Abstract
Mitochondrial biogenesis is tightly regulated in response to extracellular and intracellular signals, thereby adapting yeast cells to changes in their environment. The Hap2/3/4/5 complex is a master transcriptional regulator of mitochondrial biogenesis in yeast. Hap4 is the regulatory subunit of the complex and exhibits increased expression when the Hap2/3/4/5 complex is activated. In cells grown under glucose derepression conditions, both the HAP4 transcript level and Hap4 protein level are increased. As part of an inter-organellar signaling mechanism coordinating gene expression between the mitochondrial and nuclear genomes, the activity of the Hap2/3/4/5 complex is reduced in respiratory-deficient cells, such as ρ0 cells lacking mitochondrial DNA, as a result of reduced Hap4 protein levels. However, the underlying mechanism is unclear. Here, we show that reduced HAP4 expression in ρ0 cells is mediated through both transcriptional and post-transcriptional mechanisms. We show that loss of mitochondrial DNA increases the turnover of Hap4, which requires the 26S proteasome and ubiquitin-conjugating enzymes Ubc1 and Ubc4. Stabilization of Hap4 in the ubc1 ubc4 double mutant leads to increased expression of Hap2/3/4/5-target genes. Our results indicate that mitochondrial biogenesis in yeast is regulated by the functional state of mitochondria partly through ubiquitin/proteasome-dependent turnover of Hap4.
1. Introduction
Mitochondria are the power plants of eukaryotic cells and play important roles in intermediary metabolism. Except for a small number of respiratory chain components encoded in the mitochondrial genome, most mitochondrial proteins are encoded in the nuclear genome. Mitochondrial biogenesis requires coordinated expression of genes from both the nuclear and mitochondrial genomes [1,2]. Multiple signaling pathways between mitochondria and the nucleus exist to coordinate gene expression in the nucleus in response to changes in the functional state of mitochondria as well as changes in the cellular environment [3,4,5,6,7,8]. The prototypal pathway of this sort is the mitochondria-to-nucleus retrograde signaling pathway, also known as the RTG pathway. Saccharomyces cerevisiae maintains viability after losing mitochondrial DNA (mtDNA) and becomes a ρ0 “petite”. Mitochondrial dysfunction due to loss of mtDNA activates two basic helix-loop-helix leucine zipper transcription factors, Rtg1 and Rtg3, leading to changes in nuclear gene expression and a reconfiguration of metabolic pathways that adapt cells to mitochondrial defects [5]. Cells lacking their mitochondrial genome also induce the expression of PDR5, encoding an ATP-binding cassette transporter involved in multidrug resistance, through the activation of the zinc finger transcription factor Pdr3 [6,7].
The Hap2/3/4/5 transcription factor is a multi-subunit master regulator of mitochondrial biogenesis and an essential part of yeast’s metabolic remodeling when cells switch from glycolysis to respiratory metabolism for generating ATP [9,10,11,12,13]. The Hap2/3/5 trimer binds to CCAAT sequence elements in the promoters of target genes and requires Hap4 to provide transcriptional activation domain activity [14]. When yeast cells exhaust the supply of glucose, there is an increase in HAP4 mRNA and Hap4 protein levels while the other members of the complex are constitutively expressed [12,14,15]. An elevation in Hap4 protein level results in the formation of the Hap2/3/4/5 complex to activate the expression of its target genes, including those encoding the enzymes of the tricarboxylic acid cycle and components of the mitochondrial respiratory complexes [13,16,17]. Apart from the Hap2/3/4/5 complex, a heme-activated protein, Hap1 also contributes to mitochondrial biogenesis by activating expression of genes under aerobic conditions, including those encoding components of Complexes III and IV of the electron transport chain [18].
Hap4’s importance in the regulation of the Hap2/3/4/5 complex and mitochondrial biogenesis has led to studies on the regulation of its expression. A HAP4 promoter analysis reveals a 30-bp activating sequence at position −869 bp upstream of the ATG start codon that is important for its expression [19]. Cat8 is implicated in the regulation of HAP4 expression by this activating sequence, but the effect has been proposed to be indirect. Zhang et al. reported that increased heme synthesis induces transcription of both HAP4 and genes involved in respiratory metabolism [20]. They further showed that both Hap1 and Hap2 are positive regulators of HAP4 expression in galactose-grown cells, suggesting that HAP4 is subject to autoregulatory control. In glucose-grown cells, the role of Hap1 and Hap2 in HAP4 expression is less clear: although hap1 and hap2 mutations increase HAP4 expression in wild-type HEM1 cells, they decrease HAP4 expression in hem1 mutant cells [20]. Rox1, a transcriptional repressor of hypoxic genes [21], negatively regulates HAP4 expression in glucose-grown cells [20]. We recently reported that protein kinase A (PKA) is a potent inhibitor of HAP4 expression. In mutants with reduced PKA activity such as ira1, ira2, bcy1, gpb1 gpb2, pde1 pde2 mutants, HAP4 expression is reduced. On the other hand, a complete lack of PKA activity due to a triple deletion mutation in the three genes encoding the catalytic subunits of PKA in a yak1 mutant strain dramatically increases HAP4 expression [22].
We have previously reported a transcriptional factor switch from the Rtg1/3 complex to the Hap2/3/4/5 complex for the expression of early tricarboxylic acid cycle genes, CIT1, ACO1, IDH1, and IDH2, in response to mitochondrial dysfunction [23]. In ρ0 cells, the expression of the rest of the tricarboxylic acid cycle is reduced, suggesting that the activity of the Hap2/3/4/5 complex is suppressed. Genome-wide transcriptional profiling reveals reduced expression of many Hap2/3/4/5-target genes as a result of reduced Hap4 protein level [24]. Other studies have shown that Hap4 is a highly unstable protein, whose turnover is affected by the cellular environment [25,26,27]. Elevated production of reactive oxygen species increases the turnover of Hap4 and consequently reduces mitochondrial biogenesis [25]. Conversely, an increase in the cellular heme level and glutathione redox state stabilizes Hap4, resulting in increased mitochondrial biogenesis [27]. Based on the findings that the HAP4 transcript level does not change in ρ0 cells and that Hap4 overexpression leads to its localization in the vacuole, Bourges and colleagues have proposed that a vacuolar degradation mechanism might be behind reduced Hap4 protein levels in ρ0 cells [24]. It has also been suggested that reactive oxygen species may not be involved in the downregulation of Hap4 in response to mitochondrial dysfunction.
In an aim to uncover the mechanism behind reduced Hap4 protein levels in ρ0 cells, we analyzed the expression of a lacZ reporter gene under the control of the HAP4 promoter using β-galactosidase activity assays and Hap4 stability using a cycloheximide chase assay. We found that reduced Hap4 protein level in ρ0 cells is due to the combined effect of reduced promoter activity of the HAP4 gene and increased turnover of Hap4 protein. We show that two ubiquitin-conjugating enzymes, Ubc1 and Ubc4, are required for Hap4 turnover. Our data suggest that ubiquitin-dependent turnover of Hap4 mediates mitochondria-to-nucleus signaling and plays a negative regulatory role in mitochondrial biogenesis.
