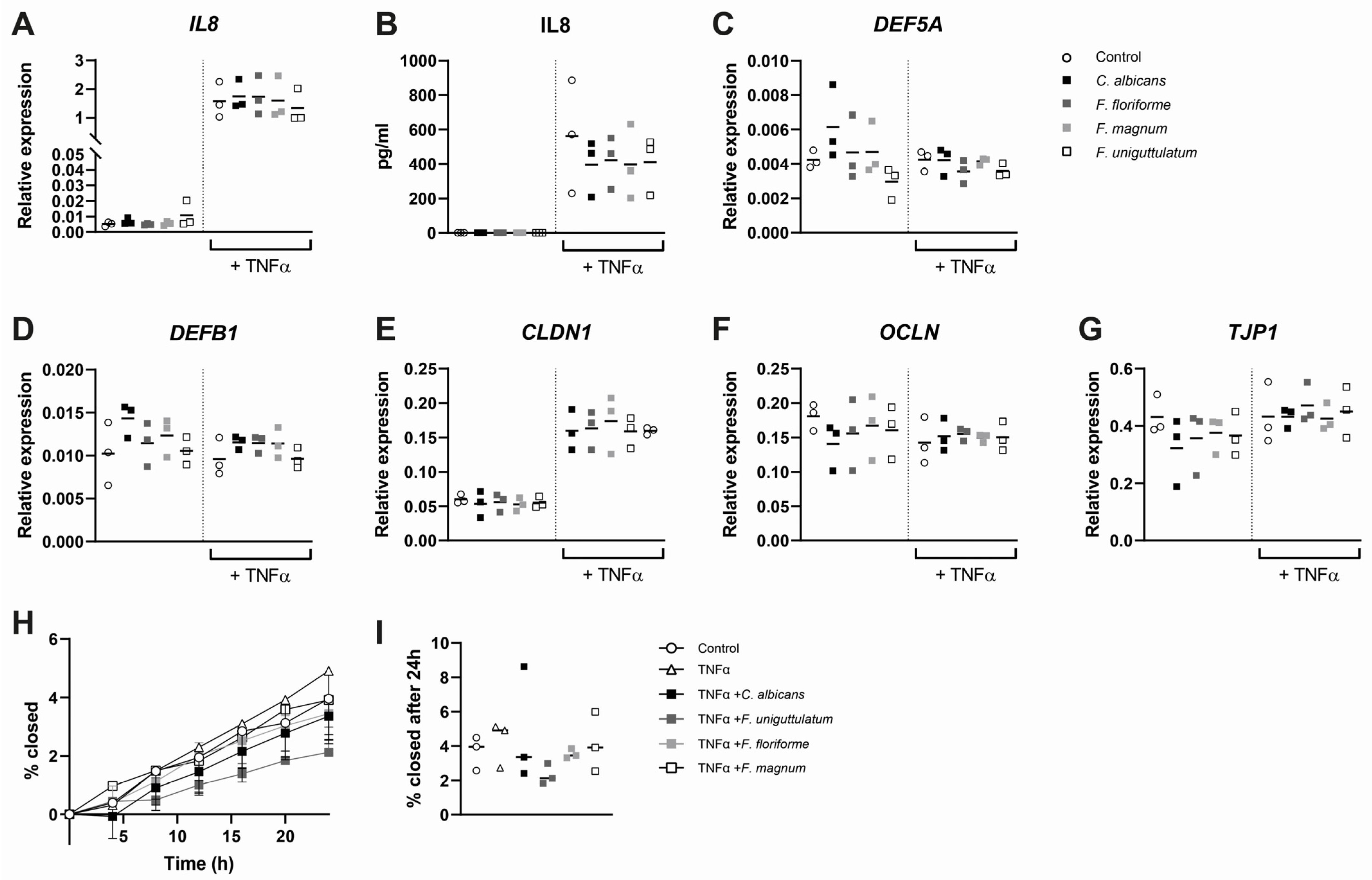
3.4. Epithelial Cytokine Release, Gene Expression, and Wound Healing Are Not Affected by Stimulation with Filobasidium spp.
