Torque Teno Sus Virus (TTSuV) Prevalence in Wild Fauna of Northern Italy
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Sample Preparation and DNA Extraction
2.3. Typing PCR
2.4. Sequencing
2.5. Statistical Analysis
2.6. Phylogenetic Analysis
3. Results
3.1. TTSuV Prevalence in Wild Ungulates and Associated Risk Factors
3.2. TTSuV1a and TTSuVk2a Prevalence
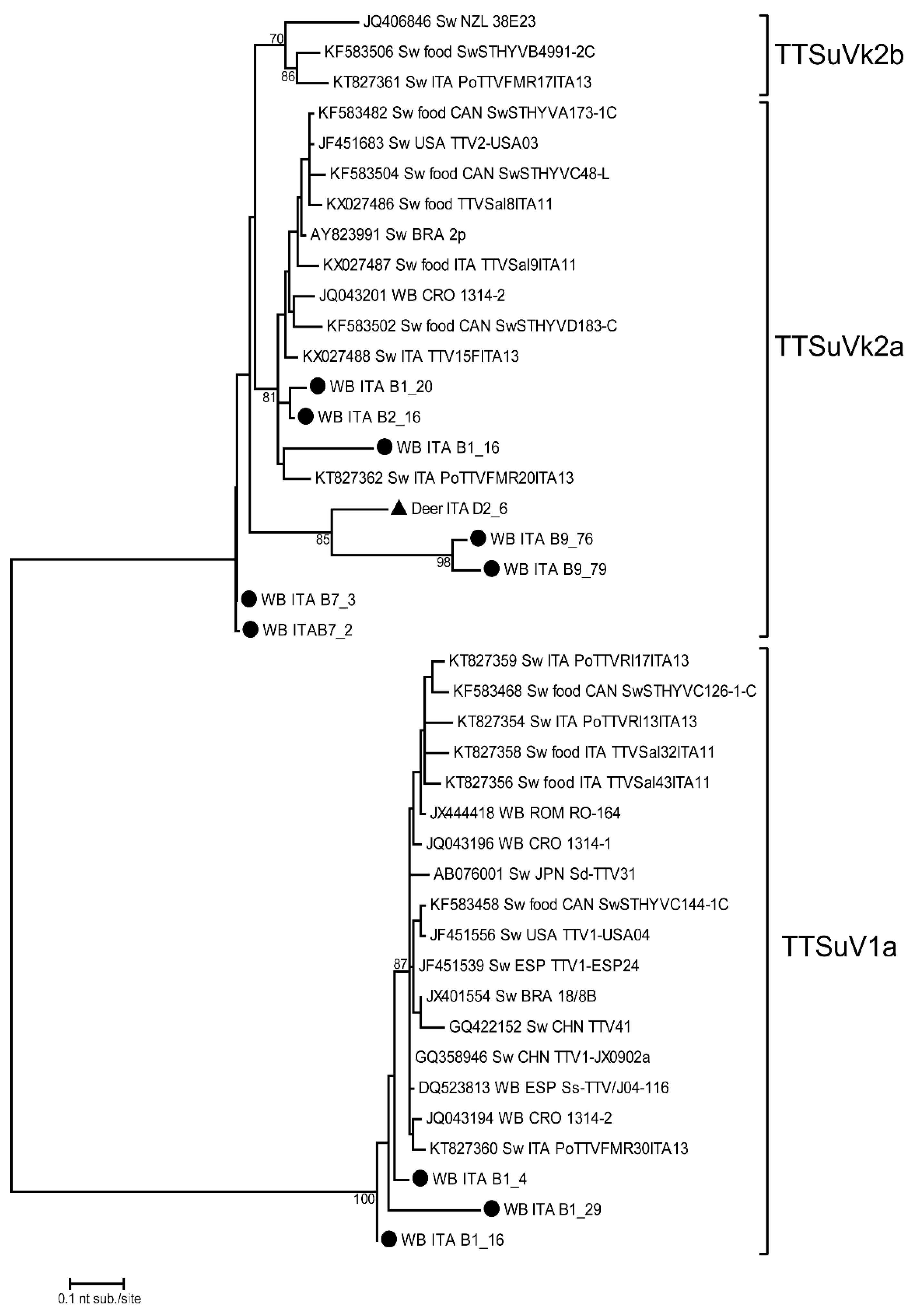
3.3. Phylogenetic Tree
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gallian, P.; Biagini, P.; Zhong, S.; Touinssi, M.; Yeo, W.; Cantaloube, J.F.; Attoui, H.; de Micco, P.; Johnson, P.J.; de Lamballerie, X. TT Virus: A Study of Molecular Epidemiology and Transmission of Genotypes 1, 2 and 3. J. Clin. Virol. 2000, 17, 43–49. [Google Scholar] [CrossRef]
- International Committee on Taxonomy of Viruses. Available online: https://talk.ictvonline.org/ictv-reports/ictv_9th_report/ssdna-viruses-2011/w/ssdna_viruses/139/anelloviridae (accessed on 4 December 2021).
