Abstract
Thermophiles that produce extracellular hydrolases are of great importance due to their applications in various industries. Thermophilic enzymes are of interest for industrial applications due to their compatibility with industrial processes, and the availability of the organisms is essential to develop their full potential. In this study, a culture-dependent approach was used to identify thermophilic bacteria from five hot springs in Republic of Korea. Characterization, taxonomic identification, and extracellular hydrolase (amylase, lipase, and protease) activity of 29 thermophilic bacterial isolates from the Neungam carbonate, Mungang sulfur, Deokgu, Baegam, and Dongnae hot springs were investigated. Identification based on the full-length 16S rRNA gene sequence revealed that strains belonged to the phylum Bacillota and were classified as Aeribacillus, Bacillus, Caldibacillus, Geobacillus, and Thermoactinomyces genera. It was found that 22 isolates could produce at least one extracellular enzyme. Geobacillus, representing 41.4% of the isolates, was the most abundant. The highest amount of proteolytic and lipolytic enzymes was secreted by strains of the genus Geobacillus, whereas Caldibacillus species produced the highest amount of amylolytic enzyme. The Geobacillus species producing hydrolytic extracellular enzymes appeared to be the most promising.
1. Introduction
Extremophiles, including thermophilic, psychrophilic, acidophilic, alkaliphilic, and halophilic microorganisms, have been studied in various fields because of their capacity to survive and function under harsh environments [1]. Among the extremophiles, thermophilic microorganisms have been generally isolated from hot springs and the deep sea of the Earth. Thermophiles are divided into moderate thermophiles (optimum growth temperature 50–60 °C), extreme thermophiles (optimum growth temperature 60–80 °C), and hyperthermophiles (optimum growth temperature 80–110 °C) [2]. They are the reservoirs for production of industrially valuable thermostable enzymes that can be used for biotechnological processes at high temperatures [3,4,5,6].
Thermostable bacterial hydrolytic enzymes have been developed and used extensively in biological processes for production, meaning that thermostability makes them highly demandable for eco-friendly industrial purposes, and extensive manufacturing and usage of thermostable enzymes has resulted from continued industrial demand [7,8,9]. Among the commercially valuable enzymes, α-amylases, lipases, and proteases are the major enzymes produced by thermophiles [7,10]. Their importance in biotechnological applications is significant; for example, proteases and amylases are used together in the food industry, detergent industry, and pharmaceuticals [11]. Due to their increased importance, many scientists have focused on their research to discover new thermophilic microorganisms [12,13,14].
Republic of Korea’s Hot Spring Act defines a hot spring as groundwater with temperature of 25 °C or higher with qualified standards. It is difficult to find volcanic-oriented hot springs as the Korean peninsula is known to be distant from high enthalpy geothermal energy. The area of distribution of the hot springs coincides with areas of granite, and the heat is derived from the process of radioactive decay of the granite. Hot springs with temperatures ranging from 25 °C to 78 °C are found in different regions of Republic of Korea. The low-temperature hot springs demonstrate 85.4%. The physical and chemical characteristics of Republic of Korean hot springs are the low proportion of high temperature, low mineral contents, and alkaline pH [15,16,17]. The physiochemical features of hot spring water facilitate growth of thermophiles. Consequently, research on assessment of the possibility of industrial application by isolation of thermophiles from hot spring water in Republic of Korea is necessary. This study aims to develop a continuous approach for screening, isolating, and characterizing novel thermophilic microorganisms with great biotechnological and environmental potential.
2. Materials and Methods
2.1. Sample Collection
Five hot springs in Republic of Korea were chosen to collect the sample, including the Neungam carbonate, Mungang sulfur, Deokgu, Baegam, and Dongnae Yangtangjang. The samples were collected using a pump in a sterilized sampling bottle and immediately screened for bacterial isolation. Additionally, after screening, samples were stored at 4 °C for up to 5 days.