2. Materials and Methods
2.1. Strains, Plasmids, and Growth Media and Growth Conditions
Yeast strains and plasmids used in this study are listed in Table 1 and Table 2, respectively. Yeast cells were grown at 30 °C in YNBcas5%D (0.67% yeast nitrogen base, 1% casamino acids, and 5% D-glucose), YNBcasR (0.67% yeast nitrogen base, 1% casamino acids, and 2% raffinose), YPGlycerol (1% bacto yeast extract, 2% peptone, and 3% glycerol [v/v]), and YPD (1% bacto yeast extract, 2% peptone, and 2% D-glucose). When required, uracil, adenine, and tryptophan were added to YNBcas5D and YNBcasR at standard concentrations to meet auxotrophic requirements [28]. The plate medium contains 2% agar. When indicated, 50 µM carbobenzoxyl-leucinyl-leucinyl-leucinal (MG132) was added to the growth media to inhibit proteasome activity.

Table 1.
S. cerevisiae strains used in this study.

Table 2.
Plasmids used in this study.
2.2. Creation of ρ0 Petites
To induce loss of mitochondrial DNA, ρ+ cells were grown to saturation in YPD liquid medium supplemented with 15 µg/mL ethidium bromide and then plated on YPD medium to obtain single colonies. Colonies were individually picked and grown overnight in YPD medium and cells were stained with DAPI as described [28]. ρ0 petites without mitochondrial DNA were confirmed by both fluorescence microscopy and inability to grow on YPGlycerol plate medium.
2.3. Yeast Transformations and β-Galactosidase Activity Assays
Yeast strains were transformed with plasmids using the high-efficiency transformation method as described [28]. Cells were cultured in specified liquid medium overnight to reach OD600 0.6~0.8 after undergoing a minimum of six cell divisions and collected for β-galactosidase assays using cellular extracts as described. For each plasmid-strain combination, β-galactosidase activities (in nanomoles of hydrolyzed o-nitrophenyl-β-D-galactopyranoside per milligram of protein per minute) were averages of duplicate assays of two to six independent cultures. Specific activity was calculated in relation to the total protein amount in the cellular extracts, which was determined using the Bradford assay with bovine serum albumin as the standard. When indicated, the means of β-galactosidase activities from two groups were compared by a t-test using Graphpad software (San Diego, CA, USA).
2.4. Cellular Extract Preparation and Immunoblotting
Total cellular proteins were prepared in extraction buffer (1.85 N NaOH-7.5% β-mercaptoethanol) followed by precipitation with trichloroacetic acid as described [33]. Trichloroacetic acid pellets were neutralized with 1M unbuffered Tris and resuspended in 1x SDS-PAGE loading buffer. Equivalent amounts of protein samples based on OD600 readings of the collected cell cultures were loaded into the lanes of the same gel. Immunoblotting of Hap4 with a C-terminal 3xHA epitope tag was carried out by incubating nitrocellulose membranes with rat monoclonal anti-HA antibody 3F10 (Roche Diagnostics GmbH, Mannheim, Germany), followed by goat anti-rat HRP-conjugated polyclonal secondary antibody (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA). For loading controls, blots were first deprobed in stripping buffer (2% sodium dodecyl sulfate, 100 mM β-mercaptoethanol, 62.5 mM Tris-HCl pH 6.7) for 45 min at 60 °C with agitation and then reprobed with anti-Ilv5 (acetohydroxyacid reductoisomerase) or anti-Pgk1 (3-phosphoglycerate kinase) rabbit polyclonal antibodies. Chemiluminescence images of Western blots were captured using the Bio-Rad Chemi-Doc photo documentation system (Bio-Rad Life Science, Hercules, CA, USA). A t-test was carried out to determine whether there was a significant difference in Hap4-HA protein levels from two groups of data.
2.5. Cycloheximide Chase Assay
Cells expressing HA epitope-tagged Hap4 were grown in YNBcas5%D or YNBcasR medium to OD600 0.6~0.8. Protein synthesis was then inhibited by the addition of 50 µg/mL cycloheximide to initiate chase. At 5 min intervals for a total of 15 min, 1 mL aliquots of cell cultures were withdrawn and immediately subject to cellular extract preparation. Immunoblotting of Hap4-HA was carried out as described above. For the MG132 treatment to inhibit the function of the 26S proteasomes, erg6∆ mutants expressing HAP4-HA were grown in YNBcas5%D or YNBcasR medium overnight to reach OD600 ~0.4 and MG132 was added at a final concentration of 50 μM. Cycloheximide chase assays were then performed after cultures reached OD600 0.6~0.8. For the determination of the half-life of Hap4-HA, band intensities on Western blots over the course of cycloheximide chase were quantified using the Bio-Rad QuantityOne software and used to fit an exponential curve, y = Ae−kt. The half-life was calculated from ln(2)/k. A t-test was carried out to determine whether there was a significant difference in the Hap4 half-lives from two groups of data.
3. Results
3.1. HAP4 Expression Is Affected by the Functional State of Mitochondria and the Quality of Carbon Sources
Yeast cells become ρ0 petites after losing mitochondrial DNA (mtDNA). It was reported that the Hap4 protein level is reduced by 2-fold in ρ0 cells grown in rich galactose media in comparison to that in ρ+ cells containing wild-type mtDNA [24]. Although it was suggested that transcriptional regulation was not involved, the underlying mechanism was unclear. To facilitate the analysis of transcriptional regulation of HAP4, we generated a HAP4-lacZ reporter gene by fusing a 1.8 kbp promoter sequence of HAP4 to the lacZ coding sequence. The reporter gene was then introduced into otherwise wild-type ρ+ and ρ0 cells. Since Hap4 expression is also subject to transcriptional regulation in response to changes in carbon sources, transformants were grown in media with two different carbon sources, D-glucose (dextrose) and raffinose, and β-galactosidase assays on HAP4-lacZ expression were conducted. Raffinose is a fermentable trisaccharide composed of galactose, glucose, and fructose. When it is used as the sole carbon source in growth media, raffinose leads to glucose derepression and has been used extensively in the study of the mitochondria-to-nucleus retrograde signaling pathway [23,34,35]. Figure 1A shows that HAP4-lacZ expression is greatly increased in ρ+ cells grown in raffinose medium compared to dextrose, consistent with published results [12]. In ρ0 cells compared to ρ+ cells, the expression of the HAP4-lacZ reporter gene was reduced in both dextrose- and raffinose-grown cells, suggesting that HAP4 transcription is reduced in response to mitochondrial DNA loss.
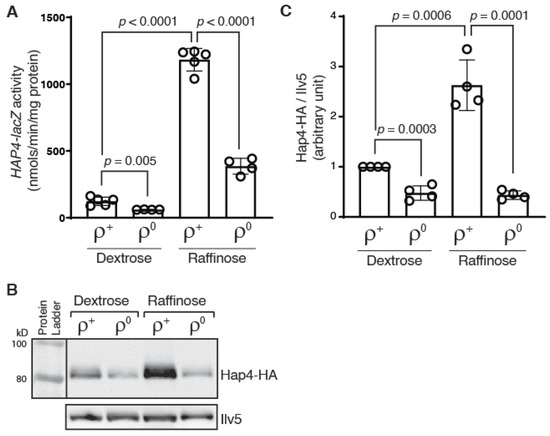
Figure 1.