- Nishizawa, T.; Okamoto, H.; Konishi, K.; Yoshizawa, H.; Miyakawa, Y.; Mayumi, M. A Novel DNA Virus (TTV) Associated with Elevated Transaminase Levels in Posttransfusion Hepatitis of Unknown Etiology. Biochem. Biophys. Res. Commun. 1997, 241, 92–97. [Google Scholar] [CrossRef]
- Prasetyo, A.A.; Desyardi, M.N.; Tanamas, J.; Suradi; Reviono; Harsini; Kageyama, S.; Chikumi, H.; Shimizu, E. Respiratory Viruses and Torque Teno Virus in Adults with Acute Respiratory Infections. Intervirology 2015, 58, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Brassard, J.; Gagné, M.-J.; Leblanc, D.; Poitras, É.; Houde, A.; Boras, V.F.; Inglis, G.D. Association of Age and Gender with Torque Teno Virus Detection in Stools from Diarrheic and Non-Diarrheic People. J. Clin. Virol. 2015, 72, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Kasirga, E.; Sanlidag, T.; Akcali, S.; Keskin, S.; Aktas, E.; Karakoç, Z.; Helvaci, M.; Sözen, G.; Kuzu, M. Clinical Significance of TT Virus Infection in Children with Chronic Hepatitis B. Pediatrics Int. Off. J. Jpn. Pediatric Society 2005, 47, 300–304. [Google Scholar] [CrossRef] [PubMed]
- Gergely, P.; Perl, A.; Poór, G. Possible Pathogenic Nature of the Recently Discovered TT Virus: Does It Play a Role in Autoimmune Rheumatic Diseases? Autoimmun. Rev. 2006, 6, 5–9. [Google Scholar] [CrossRef]
- Spandole, S.; Cimponeriu, D.; Berca, L.M.; Mihăescu, G. Human Anelloviruses: An Update of Molecular, Epidemiological and Clinical Aspects. Arch. Virol. 2015, 160, 893–908. [Google Scholar] [CrossRef] [PubMed]
- Leary, T.P.; Erker, J.C.; Chalmers, M.L.; Desai, S.M.; Mushahwar, I.K. Improved Detection Systems for TT Virus Reveal High Prevalence in Humans, Non-Human Primates and Farm Animals. J. Gen. Virol. 1999, 80(Pt. 8), 2115–2120. [Google Scholar] [CrossRef]
- Cong, M.; Nichols, B.; Dou, X.; Spelbring, J.E.; Krawczynski, K.; Fields, H.A.; Khudyakov, Y.E. Related TT Viruses in Chimpanzees. Virology 2000, 274, 343–355. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Okamoto, H.; Takahashi, M.; Nishizawa, T.; Tawara, A.; Fukai, K.; Muramatsu, U.; Naito, Y.; Yoshikawa, A. Genomic Characterization of TT Viruses (TTVs) in Pigs, Cats and Dogs and Their Relatedness with Species-Specific TTVs in Primates and Tupaias. J. Gen. Virol. 2002, 83(Pt. 6), 1291–1297. [Google Scholar] [CrossRef]
- Martínez, L.; Kekarainen, T.; Sibila, M.; Ruiz-Fons, F.; Vidal, D.; Gortázar, C.; Segalés, J. Torque Teno Virus (TTV) Is Highly Prevalent in the European Wild Boar (Sus Scrofa). Vet. Microbiol. 2006, 118, 223–229. [Google Scholar] [CrossRef]
- Brassard, J.; Gagné, M.-J.; Lamoureux, L.; Inglis, G.D.; Leblanc, D.; Houde, A. Molecular Detection of Bovine and Porcine Torque Teno Virus in Plasma and Feces. Vet. Microbiol. 2008, 126, 271–276. [Google Scholar] [CrossRef]
- Águeda-Pinto, A.; Kraberger, S.; Lund, M.C.; Gortázar, C.; McFadden, G.; Varsani, A.; Esteves, P.J. Coinfections of Novel Polyomavirus, Anelloviruses and a Recombinant Strain of Myxoma Virus-MYXV-Tol Identified in Iberian Hares. Viruses 2020, 12, 340. [Google Scholar] [CrossRef]