2.2. Isolation of Thermophilic Bacteria
The hot spring water samples were serially diluted using a sterile 0.85% saline buffer. An aliquot of each suspension was spread on marine broth 2216 (MB; BD Difco) supplemented with 1.5% (w/v) agar (MA) plates and incubated at 60 °C for five days. Morphologically different colonies were selected, and, after several transfers using the same solid medium, pure colonies were obtained. Obtained isolates were suspended in marine broth 2216 supplemented with 10% (v/v) dimethyl sulfoxide (DMSO) and stored at −80 °C for long-time preservation.
2.3. Taxonomic Identification of the Isolates
The isolated strains were identified by 16S rRNA gene sequencing. The 16S rRNA identification was performed by the BIOFACT Co., Ltd. (Daejeon, Republic of Korea) using ABI PRISM 3730XL DNA analyzer (Applied Biosystems, Foster City, CA, USA). Genomic DNA of the strains was isolated using Chelex 100 Boiling Resin (BIO-RAD, Hercules, CA, USA), pro-K, and S-Taq Buffer (BIOFACT, Daejeon, Republic of Korea), followed by PCR amplification of the 16S rRNA gene by Hush Run™ cycler (BIOFACT, Daejeon, Republic of Korea) using universal primers 27F (5′-AGA GTT TGA TCC TGG CTC AG-3′) and 1492R (5′-GGT TAC CTT GTT ACG ACT T-3′). A 50 µL reaction mixture contained 5 U S-Taq DNA polymerase (BIOFACT, Daejeon, Republic of Korea), 10 mM dNTPs, and 10X S-Taq Reaction Buffer (BIOFACT, Daejeon, Republic of Korea). Identification based on the 16S rRNA sequence of the strains was analyzed using EzBioCloud Database (https://www.ezbiocloud.net/) accessed on 25 March 2022. Multiple sequence alignment was completed by the BioEdit program, and the phylogenetic tree was constructed using the Maximum Likelihood algorithm by the MEGA 6.0 program [18,19].
2.4. Determination of Growth Characteristics
The growth temperature range of the isolates was determined by cultivating at 45, 50, 55, 60, and 65 °C, respectively. The salt tolerance of the isolates was determined using MA medium supplemented with NaCl (w/v) concentration at 3, 6, 9, 12, and 15% incubating the plates at 60 °C. The pH tolerance was determined using MA medium with a pH range of 4.0, 7.0, and 9.0 adjusted with 1N NaOH and 1M HCl. The growth of the isolated bacteria at 60 °C on complex media was tested using nutrient agar (Difco, Sparks, MD, USA), R2A agar (Difco, Sparks, MD, USA), and tryptic soy agar (Difco, Sparks, MD, USA).
2.5. Determination of Hydrolytic Enzyme Activities
The hydrolytic enzyme activity of the isolated strains was tested on a marine agar medium supplemented with specific substrates. After several days of incubation at 60 °C, the enzyme activities were determined by the agar diffusion method. The amount of produced enzymes was determined as enzyme intensity (EI). EI was calculated as follows: (colony diameter + halo zone diameter)/colony diameter [20,21,22]. Each experiment was performed in triplicate.
2.5.1. Amylase Activity
The amylolytic property was identified using the starch hydrolysis method. The strains were inoculated onto marine agar supplemented with 0.2% (w/v) soluble starch (Difco, USA). After incubation for 3 days at 60 °C, agar plates were treated with 1% iodine solution for 2–3 min and presence of transparent zones around the colony indicated amylase activity [23].
2.5.2. Lipase Activity
Lipase activity was determined by the opaque zone around the colony as described by [24]. The strains were inoculated onto marine agar (Difco, Sparks, MD, USA) medium supplemented with 1% (v/v) Tween 80 (Sigma, St. Louis, MO, USA). Tween 80 was added separately after sterilization. After incubation at 60 °C for 3 days, lipase activity was evaluated by an opaque zone around the colony.