HAP4 expression is reduced in ρ0 cells. (A) A β-galactosidase activity assay of the expression of a HAP4-lacZ reporter gene in wild-type ρ+ strain (BY4741) and its ρ0 derivative grown in dextrose and raffinose media. β-galactosidase activities were determined as described in Materials and Methods. The data is presented as the mean ± standard deviation. A t-test was carried out and the p values indicate a significant difference between two groups of data. (B) Immunoblotting of Hap4-HA in a ρ+ hap4Δ mutant (BY4741 hap4) and its ρ0 derivative grown in dextrose and raffinose media. Equivalent amounts of protein samples based on OD600 readings of the collected cell cultures were loaded into the lanes of the same gel. Ilv5 was included as a loading control. The result was representative of four independent sets of results. (C) Quantification of Hap4-HA/Ilv5 from Western blotting data.
We next examined Hap4 protein levels in ρ+ and ρ0 cells grown in dextrose and raffinose media. We generated a HAP4-HA fusion gene encoding a 3x human influenza hemagglutinin (HA) epitope tag at the C-terminus of Hap4. The fusion protein was determined to be functional by its ability to rescue the growth defect of a hap4Δ mutant in growth medium with nonfermentable carbon sources (Supplemental Figure S1). When the protein level of Hap4-HA was determined by Western blotting, we found that Hap4 protein level is largely consistent with the promoter activity of HAP4: there is an increased Hap4 protein level in cells grown in raffinose medium compared to dextrose medium and a reduction in Hap4 protein level due to the loss of mtDNA (Figure 1B,C). However, the Hap4 protein level in ρ0 cells become largely independent of the carbon sources. The discrepancy between HAP4-lacZ reporter gene activities and Hap4-HA protein levels in ρ0 cells grown in dextrose versus raffinose medium suggest that a post-transcriptional mechanism(s) exists to modulate Hap4 protein levels.
3.2. Transcriptional Regulation of KGD1, a Target of the Hap2/3/4/5 Complex, Correlates with Protein Levels of Hap4
To help dissect the regulation of Hap4, we generated a KGD1-lacZ reporter gene as a readout of the activity of the Hap2/3/4/5 complex. KGD1 encodes a subunit of the mitochondrial α-ketoglutarate dehydrogenase complex and is a target of the Hap2/3/4/5 complex [36]. Consistently, KGD1-lacZ has a low, basal expression in ρ+ cells grown in dextrose medium (glucose repression), and its expression is increased by 21-fold in ρ+ cells grown in raffinose medium (glucose derepression) (Figure 2A). ρ0 reduces KGD1-lacZ expression by 1.6- and 6.0-fold in dextrose- and raffinose-grown cells, respectively, consistent with reduced expression of Hap4 in ρ0 cells compared to ρ+ cells. In ρ+ cells grown in dextrose and raffinose media, hap4Δ reduces KGD1-lacZ expression by 2- and 6-fold, respectively. In contrast, hap4Δ has little to no effect on KGD1-lacZ expression in ρ0 cells grown in either dextrose or raffinose medium, suggesting that the Hap2/3/4/5 complex is largely inactive in ρ0 cells, likely due to a low expression level of Hap4.
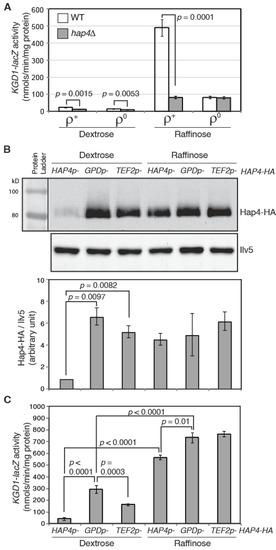
Figure 2.
Expression of KGD1-lacZ reporter gene correlates with Hap4-HA protein levels. (A) A β-galactosidase activity assay of KGD1-lacZ expression in ρ+ and ρ0 cells of wild-type strain (ZLY3440) and its isogenic hap4Δ mutant (DCY247). (B) (upper panel) Immunoblotting of Hap4-HA in hap4Δ cells (DCY247) carrying centromeric plasmids encoding HAP4-HA under the control of its endogenous promoter HAP4p, a heterologous promoter GPDp or TEF2p. Equivalent amounts of protein samples based on OD600 readings of the collected cell cultures were loaded into the lanes of the same gel. Ilv5 was included as a loading control. (lower panel) Quantification of Hap4-HA/Ilv5 from two sets of Western blotting results. (C) A β-galactosidase activity assay of KGD1-lacZ expression in hap4Δ mutant cells carrying plasmids as described for panel (B).
Overexpression of Hap4 leads to increased expression of Hap2/3/4/5-target genes [16,17,37]. Hap4 protein levels can be modulated by both the carbon sources and functional states of mitochondria. Mitochondria have many different functions. It is conceivable that mitochondrial biogenesis can be fine-tuned through different levels of Hap4. However, to our knowledge, this possibility has not been reported previously. We sought to examine KGD1-lacZ expression in cells expressing different levels of Hap4. Accordingly, we overexpressed Hap4 under the control of two strong heterologous promoters, TEF2 and GPD (encoding translation elongation factor lα and glyceraldehyde-3-phosphate dehydrogenase, respectively) [38]. Plasmids encoding 3xHA epitope-tagged Hap4 under the control of the promoter of TEF2, GPD, or its own were introduced separately into a hap4Δ mutant strain carrying an integrated KGD1-lacZ reporter gene. Hap4-HA levels were detected by Western blotting. Figure 2B shows that in glucose-grown cells where HAP4 is under the control of the heterologous promoter TEF2 or GPD in comparison to its native promoter, Hap4 protein levels are much higher. Consistent with GPD being a stronger promoter than TEF2 in dextrose-grown cells [38], the Hap4 protein level is also higher in cells expressing GPDp-HAP4 than cells expressing TEF2p-HAP4. In raffinose-grown cells, HAP4 expression under the control of either of the heterologous promoters is only marginally higher than its native promoter due to glucose derepression of HAP4 expression.
We then determined KGD1-lacZ expression in cells expressing different levels of Hap4 via β-galactosidase activity assays. In dextrose-grown cells, overexpression of Hap4 under the control of the GPD or TEF2 promoter increases KGD1-lacZ expression, and there is a positive correlation between Hap4 protein levels and KGD1-lacZ activity (Figure 2B,C). In raffinose-grown cells, the marginal overexpression of HAP4 under the control of GPD and TEF2 promoters slightly increases KGD1-lacZ expression. Significantly, in cells in which HAP4 is under the control of the GPD or TEF2 promoter, the expression of KGD1 is significantly lower in cells grown in dextrose medium compared to raffinose medium even though Hap4-HA levels are comparable in those cells. This indicates that a regulatory mechanism other than the Hap4 protein level exists to achieve maximal induction of KGD1-lacZ expression under glucose derepression conditions. Together, these data support the notion that varying Hap4 protein levels lead to a graded transcriptional response of the target genes of the Hap2/3/4/5 complex, rather than an all-or-none effect. Since the Hap2/3/4/5 complex mediates the expression of many mitochondrial proteins [13,17,24], by modulating Hap4 protein levels, yeast cells are able to fine-tune mitochondrial biogenesis to meet changing energetic and metabolic requirements in cells.