- Ssemadaali, M.A.; Effertz, K.; Singh, P.; Kolyvushko, O.; Ramamoorthy, S. Identification of Heterologous Torque Teno Viruses in Humans and Swine. Sci. Rep. 2016, 6, 26655. [Google Scholar] [CrossRef]
- Varsani, A.; Opriessnig, T.; Celer, V.; Maggi, F.; Okamoto, H.; Blomström, A.L.; Cadar, D.; Harrach, B.; Biagini, P.; Kraberger, S. Taxonomic update for mammalian anelloviruses (family Anelloviridae). Arch. Virol. 2021, 166, 2943–2953. [Google Scholar] [CrossRef] [PubMed]
- Manzin, A.; Mallus, F.; Macera, L.; Maggi, F.; Blois, S. Global Impact of Torque Teno Virus Infection in Wild and Domesticated Animals. J. Infect. Dev. Ctries. 2015, 9, 562–570. [Google Scholar] [CrossRef]
- Liu, J.; Guo, L.; Zhang, L.; Wei, Y.; Huang, L.; Wu, H.; Liu, C. Three New Emerging Subgroups of Torque Teno Sus Viruses (TTSuVs) and Co-Infection of TTSuVs with Porcine Circovirus Type 2 in China. Virol. J. 2013, 10, 189. [Google Scholar] [CrossRef]
- Cortey, M.; Pileri, E.; Segalés, J.; Kekarainen, T. Globalisation and Global Trade Influence Molecular Viral Population Genetics of Torque Teno Sus Viruses 1 and 2 in Pigs. Vet. Microbiol. 2012, 156, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Krakowka, S.; Ellis, J.A. Evaluation of the Effects of Porcine Genogroup 1 Torque Teno Virus in Gnotobiotic Swine. Am. J. Vet. Res. 2008, 69, 1623–1629. [Google Scholar] [CrossRef]
- Krakowka, S.; Hartunian, C.; Hamberg, A.; Shoup, D.; Rings, M.; Zhang, Y.; Allan, G.; Ellis, J.A. Evaluation of Induction of Porcine Dermatitis and Nephropathy Syndrome in Gnotobiotic Pigs with Negative Results for Porcine Circovirus Type 2. Am. J. Vet. Res. 2008, 69, 1615–1622. [Google Scholar] [CrossRef] [PubMed]
- Ellis, J.A.; Allan, G.; Krakowka, S. Effect of Coinfection with Genogroup 1 Porcine Torque Teno Virus on Porcine Circovirus Type 2-Associated Postweaning Multisystemic Wasting Syndrome in Gnotobiotic Pigs. Am. J. Vet. Res. 2008, 69, 1608–1614. [Google Scholar] [CrossRef]
- Li, G.; Wang, R.; Cai, Y.; Zhang, J.; Zhao, W.; Gao, Q.; Franzo, G.; Su, S. Epidemiology and Evolutionary Analysis of Torque Teno Sus Virus. Vet. Microbiol. 2020, 244, 108668. [Google Scholar] [CrossRef] [PubMed]
- Sibila, M.; Martínez-Guinó, L.; Huerta, E.; Llorens, A.; Mora, M.; Grau-Roma, L.; Kekarainen, T.; Segalés, J. Swine Torque Teno Virus (TTV) Infection and Excretion Dynamics in Conventional Pig Farms. Vet. Microbiol. 2009, 139, 213–218. [Google Scholar] [CrossRef]
- Aramouni, M.; Segalés, J.; Cortey, M.; Kekarainen, T. Age-Related Tissue Distribution of Swine Torque Teno Sus Virus 1 and 2. Vet. Microbiol. 2010, 146, 350–353. [Google Scholar] [CrossRef]