2.5.3. Protease Activity
Protease activity was detected by inoculating the strains onto 2% (w/v) skim milk agar (Difco, Sparks, MD, USA) medium. Skim milk was added separately after sterilization. After incubation at 60 °C for 3 days, protease activity was evaluated by clear zone around the colony [25].
2.6. Accession Numbers
The 16S rRNA gene sequences of strains were deposited to the GenBank/EMBL/DDBJ database.
2.7. Deposition of Strains
All strains isolated through this study were deposited in the Microbial Value Enhancement Project, Korea Research Institute of Bioscience and Biotechnology.
3. Results
3.1. Sample Characteristics
The samples from the five hot springs were collected in January 2017. The five hot spring sites were located in different cities in Republic of Korea (Figure 1), with a temperature range between 26 and 60 °C and pH 6.5–9.1 (Table 1).
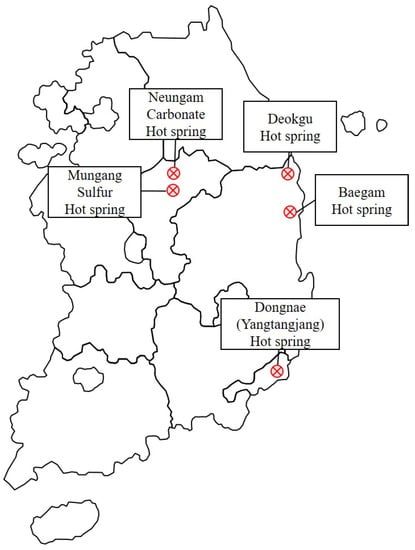
Figure 1.
Map of the hot spring area in Korea where the samples were collected for this study. Neungam and Mungang hot springs are located in Chungju, Deokgu and Baegam hot springs are located in Uljin, and Dongnae (Yangtangjang) hot spring is located in Busan, respectively.

Table 1.
Hot spring properties.
3.2. Isolation and Growth Characteristics of Thermophilic Bacteria
Within the scope of this study, a total of 29 strains were isolated from hot spring water samples. The screening results showed that three strains were isolated from the Neungam Carbonate, sixteen strains from the Mungang Sulfur, two strains were isolated from the Deokgu, six strains were separated from the Baegam, and two strains from the Dongnae hot springs, respectively. All the strains showed growth on one of the complex media (nutrient agar, R2A, tryptic soy agar), which are widely used for massive cultivation of bacteria in industrial processing, meaning that newly isolated 29 thermophilic bacteria can be cultivated more easily using industrially applied commercial media. Growth of all the strains occurred between 45 °C and 65 °C, indicating that the isolated strains are thermophiles. All the strains showed growth at pH 7.0. On the other hand, any of the strains could grow at acidic (pH 4.0) and alkaline (pH 9.0) conditions. The halophilic nature of the strains showed that all the strains could tolerate 3% (w/v) NaCl concentration and 10 strains could tolerate 6% (w/v) NaCl concentration. Moreover, isolates EF60115, EF60123, and EF60155 could tolerate up to 9% (w/v) NaCl concentration. Any of the strains could tolerate up to 15% (w/v) NaCl condition (Table 2).

Table 2.
Growth characteristics of aerobically cultured thermophilic bacteria isolated from the diverse hot springs in Republic of Korea.
3.3. Taxonomic Identification Based on 16S rRNA Gene Sequencing and Phylogenetic Analysis
Taxonomic identification and phylogenetic analysis of the isolates were assessed by 16S rRNA sequencing. The complete 16S rRNA gene sequences of 29 isolates were successfully amplified. A pairwise comparison result of the 16S rRNA gene sequences on the EzBioCloud server showed that there were eight different types of species (Table 3 and Table 4). As shown in Table 3, the strains belong to phylum Bacillota, class Bacilli, and order Bacillales. Further, twenty-eight strains belong to Bacillaceae (96.6%) and one strain belongs to Thermoactinomycetaceae (3.4%). Among the Bacillaceae, four genera were confirmed with five strains of Aeribacillus (17.2%), one strain of Bacillus (3.4%), eleven strains of Caldibacillus (37.9%), and twelve strains of Geobacillus (41.4%). On the other hand, one strain of Thermoactinomyces was identified. Phylogenetic analysis was conducted to evaluate the relationships of the isolated strains with closely related type strains (Figure 2).