3.3. Hap4 Has a Shorter Half-Life in ρ0 Cells than in ρ+ Cells
HAP4 expression correlates with respiratory metabolism. Hap4 is needed when cells have functional mtDNA, which encodes several proteins essential for oxidative phosphorylation. When cells lose mitochondrial DNA and become unable to use nonfermentable carbon sources, it may be advantageous for cells to reduce the Hap4 protein level to down-regulate the expression of genes involved in respiratory metabolism. This is partly achieved by reducing the promoter activity of HAP4 in ρ0 cells (Figure 1A). The discrepancy between the Hap4 protein level and the HAP4-lacZ reporter gene activity in ρ0 grown in raffinose medium in Figure 1 prompted us to conduct a comparative analysis on Hap4 stability in ρ+ and ρ0 cells.
When we tried to transform ρ0 cells with a plasmid encoding TEF2p-HAP4 or GPDp-HAP4, it was difficult to get transformants, suggesting that overexpression of HAP4 in ρ0 cells is toxic. The fewer transformants we obtained may contain suppressor mutations. Nevertheless, we examined Hap4-HA expression in ρ+ and ρ0 cells expressing GPDp-HAP4 by Western blotting. Figure 3A shows that the Hap4-HA protein level was significantly lower in ρ0 cells compared to ρ+ cells grown in both dextrose and raffinose medium. GPD encodes isozyme 3 of glyceraldehyde-3-phosphosphate dehydrogenase, a glycolytic enzyme. A genome-wide transcriptome analysis in response to mitochondrial dysfunctions by Epstein et al. shows that ρ0 cells slightly increase the expression of most genes encoding glycolytic enzymes, including GPD [39]. The metabolic reconfiguration in ATP production in ρ0 cells is expected since increased metabolic influx into glycolysis can compensate for the lack of ATP production from mitochondria. Together, these data suggest that loss of mitochondrial DNA may increase instability of Hap4-HA.
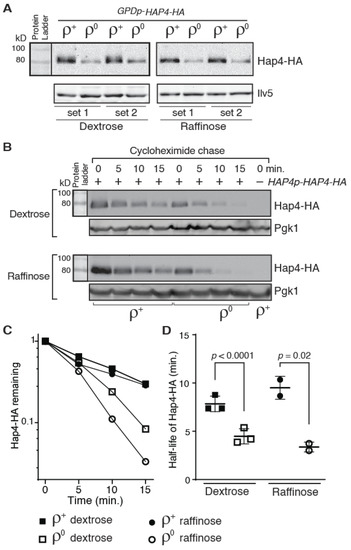
Figure 3.
Hap4-HA is less stable in ρ0 cells than in ρ+ cells. (A) Western blot analysis of Hap4-HA expression in hap4Δ mutant cells (DCY247) carrying a centromeric plasmid encoding GPDp-HAP4-HA (pDC216). Equivalent amounts of protein samples based on OD600 readings of the collected cell cultures were loaded into the lanes of the same gel. (B) A cycloheximide chase assay of Hap4-HA stability in ρ+ and ρ0 cells of strain DCY247 grown in dextrose and raffinose media. Expression of HAP4-HA was under the control of its own promoter and the cycloheximide chase assay was conducted as described in Materials and Methods. Pgk1 was included as a loading control. (C) Quantification of Hap4-HA levels in panel (B). (D) The half-lives of Hap4-HA were determined from Western blots of Hap4-HA from ρ+ and ρ0 cells as described for panel (B) and plotted in the graph.
We performed a cycloheximide chase assay to investigate whether Hap4 stability was different in ρ+ versus ρ0 cell. After the addition of cycloheximide to cell cultures to inhibit protein translation, aliquots of samples were collected every five minutes for a total of 15 min and Hap4 protein levels in cells during the time course were determined by Western blotting. We found that there is an increased turnover of Hap4-HA in ρ0 cells compared to ρ+ cell: In dextrose-grown cells, the half-life of Hap4-HA in ρ+ and ρ0 cells is 7.8 min and 4.5 min, respectively; in raffinose-grown cells, the half-life of Hap4-HA in ρ+ and ρ0 cells is 9.5 min and 3.4 min, respectively (Figure 3B–D). Faster turnover of Hap4 in ρ0 cells in comparison to ρ+ cells was also observed in cells expressing HA-tagged Hap4 from the GPD promoter (Supplemental Figure S2). These data suggest that yeast cells can sense the functional state of mitochondria and regulate Hap4 stability accordingly. Together with data in Figure 1, our results suggest that a lower level of the Hap4-HA protein in ρ0 cells compared to ρ+ cells results from the combined effect of reduced transcription from the HAP4 promoter and increased turnover of Hap4 protein.
3.4. Hap4 Turnover Requires the 26S Proteasome
Hap4 has been reported to be quickly turned over [25,26,27]. However, the mechanism behind Hap4 instability is not clear. A genome-wide analysis of ubiquitylated proteins uncovered Hap4 as a potential candidate [40]. To test whether Hap4-HA turnover requires the ubiquitin/proteasome system, we treated HAP4-HA expressing cells with the proteasome inhibitor MG132. An erg6Δρ mutant was used to facilitate the diffusion of MG132 into cells [41]. Figure 4A shows that MG132 treatment increases the stability of Hap4 in ρ+ and ρ0 cells grown in both dextrose and raffinose medium, suggesting that the proteasome/ubiquitin system is required for Hap4 turnover. We calculated the half-life of Hap4-HA in erg6Δ mutant cells without MG132 treatment and confirmed that there was also faster turnover of Hap4 in ρ0 cells in comparison to ρ+ cells grown in both dextrose medium and raffinose medium (Supplemental Figure S3). The strains used to generate the data on Hap4-HA half-lives in Figure 3D and Figure S3 are from different strain backgrounds, suggesting that increased Hap4-HA turnover in response to mitochondrial DNA loss is not strain specific.
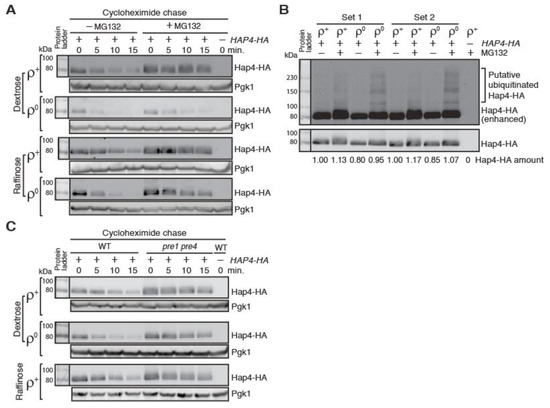
Figure 4.