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Actis, G.C.; Pellicano, R.; Fagoonee, S.; Ribaldone, D.G. History of Inflammatory Bowel Diseases. J. Clin. Med. 2019, 8, 1970. [Google Scholar] [CrossRef] [Green Version]
- Qiu, P.; Ishimoto, T.; Fu, L.; Zhang, J.; Zhang, Z.; Liu, Y. The Gut Microbiota in Inflammatory Bowel Disease. Front. Cell. Infect. Microbiol. 2022, 12, 733992. [Google Scholar] [CrossRef]
- Caruso, R.; Lo, B.C.; Nunez, G. Host-microbiota interactions in inflammatory bowel disease. Nat. Rev. Immunol. 2020, 20, 411–426. [Google Scholar] [CrossRef] [PubMed]
- Sokol, H.; Leducq, V.; Aschard, H.; Pham, H.P.; Jegou, S.; Landman, C.; Cohen, D.; Liguori, G.; Bourrier, A.; Nion-Larmurier, I.; et al. Fungal microbiota dysbiosis in IBD. Gut 2017, 66, 1039–1048. [Google Scholar] [CrossRef] [Green Version]
- Imai, T.; Inoue, R.; Kawada, Y.; Morita, Y.; Inatomi, O.; Nishida, A.; Bamba, S.; Kawahara, M.; Andoh, A. Characterization of fungal dysbiosis in Japanese patients with inflammatory bowel disease. J. Gastroenterol. 2019, 54, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Leonardi, I.; Paramsothy, S.; Doron, I.; Semon, A.; Kaakoush, N.O.; Clemente, J.C.; Faith, J.J.; Borody, T.J.; Mitchell, H.M.; Colombel, J.F.; et al. Fungal Trans-kingdom Dynamics Linked to Responsiveness to Fecal Microbiota Transplantation (FMT) Therapy in Ulcerative Colitis. Cell Host Microbe 2020, 27, 823–829.e3. [Google Scholar] [CrossRef] [PubMed]
- Underhill, D.M.; Braun, J. Fungal microbiome in inflammatory bowel disease: A critical assessment. J. Clin. Investig. 2022, 132, e155786. [Google Scholar] [CrossRef]
- Limon, J.J.; Tang, J.; Li, D.; Wolf, A.J.; Michelsen, K.S.; Funari, V.; Gargus, M.; Nguyen, C.; Sharma, P.; Maymi, V.I.; et al. Malassezia Is Associated with Crohn’s Disease and Exacerbates Colitis in Mouse Models. Cell Host Microbe 2019, 25, 377–388.e6. [Google Scholar] [CrossRef] [Green Version]
- Jain, U.; Ver Heul, A.M.; Xiong, S.; Gregory, M.H.; Demers, E.G.; Kern, J.T.; Lai, C.W.; Muegge, B.D.; Barisas, D.A.G.; Leal-Ekman, J.S.; et al. Debaryomyces is enriched in Crohn’s disease intestinal tissue and impairs healing in mice. Science 2021, 371, 1154–1159. [Google Scholar] [CrossRef]
- Costello, S.P.; Soo, W.; Bryant, R.V.; Jairath, V.; Hart, A.L.; Andrews, J.M. Systematic review with meta-analysis: Faecal microbiota transplantation for the induction of remission for active ulcerative colitis. Aliment. Pharmacol. Ther. 2017, 46, 213–224. [Google Scholar] [CrossRef] [PubMed]
- Wilson, B.C.; Vatanen, T.; Cutfield, W.S.; O’Sullivan, J.M. The Super-Donor Phenomenon in Fecal Microbiota Transplantation. Front. Cell. Infect. Microbiol. 2019, 9, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossen, N.G.; Fuentes, S.; van der Spek, M.J.; Tijssen, J.G.; Hartman, J.H.; Duflou, A.; Lowenberg, M.; van den Brink, G.R.; Mathus-Vliegen, E.M.; de Vos, W.M.; et al. Findings From a Randomized Controlled Trial of Fecal Transplantation for Patients With Ulcerative Colitis. Gastroenterology 2015, 149, 110–118.e4. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, S.; Rossen, N.G.; van der Spek, M.J.; Hartman, J.H.; Huuskonen, L.; Korpela, K.; Salojärvi, J.; Aalvink, S.; de Vos, W.M.; D’Haens, G.R.; et al. Microbial shifts and signatures of long-term remission in ulcerative colitis after faecal microbiota transplantation. ISME J. 2017, 11, 1877–1889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahman, S.; Davids, M.; van Hamersveld, P.H.P.; Welting, O.; Rahaoui, H.; Schuren, F.; Meijer, S.L.; van den Wijngaard, R.M.; Hakvoort, T.B.M.; de Jonge, W.J.; et al. Dietary Curdlan Enhances Bifidobacteria and Reduces Intestinal Inflammation in Mice. Nutrients 2021, 13, 1305. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [Green Version]