Table 3.
Phylum analysis of aerobically cultured thermophilic bacteria isolated from the diverse hot springs in Republic of Korea.

Table 4.
Taxonomic identification of the isolates based on 16S rRNA gene sequences.
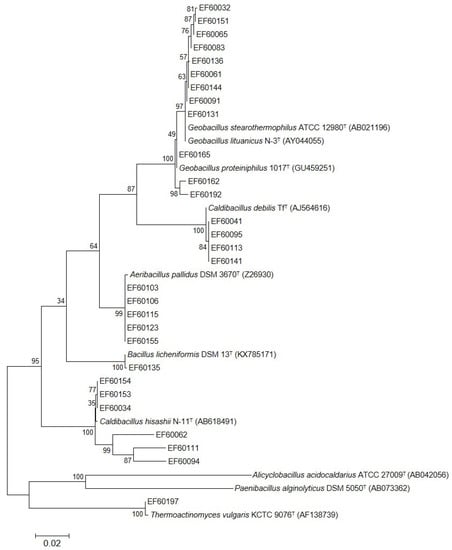
Figure 2.
Phylogenetic analysis of 29 isolates and their closely related type strains. The phylogenetic tree was constructed based on 16S rRNA gene sequence alignments using the Maximum Likelihood method in MEGA 6.0. Bootstrap values are indicated at the branch points. The scale bar indicates a branch length equivalent to 0.02 changes per nucleotide. Alicyclobacillus acidocaldarius ATCC 27009T and Paenibacillus alginolyticus DSM 5050T served as outgroup.
3.4. Production of Extracellular Hydrolytic Enzymes
Bacterial isolates were screened for extracellular amylase, protease, and lipase activity. Among the isolates, at least one extracellular enzyme activity was detected in 22 strains (75.9%). Amylase activity was detected in nineteen strains (65.5%), protease activity in ten strains (34.5%), and lipase activity in five strains (17.24%) (Figure 3). The degree of hydrolytic capacity for degrading a certain substrate was evaluated and shown as enzyme intensity (EI) to show which of the isolates produced larger amounts of the enzymes (Table 5). The amylolytic activity was the most common among isolates with the highest amylase activity in strains EF60154 (EI 4.02), EF60094 (EI 3.33), EF60111 (EI 3.16), and EF60034 (EI 3.1) and strains EF60165 (EI 3.67), EF60131 (EI 3.12), EF60136 (EI 3.09), and EF60061 (EI 3.08). The highest protease and lipase activity was detected in strains EF60192 (EI 10.71) and EF60131 (EI 3.46), respectively. Two strains, EF60083 and EF60192, produced all the tested hydrolytic enzymes.
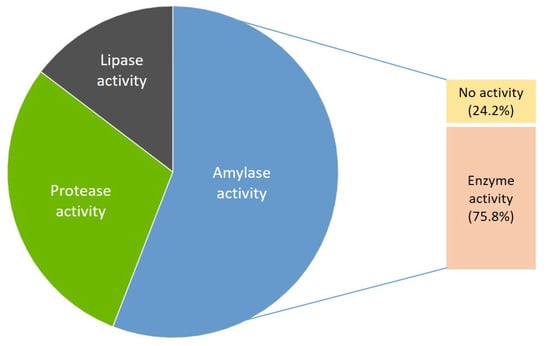
Figure 3.
Production of extracellular hydrolase among isolates.

Table 5.
Amount of extracellular hydrolases produced by thermophilic isolates expressed as EI.
4. Discussion
The region of hot springs that are habitats for thermophiles is limited to a restricted number of sites. In Korea, there are about 400 hot springs, many recognized as having therapeutic and medicinal effects. In these environmental conditions, living organisms have to cope with harsh temperatures and the low availability of nutritional compounds. Biodiversity in these environments is low, but some microorganisms can adapt to these conditions by developing survival strategies.