Inhibition of the activity of the 26S proteasome leads to increased stability of Hap4-HA. (A) A cycloheximide chase assay on Hap4-HA in ρ+ and ρ0 cells of an erg6Δ mutant strain (ZLY4531) with or without the treatment of the proteasome inhibitor MG132. Hap4-HA was detected using immunoblotting. Pgk1 was a loading control. (B) Increased formation of slower mobility forms of Hap4-HA in ρ0 cells compared to ρ+ cells when the proteasomal function is inhibited. ρ+ and ρ0 cells of an erg6Δ mutant strain expressing HAP4-HA from its own promoter were grown in raffinose medium with or without MG132 treatment. Protein samples were separated by SDS-PAGE and Hap4-HA was detected by Western blotting. The gel picture in the upper panel was enhanced to visualize the putative ubiquitinated forms of Hap4-HA. (C) Hap4-HA is stabilized in a pre1 pre4 mutant, which is defective in proteasome protease function. ρ+ and ρ0 cells of wild-type strain (15Daub) and its isogenic pre1 pre4 double mutant (PY555) were grown in dextrose or raffinose medium as indicated. Hap4-HA was detected by immunoblotting. Pgk1 was a loading control.
In cells treated with MG132, we detected slower mobility forms of Hap4-HA on Western blots over a wide range, which are putative ubiquitinated forms of Hap4-HA based on the migration pattern (Figure 4B). Faster turnover of Hap4-HA in ρ0 cells compared to ρ+ cells suggest that there may be increased ubiquitination of Hap4 in response to loss of mitochondrial DNA. To test this possibility, we compared Hap4-HA on Western blot from protein samples generated from ρ+ and ρ0 erg6Δ mutant cells grown in raffinose medium and treated without or with MG132 for 4 hours. We loaded 2.3 times as much protein extracts from ρ0 cells as from ρ+ cells onto the gel to have comparable levels of non-ubiquitinated Hap4. Figure 4B shows that there is increased ubiquitination of Hap4-HA in ρ0 cells compared to ρ+ cells. Together, our data suggest that faster turnover of Hap4 in ρ0 cells results from its increased ubiquitination.
The function of the 26S proteasome is essential for cell viability [42]. erg6Δ mutant cells treated with 50 μM MG132 are slow-growing but still viable due to the residual activity of the proteasome (20–30% full activity) [41]. Therefore, it is not surprising that MG132 treatment does not completely abolish the turnover of Hap4-HA, especially in ρ0 cells (Figure 4A). Although the effect of MG132 on Hap4-HA stability was variable, we observed that MG132 was least effective in stabilizing Hap4-HA in ρ0 cells grown in dextrose medium (Figure 4A). This may be explained by increased expression of PDR5 in ρ0 cells grown in dextrose medium [43]. Pdr5 is a ATP-binding cassette drug efflux pump that reduces the efficacy of proteasome inhibitors [44]. It is also likely that Hap4 may be subjected to proteosome-independent turnover in ρ0 cells. To differentiate these two possibilities, we examined Hap4 stability in a pre1 pre4 double mutant, which is defective in the proteasomal function [29]. Figure 4C shows that the pre1 pre4 double mutation increases the stability of Hap4-HA to a similar extent in ρ+ and ρ0 cells grown in dextrose medium and in ρ+ cells grown in raffinose medium. ρ0 ubc1 ubc4 mutant cells did not grow in raffinose medium and thus were not included in the analysis in Figure 4C. Altogether, our data suggest that Hap4 turnover requires the 26S proteasome and loss of mitochondrial DNA increases Hap4 ubiquitination and turnover.
3.5. Hap4-HA Is Stabilized in a ubc1Δ ubc4Δ Double Mutant
Protein ubiquitination requires the activity of a cascade of three enzymes: an E1 ubiquitin-activating enzyme, an E2 ubiquitin-conjugating enzyme (Ubc), and an E3 ubiquitin ligase [45,46]. The yeast genome encodes a family of 13 ubiquitin-conjugating enzymes, two of which, Ubc3/Cdc34 and Ubc9, are essential [42]. Despite its sequence similarity to other ubiquitin-conjugating enzymes, Ubc9 is a SUMO-conjugating enzyme [47]. To identify the E2 enzyme(s) responsible for Hap4 turnover, Hap4-HA stability was examined using cycloheximide chase in 11 mutants each carrying a deletion mutation of a non-essential UBC gene as well as two mutants carrying temperature-sensitive alleles of CDC34 or UBC9. Since Hap4-HA is least stable in ρ0 cells grown in raffinose medium (Figure 3D), we generated ρ0 derivatives of the 13 ubc single mutants and grew them in raffinose medium to test Hap4-HA stability to maximize the chance of identifying the E2 enzyme responsible for Hap4 turnover. Additionally, cdc34-2 and ubc9-1 mutant cells were switched to 37 °C, a non-permissive temperature for both mutants, before cycloheximide chase was initiated. Figure 5A,B show that Hap4-HA is somewhat stabilized in ubc1 and ubc5 single deletion mutants. Ubc1, 4 and 5 are known to have partial overlapping functions and constitute an enzyme sub-family that is essential for cell growth and viability [48,49]. To test whether these three enzymes have a redundant function in mediating Hap4 turnover, we determined Hap4-HA stability using cycloheximide chase assay in ρ0 derivatives of ubc1/4Δ, ubc1/5Δ, and ubc4/5Δ double mutants grown in raffinose medium. Figure 5B shows that ubc1/4Δ significantly increases Hap4-HA stability. ubc4/5Δ seems to slightly increase Hap4-HA stability. Hap4-HA stability in ubc1/5Δ mutant cells is similar to what is observed in the ubc1Δ single mutant. We then determined Hap4-HA stability in ubc1/4Δ ρ+ cells grown in raffinose medium and ubc1/4Δ ρ+ and ρ0 cells grown in dextrose medium. We found that Hap4-HA also shows increased stability under these conditions (Figure 5C,D). Together, our data indicate that Ubc1 and Ubc4 are the primary ubiquitin-conjugating enzymes that mediate Hap4 turnover. Surprisingly, quantitative analysis of Hap4-HA stability shows that there is no significant difference in its half-lives between ρ+ and ρ0 cells of the wild type Y0002 strain grown in dextrose medium (Figure 5D, left panel and Figure S4). Nevertheless, Hap4-HA is still less stable in ρ0 cells than ρ+ cells of the Y0002 strain when they are grown in raffinose medium (Figure 5D, right panel and Figure S4). The strains used to generate the data in Figure 5C,D are from Y0002 background, which is different from PSY142 background for generating the data in Figure 3D and BY4741 background in Figure S3. A comparison of Hap4-HA stability in these three strains shows that ρ0 cells have similar Hap4 half-lives in dextrose- and raffinose-grown cells, respectively (Figure 3D, Figures S3 and S4). The lack of a difference in Hap4-HA half-lives between Y0002 ρ+ and ρ0 cells grown in dextrose medium appears to be due to its increased turnover in ρ+ cells. ρ+ Y0002 strain grown in raffinose medium also shows increased turnover of Hap4-HA compared to ρ+ cells of PSY142 and BY4741 background strains grown in the same medium. We hypothesize that an unknown mitochondrial defect in Y0002 cells might account for increased Hap4 instability in ρ+ cells, especially in cells grown in dextrose medium. It is also possible that other strain background differences might be responsible.