- Gallei, A.; Pesch, S.; Esking, W.S.; Keller, C.; Ohlinger, V.F. Porcine Torque Teno Virus: Determination of Viral Genomic Loads by Genogroup-Specific Multiplex Rt-PCR, Detection of Frequent Multiple Infections with Genogroups 1 or 2, and Establishment of Viral Full-Length Sequences. Vet. Microbiol. 2010, 143, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Cadar, D.; Kiss, T.; Ádám, D.; Cságola, A.; Novosel, D.; Tuboly, T. Phylogeny, Spatio-Temporal Phylodynamics and Evolutionary Scenario of Torque Teno Sus Virus 1 (TTSuV1) and 2 (TTSuV2) in Wild Boars: Fast Dispersal and High Genetic Diversity. Vet. Microbiol. 2013, 166, 200–213. [Google Scholar] [CrossRef] [PubMed]
- Blois, S.; Mallus, F.; Liciardi, M.; Pilo, C.; Camboni, T.; Macera, L.; Maggi, F.; Manzin, A. High Prevalence of Co-Infection with Multiple Torque Teno Sus Virus Species in Italian Pig Herds. PLoS ONE 2014, 9, e113720. [Google Scholar] [CrossRef]
- Martelli, F.; Caprioli, A.; Di Bartolo, I.; Cibin, V.; Pezzotti, G.; Ruggeri, F.M.; Ostanello, F. Detection of Swine Torque Teno Virus in Italian Pig Herds. J. Vet. Med. B Infect. Dis. Vet. Public Health 2006, 53, 234–238. [Google Scholar] [CrossRef]
- Monini, M.; Vignolo, E.; Ianiro, G.; Ostanello, F.; Ruggeri, F.M.; Di Bartolo, I. Detection of Torque Teno Sus Virus in Pork Bile and Liver Sausages. Food Environ. Virol. 2016, 8, 283–288. [Google Scholar] [CrossRef]
- Cicognani, L.; Quesada, P.M.; Monti, F.; Gellini, S.; Baldassarri, F. Preliminary Data on the Density and Structure of a Fallow Deer (Cervus Dama) Population in the Foreste Casentinesi M. Falterona and Campigna National Park. Hystrix It. J. Mamm. 2000, 11. [Google Scholar] [CrossRef]
- Istituto Superiore per la Protezione e la Ricerca Ambientale. Available online: https://www.isprambiente.gov.it/it (accessed on 1 December 2020).
- Engeman, R.M.; Massei, G.; Sage, M.; Gentle, M.N. Monitoring Wild Pig Populations: A Review of Methods. Environ. Sci. Pollut. Res. Int. 2013, 20, 8077–8091. [Google Scholar] [CrossRef] [PubMed]
- Segalés, J.; Martínez-Guinó, L.; Cortey, M.; Navarro, N.; Huerta, E.; Sibila, M.; Pujols, J.; Kekarainen, T. Retrospective Study on Swine Torque Teno Virus Genogroups 1 and 2 Infection from 1985 to 2005 in Spain. Vet. Microbiol. 2009, 134, 199–207. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- MEGA Software. Available online: https://www.megasoftware.net (accessed on 27 July 2021).
- R Statistical Software. R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.R-project.org (accessed on 8 February 2021).
- Ramos, N.; Mirazo, S.; Botto, G.; Teixeira, T.F.; Cibulski, S.P.; Castro, G.; Cabrera, K.; Roehe, P.M.; Arbiza, J. High Frequency and Extensive Genetic Heterogeneity of TTSuV1 and TTSuVk2a in PCV2- Infected and Non-Infected Domestic Pigs and Wild Boars from Uruguay. Vet. Microbiol. 2018, 224, 78–87. [Google Scholar] [CrossRef]
- Luka, P.D.; Erume, J.; Yakubu, B.; Owolodun, O.A.; Shamaki, D.; Mwiine, F.N. Molecular Detection of Torque Teno Sus Virus and Coinfection with African Swine Fever Virus in Blood Samples of Pigs from Some Slaughterhouses in Nigeria. Adv. Virol. 2016, 2016, 6341015. [Google Scholar] [CrossRef]