Bacterial strains isolated in this study are thermophiles that showed growth at relatively high temperatures. One of the important potentials of thermophiles is their productive capacity of thermostable enzymes. Applications of enzymes in industries are numerous; for example, hydrolases (amylases, lipases, and proteases, etc.) account for over 75% of all enzymes produced on a commercial scale. These thermostable hydrolytic enzymes can be extracted from thermophiles because they have many desirable properties, including the capacity to reduce the probability of microbial contamination in large-scale fermentation and the capacity to function for extended periods of time [26,27]. This reason leads to intensive and extended studies to perform fully for investigating such promising microorganisms. The majority (75.8%) of the thermophilic strains produced at least one extracellular hydrolase (Table 4). The amylolytic activity was the most common in isolates with 19 (65.5%) strains. The highest amylase activity was produced by isolate EF60154 from the Baegam hot spring, which was identified as Caldibacillus hisashii. Proteolytic activity was detected in 10 (34.5%) strains, with the highest protease activity in isolates EF60192 and EF60165, which were identified as Geobacillus proteiniphilus. Lipolytic activity was less common among isolates with 5 (17.2%) strains, producing very small amounts. The highest lipase activity was detected in the EF60131 strain, which was identified as Geobacillus lituanicus. Isolates EF60192 and EF60083 that were identified as Geobacillus proteiniphilus and Geobacillus stearothermophilus, respectively, produced all the tested enzymes. Among the isolates, eight strains could produce two extracellular hydrolases. On the other hand, seven strains did not show any enzyme activity. There are numerous biotechnological uses for hydrolases; each one necessitates certain enzyme properties in terms of specificity and thermostability [28]. There may be differences in enzyme production depending on the environmental factors and chemical characteristics of the water samples [29]. Most of these promising thermophilic bacterial isolates belong to the genus Geobacillus. Adaptation of Geobacillus spores to environmental stress and meeting their basic nutritional requirements for growth may be the reason for the occurrence of these species in all five hot springs [30].
The results of this study will achieve great significance in terms of securing the diversity of microbiological resources in Korea, and it is expected that a new strain can be found for production of hydrolytic enzymes in water samples around domestic hot spring regions and basic biological materials for research on bioengineering in food and cosmetics industries.
Author Contributions
Conceptualization, Y.-J.L., S.-B.K. and S.-J.L.; methodology, Y.-J.L. and S.-J.L.; software, D.G.; validation, S.-B.K. and G.N.; formal analysis, H.K.; investigation, Y.-J.L., D.G., D.O., G.E.J. and S.-J.L.; resources, D.G., D.O., G.E.J., H.K. and S.-J.L.; data curation, Y.-J.L., D.G., I.-T.C., Y.-J.J., G.N. and S.-J.L.; writing—original draft preparation, Y.-J.L., D.G. and S.-J.L.; writing—review and editing, Y.-J.L., S.-B.K., G.N., I.-T.C., Y.-J.J. and S.-J.L.; visualization, Y.-J.L. and D.G.; supervision, S.-J.L.; project administration, S.-J.L.; funding acquisition, Y.-J.L., S.-B.K., I.-T.C. and S.-J.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Institute of Biological Resources funded by the Ministry of Environment (NIBR202102109), Basic Research of the NRF (NRF-2020R1F1A1076624, NRF-2021R1F1A1064036), grant from the Ministry of Ocean and Fisheries (PM62830), and grant from the Technology Innovation Program (20015807) funded by the Ministry of Trade, Industry & Energy.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no potential conflicts of interest.