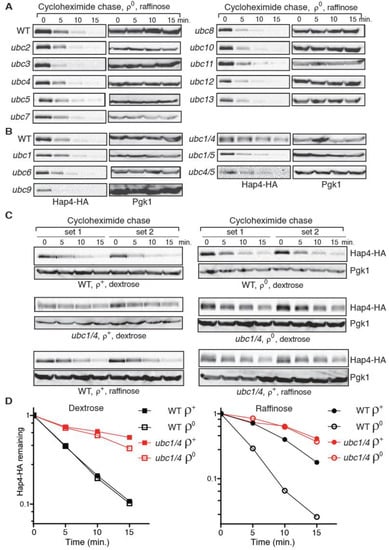
Figure 5.
A ubc1Δ ubc4Δ double mutation stabilizes Hap4-HA. (A,B) A cycloheximide chase assay on Hap4-HA stability in ρ0 cells of wild type (BY4741 in panel A; Y0002 in panel B) and respective isogenic ubc mutant strains as indicated grown in raffinose medium. Hap4-HA was detected using Western blotting. (C) Hap4-HA is stabilized in ρ+ and ρ0 cells of a ubc1/4Δ mutant (ZLY3359) grown in dextrose or raffinose medium as indicated. (D) Quantification of Hap4-HA levels from ρ+ and ρ0 cells of wild type (WT, Y0002) and isogenic ubc1/4Δ mutant (ZLY3359) grown in dextrose and raffinose medium over the cycloheximide chase period. The means of remaining Hap4-HA levels during the chase period from two independent sets of experiments were plotted in the graphs.
We observed slower mobility forms of Hap4-HA on Western blots from protein samples of erg6 mutant cells treated with MG132 to inhibit the proteasome function (Figure 4B and Figure S5A). They are putative ubiquitinated forms of Hap4-HA. Consistently, these slower mobility forms are also observed in pre1 pre4 mutant cells with reduced proteasomal function, without MG132 treatment (Figure S5B). Our data in Figure 5 suggest that Ubc1 and Ubc4 are primary ubiquitin-conjugating enzymes responsible Hap4-HA turnover. A double mutation in UBC1 and UBC4 is expected to reduce ubiquitination of Hap4-HA. To test this possibility, we generated an erg6 ubc1 ubc4 triple mutant, grew cells in the presence of MG132, and examined Hap4-HA by Western blotting. Figure S5C shows that slower mobility forms of putative ubiquitinated Hap4-HA are barely detectable. Our data suggest that stability of Hap4-HA can be increased by reducing either the cellular proteasomal function or Hap4 ubiquitination.
3.6. Hap4 Stabilization Due to ubc1Δ ubc4Δ Increases Expression of Hap2/3/4/5-Target Genes
We next asked whether Hap4 turnover via the ubiquitin/proteasome system impacts the activity of the Hap2/3/4/5 complex. To this end, the expression of the lacZ reporter gene under the control of the promoter of KGD1 or SDH1 was analyzed in wild type, hap4Δ single mutant, ubc1/4Δ double mutant, and ubc1/4Δ hap4Δ triple mutant cells (Figure 6). SDH1 encodes the A subunit of the succinate dehydrogenase complex and is another target gene of the Hap2/3/4/5 complex [13,17,24]. In ρ+ cells grown in dextrose medium, ubc1/4Δ significantly increases the expression of these two reporter genes, and these increases are largely reversed in the ubc1/4Δ hap4Δ triple mutant (Figure 6A). Similar results were obtained in ρ+ cells grown in raffinose medium (Figure 6B). Importantly, hap4Δ completely abolishes increased expression of KGD1-lacZ and SDH1-lacZ reporter genes in ubc1/4Δ mutant cells grown in raffinose medium. These data indicate that Hap4 turnover mediated by Ubc1 and Ubc4 reduces the activity of the Hap2/3/4/5 complex.
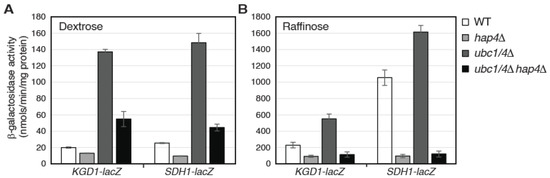
Figure 6.
Hap4 stabilization in ubc1Δ ubc4Δ double mutant cells results in increased expression of Hap2/3/4/5-target genes. β-galactosidase assays on the expression of KGD1-lacZ and SDH1-lacZ reporter genes in wild-type (WT, Y0002) and isogenic mutant strains (hap4Δ, AHY145; ubc1/4Δ, ZLY3359; ubc1/4Δ hap4Δ, ZLY3839) grown in dextrose (A) and raffinose media (B).
4. Discussion
Loss of mitochondrial DNA in yeast cells leads to reduced mitochondrial biogenesis by downregulating the protein level of Hap4. Here, we show that this downregulation is due to the combined effect of reduced transcription at the HAP4 promoter and increased turnover of the Hap4 protein. The mitochondrial genome encodes several key components of mitochondrial respiratory complexes [50]. Without mitochondrial DNA, cells cannot undergo oxidative phosphorylation. Many mitochondrial proteins involved in the tricarboxylic acid cycle and the electron transport chain are encoded in the nuclear genome and are under the control of the Hap2/3/4/5 complex. Regulation of the activity of Hap2/3/4/5 complex via modulating Hap4 expression helps to achieve mitochondrial homeostasis by coordinating gene expression from the nuclear and mitochondrial genomes. Our study reveals a negative regulatory mechanism of Hap4 through its degradation via the ubiquitin/proteasome system and provides important insights into the regulation of this important inter-organellar signaling process.
Our data is the first to describe ubiquitin/proteasome-dependent turnover of Hap4. We identified two ubiquitin-conjugating enzymes, Ubc1 and Ubc4, that are responsible for fast Hap4 turnover. Our findings echo the role of these two enzymes in the ubiquitination of fructose-1,6-bisphosphatase as well as substrates targeted by anaphase promoting complex (APC), a multi-subunit E3 ubiquitin ligase [51,52]. Ubc1, Ubc4, and Ubc5 have a redundant essential function [48,53]. All three are required for selective protein degradation, while Ubc1 seems to play a special role in the early stages of growth upon spore germination. Ubc4 and Ubc5 are closely related, with 94% sequence identity. These two enzymes and their orthologs from other species comprise the largest subfamily of E2 enzymes [54]. Therefore, it was surprising not to observe significant stabilization of Hap4 in the ubc1 ubc5 double mutant initially (Figure 5B). This is likely to be due to a higher expression level of Ubc4 compared to Ubc5. In a global analysis of protein expression in yeast, Ubc4 was found to be much more abundant than Ubc5 [55]. Consistently, a ubc1 ubc4 double mutant grows much slower than a ubc1 ubc5 double mutant [32]. Thus, in the ubc1 ubc4 double mutant, the lower level of Ubc5 may not be sufficient to carry out Hap4 ubiquitination and degradation. Nevertheless, it’s clear that Ubc1 and Ubc4 play a redundant role in Hap4 turnover. Ubc1, Ubc4, and Ubc5 are known to work with two E3 ubiquitin ligases, Rsp5 and APC [49,51]. However, we failed to detect Hap4 stabilization in an rsp5 mutant as well as in a mutant with a deletion mutation of DOC1, encoding a component of APC (our unpublished result).