- Arnaboldi, S.; Righi, F.; Carta, V.; Bonardi, S.; Pavoni, E.; Bianchi, A.; Losio, M.N.; Filipello, V. Hepatitis E Virus (HEV) Spread and Genetic Diversity in Game Animals in Northern Italy. Food Environ. Virol. 2021, 13, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Webb, B.; Rakibuzzaman, A.; Ramamoorthy, S. Torque Teno Viruses in Health and Disease. Virus Res. 2020, 285, 198013. [Google Scholar] [CrossRef]
- Chiari, M.; Zanoni, M.; Tagliabue, S.; Lavazza, A.; Alborali, L.G. Salmonella Serotypes in Wild Boars (Sus Scrofa) Hunted in Northern Italy. Acta Vet. Scand. 2013, 55, 42. [Google Scholar] [CrossRef]
- Zhang, B.; Tang, C.; Yue, H.; Ren, Y.; Song, Z. Viral Metagenomics Analysis Demonstrates the Diversity of Viral Flora in Piglet Diarrhoeic Faeces in China. J. Gen. Virol. 2014, 95(Pt. 7), 1603–1611. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Li, L.; Deng, X.; Kapusinszky, B.; Delwart, E. What Is for Dinner? Viral Metagenomics of US Store Bought Beef, Pork, and Chicken. Virology 2014, 468–470, 303–310. [Google Scholar] [CrossRef] [PubMed]
| Sampling Area | Gender | Hunting Season | Age Class 1 | Total Samples | ||
|---|---|---|---|---|---|---|
| 0 | 1 | 2 | ||||
| SO | Male | 2018/2019 | 4 | 17 | 15 | 142 |
| 2019/2020 | 22 | 57 | 27 | |||
| Female | 2018/2019 | 5 | 11 | 12 | 83 | |
| 2019/2020 | 8 | 30 | 17 | |||
| Total samples | 39 | 115 | 71 | 225 | ||
| PR | Male | 2018/2019 | 1 | 17 | 20 | 74 |
| 2019/2020 | 13 | 6 | 17 | |||
| Female | 2018/2019 | 1 | 23 (6) | 43 (32) | 101 (38) | |
| 2019/2020 | 6 | 9 | 19 | |||
| Total samples | 21 | 55 | 99 | 175 | ||
| Gender | Hunting Season | Age Class 1 | Total Samples | ||
|---|---|---|---|---|---|
| 0 | 1 | 2 | |||
| Male | 2016/2017 | 7 | 10 | 4 | 53 |
| 2017/2018 | 6 | 4 | 9 | ||
| 2018/2019 | 7 | 0 | 6 | ||
| Female | 2016/2017 | 9 | 4 | 24 | 75 |
| 2017/2018 | 3 | 4 | 6 | ||
| 2018/2019 | 4 | 4 | 17 | ||
| Total samples | 36 | 26 | 66 | 128 | |
| Target | Type | Sequence | Position | Amplicon Size (bp) |
|---|---|---|---|---|
| TTSuV1a | Forward | 5′-CGGGTTCAGGAGGCTCAAT-3′ | 8–26 | 305 |
| Reverse | 5′-GCCATTCGGAACTGCACTTACT-3′ | 291–312 | ||
| TTSuVk2a | Forward | 5′-TCATGACAGGGTTCACCGGA-3′ | 1–20 | 252 |
| Reverse | 5′-CGTCTGCGCACTTACTTATATACTCTA-3′ | 226–252 |
| Wild Ungulates (n = Samples) | TTSuV Prevalence (95% CI, n = Positive Samples) |
|---|---|
| Overall (n = 528) | 5.7% (3.9–8.0%, n = 30) |
| Species | |
| Wild ruminants (n = 128) | 9.4% (4.9–15.8%, n = 12) |
| Wild boars (n = 400) | 4.5% (2.7–7.0%, n = 18) |
| Gender | |
| Female (n = 259) | 4.2% (2.1–7.5%, n = 11) |
| Male (n = 269) | 7.1% (4.3–10.8%, n = 19) |
| Age class 1 | |
| 0 (n = 96) | 8.3% (3.7–15.8%, n = 8) |
| 1 (n = 196) | 5.1% (2.5–9.2%, n = 10) |
| 2 (n = 236) | 5.1% (2.6–8.7%, n = 12) |
| Sampling area | |
| Parma (n = 175) | 2.3% (0.6–5.7%, n = 4) |
| Sondrio (n = 353) | 7.4% (4.9–10.6%, n = 26) |
| Hunting season | |
| 2016–2017 (n = 122) | 13.9% (8.3–21.4%, n = 17) |