References
- Tango, M.S.A.; Islam, M.R. Potential of extremophiles for biotechnological and petroleum applications. Energy Sources 2002, 24, 543–559. [Google Scholar] [CrossRef]
- Gupta, G.; Srivastava, S.; Khare, S.K.; Prakash, V. Extremophiles: An overview of microorganism from extreme environment. Int. J. Agric. Environ. Biotechnol. 2014, 7, 371–380. [Google Scholar] [CrossRef]
- Singh, G.; Bhalla, A.; Kaur, P.; Capalash, N.; Sharma, P. Laccase from prokaryotes: A new source for an old enzyme. Rev. Environ. Sci. Biotechnol. 2011, 10, 309–326. [Google Scholar] [CrossRef]
- Sikdar, A.; Raziuddin, M.; Gupta, K.K. Isolation and characterization of thermophilic bacteria of a hot water spring source, Balbal. Int. J. Adv. Res. Biol. Sci. 2015, 2, 106–110. [Google Scholar]
- Bhalla, A.; Bansal, N.; Kumar, S.; Bischoff, K.M.; Sani, R.K. Improved lignocellulose conversion to biofuels with thermophilic bacteria and thermostable enzymes. Bioresour. Technol. 2013, 128, 751–759. [Google Scholar] [CrossRef] [PubMed]
- Dettmer, A.; dos Anjos, P.S.; Gutterres, M. Special review paper: Enzymes in the leather industry. J. Am. Leather Chem. Assoc. 2013, 108, 146–158. [Google Scholar]
- Baltaci, M.O.; Genc, B.; Arslan, S.; Adiguzel, G.; Adiguzel, A. Isolation and characterization of thermophilic bacteria from geothermal areas in Turkey and preliminary research on biotechnologically important enzyme production. Geomicrob. J. 2017, 34, 53–62. [Google Scholar] [CrossRef]
- Afrisham, S.; Badoei-Dalfard, A.; Namaki-Shoushtari, A.; Karami, Z. Characterization of a thermostable, CaCl2-activated and raw-starch hydrolyzing alpha-amylase from Bacillus licheniformis AT70: Production under solid state fermentation by utilizing agricultural wastes. J. Mol. Catal. B Enzym. 2016, 132, 98–106. [Google Scholar] [CrossRef]
- Demirjian, D.C.; Morís-Varas, F.; Cassidy, C.S. Enzymes from extremophiles. Curr. Opin. Chem. Biol. 2001, 5, 144–151. [Google Scholar] [CrossRef]
- Sen, S.K.; Mohapatra, S.K.; Satpathy, S.; Rao, G.T. Characterization of hot water spring source isolated clones of bacteria and their industrial applicability. Int. J. Chem. Res. 2010, 2, 1–7. [Google Scholar] [CrossRef]
- Hmidet, N.; El-Hadj Ali, N.; Haddar, A.; Kanoun, S.; Alya, S.K.; Nasri, M. Alkaline proteases and thermostable α-amylase co-produced by Bacillus licheniformis NH1: Characterization and potential application as detergent additive. Biochem. Eng. J. 2009, 47, 71–79. [Google Scholar] [CrossRef]
- Malkawi, H.I.; Al-Omari, M.N. Culture-dependent and culture-independent approaches to study the bacterial and archaeal diversity from jordanian hot springs. Afr. J. Microbiol. Res. 2010, 4, 923–932. [Google Scholar] [CrossRef]
- Vartoukian, S.R.; Palmer, R.M.; Wade, W.G. Strategies for culture of ‘unculturable’ bacteria. FEMS Microbiol. Let. 2010, 309, 1–7. [Google Scholar] [CrossRef]
- Mazzucotelli, C.A.; Ponce, A.G.; Kotlar, C.E.; Moreira, M.D. Isolation and characterization of bacterial strains with a hydrolytic profile with potential use in bioconversion of agroindustial by-products and waste. Food Sci. Technol. 2013, 33, 295–303. [Google Scholar] [CrossRef]