- Nilsson, R.H.; Larsson, K.-H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2018, 47, D259–D264. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bodenhofer, U.; Bonatesta, E.; Horejs-Kainrath, C.; Hochreiter, S. msa: An R package for multiple sequence alignment. Bioinformatics 2015, 31, 3997–3999. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T.T.-Y. ggtree: An r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 2017, 8, 28–36. [Google Scholar] [CrossRef]
- van Thiel, I.A.M.; Stavrou, A.A.; de Jong, A. Genetic and phenotypic diversity of fecal Candida albicans strains in irritable bowel syndrome. Sci. Rep. 2022, 12, 5391. [Google Scholar] [CrossRef]
- Repnik, U.; Knezevic, M.; Jeras, M. Simple and cost-effective isolation of monocytes from buffy coats. J. Immunol. Methods 2003, 278, 283–292. [Google Scholar] [CrossRef]
- Prins, M.M.C.; Giugliano, F.P.; van Roest, M.; van de Graaf, S.F.J.; Koelink, P.J.; Wildenberg, M.E. Thiopurines correct the effects of autophagy impairment on intestinal healing—A potential role for ARHGAP18/RhoA. Dis. Models Mech. 2021, 14, dmm047233. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002, 3, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruijter, J.M.; Ramakers, C.; Hoogaars, W.M.; Karlen, Y.; Bakker, O.; van den Hoff, M.J.; Moorman, A.F. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009, 37, e45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Von Elm, E.; Altman, D.G.; Egger, M.; Pocock, S.J.; Gotzsche, P.C.; Vandenbroucke, J.P.; STROBE Initiative. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: Guidelines for reporting observational studies. Lancet 2007, 370, 1453–1457. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package Version 2.5-7; R Core Team: Vienna, Austria, 2020. [Google Scholar]
- Zhu, F.; Feng, D.; Ding, C.; Zhang, T.; Chen, J.; Yu, Z.; Zhao, L.; Xu, Y.; Zhu, W.; Gong, J. Fungal Dysbiosis Aggravates Pouchitis in a Rat Model of Ileal Pouch Anal Anastomosis. Inflamm. Bowel Dis. 2020, 26, 1831–1842. [Google Scholar] [CrossRef] [PubMed]
- Heinsbroek, S.E.; Oei, A.; Roelofs, J.J.; Dhawan, S.; te Velde, A.; Gordon, S.; de Jonge, W.J. Genetic deletion of dectin-1 does not affect the course of murine experimental colitis. BMC Gastroenterol. 2012, 12, 33. [Google Scholar] [CrossRef] [PubMed]
- Chiaro, T.R.; Soto, R.; Zac Stephens, W.; Kubinak, J.L.; Petersen, C.; Gogokhia, L.; Bell, R.; Delgado, J.C.; Cox, J.; Voth, W.; et al. A member of the gut mycobiota modulates host purine metabolism exacerbating colitis in mice. Sci. Transl. Med. 2017, 9, eaaf9044. [Google Scholar] [CrossRef] [PubMed]
- Liguori, G.; Lamas, B.; Richard, M.L.; Brandi, G.; da Costa, G.; Hoffmann, T.W.; Di Simone, M.P.; Calabrese, C.; Poggioli, G.; Langella, P.; et al. Fungal Dysbiosis in Mucosa-associated Microbiota of Crohn’s Disease Patients. J. Crohn’s Colitis 2016, 10, 296–305. [Google Scholar] [CrossRef]