| 2017–2018 (n = 32) | 9.4% (2.0–25.0%, n = 3) |
| 2018–2019 (n = 225) | 2.7% (1.0–5.7%, n = 6) |
| 2019–2020 (n = 149) | 2.7% (0.7–6.7%, n = 4) |
| Wild Boars (n = Samples) | TTSuV1a Prevalence (95%CI, n = Positive Samples) | TTSuVk2a Prevalence (95%CI, n = Positive Samples) |
|---|---|---|
| Overall (n = 400) | 2.7% (1.4–4.9%, n = 11) | 2.5% (1.2–4.5%, n = 10) |
| Gender | ||
| Males (n = 216) | 3.2% (1.3–6.6%, n = 7) | 3.7% (1.6–7.2%, n = 8) |
| Females (n = 184) | 2.2% (0.6–5.5%, n = 4) | 1.1% (0.1–3.9%, n = 2) |
| Age class 1 | ||
| 0 (n = 60) | 1.7% (0–8.9%, n = 1) | 5.0% (1.0–13.9%, n = 3) |
| 1 (n = 170) | 2.9% (1.0–6.7%, n = 5) | 2.9% (1.0–6.7%, n = 5) |
| 2 (n = 170) | 2.9% (1.0–6.7%, n = 5) | 1.2% (0.1–4.2%, n = 2) |
| Hunting season | ||
| 2018–2019 (n = 169) | 5.3% (2.8–9.8%, n = 9) | 4.1% (2.0–8.3%, n = 7) |
| 2019–2020 (n = 231) | 0.9% (0.2–3.1%, n = 2) | 1.3% (0.4–3.7%, n = 3) |
| Sampling area | ||
| Sondrio (n = 225) | 3.6% (1.5–6.9%, n = 8) | 3.6% (1. 5–6.9%, n = 8) |
| Parma (n = 175) | 1.7% (0.3–4.9%, n = 3) | 1.1% (0.1–4.1%, n = 2) |
| Wild Ruminants (n = Samples) | TTSuVk2a Prevalence (95% CI, n = Positive Samples) |
|---|---|
| Overall (n = 128) | 9.4% (4.9–15.8%, n = 12) |
| Gender | |
| Males (n = 53) | 11.3% (4.3–23.0%, n = 6) |
| Females (n = 75) | 8.0% (3.0–16.6%, n = 6) |
| Age class 1 | |
| 0 (n = 36) | 11.1% (3.1–26.1%, n = 4) |
| 1 (n = 26) | 7.7% (0.9–25.1%, n = 2) |
| 2 (n = 66) | 9.1% (3.4–18.7%, n = 6) |
| Hunting season | |
| 2016–2017 (n = 58) | 13.8% (6.1–25.4%, n = 8) |
| 2017–2018 (n = 32) | 9.4% (2.0–25.0%, n = 3) |
| 2018–2019 (n = 38) | 2.6% (0.1–13.8%, n = 1) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Righi, F.; Arnaboldi, S.; Filipello, V.; Ianiro, G.; Di Bartolo, I.; Calò, S.; Bellini, S.; Trogu, T.; Lelli, D.; Bianchi, A.; et al. Torque Teno Sus Virus (TTSuV) Prevalence in Wild Fauna of Northern Italy. Microorganisms 2022, 10, 242. https://doi.org/10.3390/microorganisms10020242
Righi F, Arnaboldi S, Filipello V, Ianiro G, Di Bartolo I, Calò S, Bellini S, Trogu T, Lelli D, Bianchi A, et al. Torque Teno Sus Virus (TTSuV) Prevalence in Wild Fauna of Northern Italy. Microorganisms. 2022; 10(2):242. https://doi.org/10.3390/microorganisms10020242
Chicago/Turabian StyleRighi, Francesco, Sara Arnaboldi, Virginia Filipello, Giovanni Ianiro, Ilaria Di Bartolo, Stefania Calò, Silvia Bellini, Tiziana Trogu, Davide Lelli, Alessandro Bianchi, and et al. 2022. "Torque Teno Sus Virus (TTSuV) Prevalence in Wild Fauna of Northern Italy" Microorganisms 10, no. 2: 242. https://doi.org/10.3390/microorganisms10020242
APA StyleRighi, F., Arnaboldi, S., Filipello, V., Ianiro, G., Di Bartolo, I., Calò, S., Bellini, S., Trogu, T., Lelli, D., Bianchi, A., Bonardi, S., Pavoni, E., Bertasi, B., & Lavazza, A. (2022). Torque Teno Sus Virus (TTSuV) Prevalence in Wild Fauna of Northern Italy. Microorganisms, 10(2), 242. https://doi.org/10.3390/microorganisms10020242