Bourges et al. was unable to detect the signal of GFP-tagged Hap4 in cells with a single genomic copy using fluorescence microscopy [24]. When Hap4 was overexpressed, they found that Hap4-GFP was localized in the vacuole. Most of the data presented herein regarding the expression level and stability of Hap4 involved the use of HA-tagged Hap4 under the control of its own promoter. We did not detect vacuolar localization of GFP-tagged Hap4 under the control of its own promoter (our unpublished result). Whether vacuolar localization of Hap4 is an artifact of Hap4 overexpression is not a focus of this study. Future studies will be needed to determine potential contribution of the vacuolar degradation pathway to the regulation of Hap4.
Hap4 is stabilized in both ρ+ and ρ0 ubc1 ubc4 double mutants grown in either dextrose or raffinose medium (Figure 5). Therefore, proteasome-mediated Hap4 turnover seems to also play a housekeeping role by suppressing the activity of the Hap2/3/4/5 complex in ρ+ cells. Respiratory metabolism generates reactive oxygen species and oxidative stress [56,57]. Ubiquitin in yeast is encoded by four different genes, three of which form fusion genes with those encoding ribosomal proteins, and the fourth is UBI4, encoding a polyubiquitin chain comprising five head-to-tail repeats [58]. UBI4 expression is subject to glucose derepression mainly through the activation of the Hap2/3/4/5 complex and contributes to oxidative stress resistance in respiratory yeast cells [59,60]. Hap2/3/4/5-dependent expression of Ubi4 and ubiquitin-mediated Hap4 degradation may constitute a feedback control circuit to put a brake on respiratory metabolism. Many transcription factors are short-lived, which allows cells to respond quickly to changes in their environment. Ubiquitin-dependent degradation of transcription factors generally serves as a mechanism in down-regulating their activities. However, in some cases, ubiquitin-mediated turnover of transcription factors has been reported to play a stimulatory role in target gene expression [31]. This is unlikely to be the case for Hap4 since stabilization of Hap4 due to a ubc1 ubc4 double mutation increases the expression of Hap2/3/4/5-target genes (Figure 6).
What is the signal in ρ0 cells that leads to reduced Hap4 expression? This is unlikely to be the loss of mitochondrial DNA, per se. In a genome-wide gene expression analysis using various respiratory-deficient mutants as well as ρ0 cells, the expression of Hap2/3/4/5-target genes is reduced in all of these mutants [24]. In ρ0 cells and in an oxa1Δ mutant, which have defects in the assembly and function of three respiratory complexes, reduced expression of Hap2/3/4/5-target genes is greater compared to other respiratory-deficient mutants in which only one respiratory complex is affected. Therefore, it is likely that more than one signal in ρ0 cells may regulate Hap4 expression. Hap4 turnover has been reported to be under the control of the redox environment in cells. For example, increased production of reactive oxygen species in a tpk3Δ mutant grown in minimal lactate medium reduces the protein level of Hap4; conversely, an increased glutathione redox state due to cAMP treatment in a pde2Δ mutant grown in minimal lactate medium stabilizes Hap4 [25,27]. Since ρ0 cells have been reported to have reduced levels of reactive oxygen species and exhibit normal expression of oxidative stress responsive genes [24,61], it remains to be determined if reactive oxygen species play a role in reducing HAP4 expression in response to mitochondrial dysfunction.
The mechanism behind reduced Hap4 expression in ρ0 cells is expected to be different from that underlying intergenomic signaling, in which a set of nuclear genes are down-regulated in ρ0 cells but not in nuclear pet mutant ρ+ cells that possess mtDNA but lack respiration [3]. In the mitochondria-to-nucleus retrograde signaling pathway, we proposed that ATP might be a signaling molecule by mediating the interaction between Rtg2 and Mks1, which is a positive and negative regulator of this pathway, respectively [5,62]. Rtg2 binding to Mks1 leads to activation of retrograde signaling. Conversely, Mks1 dissociation from Rtg2 inhibits the two transcriptional activators of the pathway, Rtg1 and Rtg3. ATP, used at physiological concentrations, can dissociate Mks1 from Rtg2 from three different fungal species in an in vitro assay. Unlike vacuolar proteases, the ubiquitin/proteasome-dependent degradation of proteins is ATP-dependent [63]. ρ0 cells have similar levels of ATP to that of ρ+ cells when grown in dextrose medium and much lower ATP levels than ρ+ cells when glucose is exhausted [64,65]. Increased turnover of Hap4 in ρ0 cells is thus unlikely to be the result of direct ATP sensing via the ubiquitin/proteasome system. Future work will be directed toward the elucidation of the mechanisms underlying Hap4 turnover and transcriptional regulation of HAP4. Identification and characterization of the involved protein factors will help to understand how mitochondrial homeostasis is achieved through coordinated expression of genes encoding mitochondrial proteins from the mitochondrial and nuclear genomes, a process central to the growth and metabolism in eukaryotic cells.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms10122370/s1, Figure S1: HA epitope-tagged Hap4 is functional; Figure S2: Hap4-HA is less stable in ρ0 cells compared to ρ+ cells; Figure S3: Hap4-HA has a shorter half-life in ρ0 cells than in ρ+ cells; Figure S4: The half-lives of Hap4-HA in ρ+ and ρ0 cells of wild type Y0002 strain grown in dextrose and raffinose media; Figure S5: Reduced proteasomal function leads to the appearance of slower mobility forms of Hap4-HA on Western blot.
Author Contributions
Z.L. conceived, designed, coordinated the study, and wrote the paper; D.C. designed, performed, and analyzed the experiments shown in Figure 1, Figure 2, Figure 3 and Figure 4; A.H. carried out the screening to identify the E2 enzymes responsible for Hap4 turnover and generated data used in Figure 5; M.C. designed, performed, and analyzed the experiments shown in Figure 6; T.P. generated strains, assisted coordination of the study, provided technical assistance, and contributed to the writing of the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by US National Institutes of Health grant 1R15GM094772-01A1, Louisiana Governor’s Biotechnology Initiative, and College of Sciences Undergraduate Research Program (COSURP) from The University of New Orleans.