- Na, Y.R.; Stakenborg, M.; Seok, S.H.; Matteoli, G. Macrophages in intestinal inflammation and resolution: A potential therapeutic target in IBD. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 531–543. [Google Scholar] [CrossRef]
- Snyder, R.J.; Lantis, J.; Kirsner, R.S.; Shah, V.; Molyneaux, M.; Carter, M.J. Macrophages: A review of their role in wound healing and their therapeutic use. Wound Repair Regen. 2016, 24, 613–629. [Google Scholar] [CrossRef]
- Chikina, A.S.; Nadalin, F.; Maurin, M.; San-Roman, M.; Thomas-Bonafos, T.; Li, X.V.; Lameiras, S.; Baulande, S.; Henri, S.; Malissen, B.; et al. Macrophages Maintain Epithelium Integrity by Limiting Fungal Product Absorption. Cell 2020, 183, 411–428.e16. [Google Scholar] [CrossRef]
- Gerard, R.; Sendid, B.; Colombel, J.F.; Poulain, D.; Jouault, T. An immunological link between Candida albicans colonization and Crohn’s disease. Crit. Rev. Microbiol. 2015, 41, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Iliev, I.D.; Funari, V.A.; Taylor, K.D.; Nguyen, Q.; Reyes, C.N.; Strom, S.P.; Brown, J.; Becker, C.A.; Fleshner, P.R.; Dubinsky, M.; et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science 2012, 336, 1314–1317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leonardi, I.; Li, X.; Semon, A.; Li, D.; Doron, I.; Putzel, G.; Bar, A.; Prieto, D.; Rescigno, M.; McGovern, D.P.B.; et al. CX3CR1+ mononuclear phagocytes control immunity to intestinal fungi. Science 2018, 359, 232–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurtzman, C.P.; Fell, J.W.; Boekhout, T. The Yeasts: A Taxonomic Study; Elsevier: Amsterdam, The Netherlands, 2011; Volume 2. [Google Scholar]
- Botschuijver, S.; Roeselers, G.; Levin, E.; Jonkers, D.M.; Welting, O.; Heinsbroek, S.E.M.; de Weerd, H.H.; Boekhout, T.; Fornai, M.; Masclee, A.A.; et al. Intestinal Fungal Dysbiosis Is Associated With Visceral Hypersensitivity in Patients With Irritable Bowel Syndrome and Rats. Gastroenterology 2017, 153, 1026–1039. [Google Scholar] [CrossRef]
- Leonardi, I.; Li, X.; Iliev, I.D. Macrophage interactions with fungi and bacteria in inflammatory bowel disease. Curr. Opin. Gastroenterol. 2018, 34, 392–397. [Google Scholar] [CrossRef] [PubMed]
- Saraiva, M.; Vieira, P.; O’Garra, A. Biology and therapeutic potential of interleukin-10. J. Exp. Med. 2020, 217, e20190418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernardo, D.; Marin, A.C.; Fernández-Tomé, S.; Montalban-Arques, A.; Carrasco, A.; Tristán, E.; Ortega-Moreno, L.; Mora-Gutiérrez, I.; Díaz-Guerra, A.; Caminero-Fernández, R.; et al. Human intestinal pro-inflammatory CD11chighCCR2+CX3CR1+ macrophages, but not their tolerogenic CD11c−CCR2−CX3CR1− counterparts, are expanded in inflammatory bowel disease. Mucosal Immunol. 2018, 11, 1114–1126. [Google Scholar] [CrossRef]

| Responders | Non-Responders | |
|---|---|---|
| n = 9 | n = 22 | |
| FMT-D, n (%) | 4 (44) | 7 (32) |
| Mean age, years (SD) | 46 (12.7) | 41 (11.7) |
| Male sex, n (%) | 3 (33) | 11 (50) |
| Median disease duration, years (IQR) | 7 (20) | 8 (12) |
| Extent of disease, n (%) | ||
| E1, proctitis | 0 (0) | 0 (0) |
| E2, left-sided | 3 (33) | 10 (45) |
| E3, pancolitis | 6 (67) | 12 (55) |
| Concomitant drug treatment, n (%) | 6 (67) | 17 (77) |
| Mesalamine oral | 6 (67) | 14 (64) |
| Mesalamine rectal | 0 (0) | 1 (5) |
| Thiopurines | 0 (0) | 1 (5) |
| Systemic corticosteroids (<10 mg) | 0 (0) | 1 (5) |
| Prior anti-TNF therapy, n (%) | 0 (0) | 1 (5) |
| Median SCCAI score at inclusion (IQR) | 8 (5) | 8 (3) |