- Hann, S.K. Mineral water and spas in Korea. In Clinics in Dermatology; Parish, L.C., Crissey, J.T., Eds.; Elsevier: Amsterdam, The Netherlands, 1966; pp. 633–635. [Google Scholar]
- Kim, J.-W.; Hahn, H.J.; Woo, S.-Y.; Yun, S.-T.; Lee, J.T.; Kim, H.J. Immunoinflammatory regulation effects of Korean hot spring water. J. Jpn. Balneol. Climatol. Phys. Med. 2015, 78, 253–270. [Google Scholar] [CrossRef]
- Lee, C.-M.; Hamm, S.-Y.; Lee, C.; Choi, S.-J.; Chung, S.Y. Characteristics of south Korea’s geothermal water in relation to its geological and geochemical feature. J. Soil Groundw. Environ. 2014, 19, 25–37. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Aleem, B.; Rashid, M.H.; Zeb, N.; Saqib, A.; Ihsan, A.; Iqbal, M.; Ali, H. Random mutagenesis of super Koji (Aspergillus oryzae): Improvement in production and thermal stability of α-amylases for maltose syrup production. BMC Microbiol. 2018, 18, 200. [Google Scholar] [CrossRef]
- Ashok, A.; Doriya, K.; Rao, J.V.; Qureshi, A.; Tiwari, A.N.; Kumar, D.S. Microbes Producing L-Asparaginase free of Glutaminase and Urease isolated from Extreme Locations of Antarctic Soil and Moss. Sci. Rep. 2019, 9, 1423. [Google Scholar] [CrossRef]
- Hossain, T.J.; Chowdhury, S.I.; Mozumder, H.A.; Chowdhury, M.N.A.; Ali, F.; Rahman, N.; Dey, S. Hydrolytic Exoenzymes Produced by Bacteria Isolated and Identified from the Gastrointestinal Tract of Bombay Duck. Front. Microbiol. 2020, 11, 2097. [Google Scholar] [CrossRef] [PubMed]
- Cowan, D.A. Industrial enzymes. In Biotechnology: The Science and the Business, 2nd ed.; Springham, D.G., Moses, V., Cape, R.E., Eds.; Harwood Academic Publishers: New York, NY, USA, 1994. [Google Scholar] [CrossRef]
- Rollof, J.; Hedstrom, S.A.; Nilsson-Ehle, P. Lipolytic activity of Staphylococcus aureus strains from disseminated and localized infections. Acta Pathol. Microbiol. Scand. B 1987, 95, 109–113. [Google Scholar] [CrossRef] [PubMed]
- Burke, V.; Robinson, J.O.; Richardson, C.J.L.; Bundell, C.S. Longitudinal studies of virulence factors of Pseudomonas aeruginosa in cystic fibrosis. Pathology 1991, 23, 145–148. [Google Scholar] [CrossRef] [PubMed]
- Wati, L.; Dhamija, S.S.; Singh, D.; Nigam, P.; Marchant, R. Characterisation of genetic control of thermotolerance in mutants of Saccharomyces cerevisiae. J. Genet. Eng. Biotechnol. 1996, 16, 19–26. [Google Scholar]
- Wang, X.; Li, D.; Watanabe, T.; Shigemori, Y.; Mikawa, T.; Okajima, T.; Mao, L.; Ohsaka, T. A glucose/o-2 biofuel cell using recombinant thermophilic enzymes. Int. J. Electrochem. Sci. 2012, 7, 1071–1078. [Google Scholar]
- Rekadwad, B.N. Characterization of amylase from industrially important thermophilic microorganism: Geobacillus thermoleovorans strain rekadwadsis. Int. J. Life Sci. Biotechnol. Pharma Res. 2015, 4, 26–30. [Google Scholar]
- Khalil, A. Screening and characterization of thermophilic bacteria (lipase, cellulase and amylase producers) from hot springs in Saudi Arabia. J. Food Agric. Environ. 2011, 9, 672–675. [Google Scholar]
- Kawasaki, Y.; Aoki, M.; Makino, Y.; Sakai, H.; Tsuboi, Y.; Ueda, J. Characterization of moderately thermophilic bacteria isolated from saline hot spring in Japan. Microbiol Indones. 2011, 5, 2–12. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).