| Mayo endoscopic score at inclusion, n (%) | ||
| Mayo 1 | 2 (22) | 2 (9) |
| Mayo 2 | 6 (67) | 11 (50) |
| Mayo 3 | 1 (11) | 9 (41) |
| Site of disease at inclusion, n (%) | ||
| Rectum only | 1 (11) | 3 (14) |
| Left side of colon | 7 (78) | 13 (59) |
| Pancolitis | 1 (11) | 6 (27) |
| Gene Name | Gene Symbol | 5′-Forward Sequence | 5′-Reverse Sequence |
|---|---|---|---|
| Claudin-1 | CLDN1 | GGCAGATCCAGTGCAAAGTC | TCACTCCCAGGAGGATGC |
| Defensin Beta 1 | DEFB1 | CGCCATGAGAACTTCCTACC | CCACTGCTGACGCAATTGTA |
| Defensin Alpha 5 | DEFA5 | AAGCAGTCTGGGGAAGACAA | CTAGGAAGCTCAGCGACAGC |
| Interleukin 1β | IL1B | ACCAAACCTCTTCGAGGCAC | AGCCATCATTTCACTGGCGA |
| Interleukin 6 | IL6 | AGTGAGGAACAAGCCAGAGC | GTCAGGGGTGGTTATTGCAT |
| Interleukin 8 | IL8 | AAATTTGGGGTGGAAAGGTT | TCCTGATTTCTGCAGCTCTGT |
| Interleukin 10 | IL10 | GCCACCCTGATGTCTCAGTT | GTGGAGCAGGTGAAGAATGC |
| Occludin | OCLN | TTTGTGGGACAAGGAACACA | ATGCCATGGGACTGTCAACT |
| ZO-1 | TJP1 | AGAGCACAGCAATGGAGGAA | GACGTTTCCCCACTCTGAAA |
| Tumor Necrosis Factor Alpha | TNF | CCTGCTGCACTTTGGAGTGA | GAGGGTTTGCTACAACATGGG |
| Occludin | OCLN | TTTGTGGGACAAGGAACACA | ATGCCATGGGACTGTCAACT |
| Glyceraldehyde-30-Phosphate Dehydrogenase | GAPDH | GTCAGTGGTGGACCTGACCT | TGAGCTTGACAAAGTGGTCG |
| Hypoxanthine Phosphoribosyltransferase 1 | HPRT | CCTGGCGTCGTGATTAGTGAT | AGACGTTCAGTCCTGTCCATAA |
| Peptidylprolyl Isomerase A | PPIA | ACGGCGAGCCCTTGG | TTTCTGCTGTCTTTGGGACCT |
| Proteasome 20S Subunit Beta 6 | PSMB6 | ACCTGATGGCGGGAATCAT | ATCATACCCCCCATAGGCACT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
van Thiel, I.A.M.; Rahman, S.; Hakvoort, T.B.M.; Davids, M.; Verseijden, C.; van Hamersveld, P.H.P.; Bénard, M.V.; Lodders, M.H.; Boekhout, T.; van den Wijngaard, R.M.; et al. Fecal Filobasidium Is Associated with Clinical Remission and Endoscopic Response following Fecal Microbiota Transplantation in Mild-to-Moderate Ulcerative Colitis. Microorganisms 2022, 10, 737. https://doi.org/10.3390/microorganisms10040737
van Thiel IAM, Rahman S, Hakvoort TBM, Davids M, Verseijden C, van Hamersveld PHP, Bénard MV, Lodders MH, Boekhout T, van den Wijngaard RM, et al. Fecal Filobasidium Is Associated with Clinical Remission and Endoscopic Response following Fecal Microbiota Transplantation in Mild-to-Moderate Ulcerative Colitis. Microorganisms. 2022; 10(4):737. https://doi.org/10.3390/microorganisms10040737
Chicago/Turabian Stylevan Thiel, Isabelle A. M., Shafaque Rahman, Theodorus B. M. Hakvoort, Mark Davids, Caroline Verseijden, Patricia H. P. van Hamersveld, Mèlanie V. Bénard, Maarten H. Lodders, Teun Boekhout, René M. van den Wijngaard, and et al. 2022. "Fecal Filobasidium Is Associated with Clinical Remission and Endoscopic Response following Fecal Microbiota Transplantation in Mild-to-Moderate Ulcerative Colitis" Microorganisms 10, no. 4: 737. https://doi.org/10.3390/microorganisms10040737
APA Stylevan Thiel, I. A. M., Rahman, S., Hakvoort, T. B. M., Davids, M., Verseijden, C., van Hamersveld, P. H. P., Bénard, M. V., Lodders, M. H., Boekhout, T., van den Wijngaard, R. M., Heinsbroek, S. E. M., Ponsioen, C. Y., & de Jonge, W. J. (2022). Fecal Filobasidium Is Associated with Clinical Remission and Endoscopic Response following Fecal Microbiota Transplantation in Mild-to-Moderate Ulcerative Colitis. Microorganisms, 10(4), 737. https://doi.org/10.3390/microorganisms10040737