Acknowledgments
We thank Peter Kaiser, Stefan Jentsch, and Raymond Deshaies for yeast strains, Chen Zhang and Flavio Antonio de Oliveira Simoes for technical support, Sylvester Tumusiime for critical reading of the manuscript, the W. M. Keck Foundation for the Keck Facility, and Robin Rowe for sequencing.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Tzagoloff, A.; Myers, A.M. Genetics of mitochondrial biogenesis. Annu. Rev. Biochem. 1986, 55, 249–285. [Google Scholar] [CrossRef] [PubMed]
- Hock, M.B.; Kralli, A. Transcriptional control of mitochondrial biogenesis and function. Annu. Rev. Physiol. 2009, 71, 177–203. [Google Scholar] [CrossRef] [PubMed]
- Woo, D.K.; Phang, T.L.; Trawick, J.D.; Poyton, R.O. Multiple pathways of mitochondrial-nuclear communication in yeast: Intergenomic signaling involves ABF1 and affects a different set of genes than retrograde regulation. Biochim. Biophys. Acta 2009, 1789, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Butow, R.A.; Avadhani, N.G. Mitochondrial signaling: The retrograde response. Mol. Cell 2004, 14, 1–15. [Google Scholar] [CrossRef]
- Liu, Z.; Butow, R.A. Mitochondrial retrograde signaling. Annu. Rev. Genet. 2006, 40, 159–185. [Google Scholar] [CrossRef]
- Moye-Rowley, W.S. Retrograde regulation of multidrug resistance in Saccharomyces cerevisiae. Gene 2005, 354, 15–21. [Google Scholar] [CrossRef]
- Paul, S.; Moye-Rowley, W.S. Multidrug resistance in fungi: Regulation of transporter-encoding gene expression. Front. Physiol. 2014, 5, 143. [Google Scholar] [CrossRef]
- Miceli, M.V.; Jiang, J.C.; Tiwari, A.; Rodriguez-Quinones, J.F.; Jazwinski, S.M. Loss of mitochondrial membrane potential triggers the retrograde response extending yeast replicative lifespan. Front. Genet. 2011, 2, 102. [Google Scholar] [CrossRef]
- McNabb, D.S.; Xing, Y.; Guarente, L. Cloning of yeast HAP5: A novel subunit of a heterotrimeric complex required for CCAAT binding. Genes Dev. 1995, 9, 47–58. [Google Scholar] [CrossRef]
- Rosenkrantz, M.; Kell, C.S.; Pennell, E.A.; Devenish, L.J. The HAP2,3,4 transcriptional activator is required for derepression of the yeast citrate synthase gene, CIT1. Mol. Microbiol. 1994, 13, 119–131. [Google Scholar] [CrossRef]
- Olesen, J.T.; Guarente, L. The HAP2 subunit of yeast CCAAT transcriptional activator contains adjacent domains for subunit association and DNA recognition: Model for the HAP2/3/4 complex. Genes Dev. 1990, 4, 1714–1729. [Google Scholar] [CrossRef] [PubMed]
- Forsburg, S.L.; Guarente, L. Identification and characterization of HAP4: A third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev. 1989, 3, 1166–1178. [Google Scholar] [CrossRef] [PubMed]
- Buschlen, S.; Amillet, J.M.; Guiard, B.; Fournier, A.; Marcireau, C.; Bolotin-Fukuhara, M. The S. cerevisiae HAP complex, a key regulator of mitochondrial function, coordinates nuclear and mitochondrial gene expression. Comp. Funct. Genom. 2003, 4, 37–46. [Google Scholar] [CrossRef] [PubMed]
- McNabb, D.S.; Pinto, I. Assembly of the Hap2p/Hap3p/Hap4p/Hap5p-DNA complex in Saccharomyces cerevisiae. Eukaryot. Cell 2005, 4, 1829–1839. [Google Scholar] [CrossRef]
- DeRisi, J.L.; Iyer, V.R.; Brown, P.O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 1997, 278, 680–686. [Google Scholar] [CrossRef]
- Blom, J.; De Mattos, M.J.; Grivell, L.A. Redirection of the respiro-fermentative flux distribution in Saccharomyces cerevisiae by overexpression of the transcription factor Hap4p. Appl. Environ. Microbiol. 2000, 66, 1970–1973. [Google Scholar] [CrossRef]
- Lascaris, R.; Bussemaker, H.J.; Boorsma, A.; Piper, M.; van der Spek, H.; Grivell, L.; Blom, J. Hap4p overexpression in glucose-grown Saccharomyces cerevisiae induces cells to enter a novel metabolic state. Genome Biol. 2003, 4, R3. [Google Scholar] [CrossRef]
- Kundaje, A.; Xin, X.; Lan, C.; Lianoglou, S.; Zhou, M.; Zhang, L.; Leslie, C. A predictive model of the oxygen and heme regulatory network in yeast. PLoS Comput. Biol. 2008, 4, e1000224. [Google Scholar] [CrossRef]
- Brons, J.F.; De Jong, M.; Valens, M.; Grivell, L.A.; Bolotin-Fukuhara, M.; Blom, J. Dissection of the promoter of the HAP4 gene in S. cerevisiae unveils a complex regulatory framework of transcriptional regulation. Yeast 2002, 19, 923–932. [Google Scholar] [CrossRef]
- Zhang, T.; Bu, P.; Zeng, J.; Vancura, A. Increased heme synthesis in yeast induces a metabolic switch from fermentation to respiration even under conditions of glucose repression. J. Biol. Chem. 2017, 292, 16942–16954. [Google Scholar] [CrossRef]
- Deckert, J.; Rodriguez Torres, A.M.; Simon, J.T.; Zitomer, R.S. Mutational analysis of Rox1, a DNA-bending repressor of hypoxic genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 1995, 15, 6109–6117. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Peterson, P.P.; Liu, Z. Identification and Characterization of Rapidly Accumulating sch9Δ Suppressor Mutations in Saccharomyces cerevisiae. G3 (Bethesda) 2021, 11, jkab134. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Butow, R.A. A transcriptional switch in the expression of yeast tricarboxylic acid cycle genes in response to a reduction or loss of respiratory function. Mol. Cell. Biol. 1999, 19, 6720–6728. [Google Scholar] [CrossRef] [PubMed]
- Bourges, I.; Mucchielli, M.H.; Herbert, C.J.; Guiard, B.; Dujardin, G.; Meunier, B. Multiple defects in the respiratory chain lead to the repression of genes encoding components of the respiratory chain and TCA cycle enzymes. J. Mol. Biol. 2009, 387, 1081–1091. [Google Scholar] [CrossRef]
- Chevtzoff, C.; Yoboue, E.D.; Galinier, A.; Casteilla, L.; Daignan-Fornier, B.; Rigoulet, M.; Devin, A. Reactive oxygen species-mediated regulation of mitochondrial biogenesis in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 2010, 285, 1733–1742. [Google Scholar] [CrossRef]
- Bouchez, C.L.; Yoboue, E.D.; de la Rosa Vargas, L.E.; Salin, B.; Cuvellier, S.; Rigoulet, M.; Duvezin-Caubet, S.; Devin, A. “Labile” heme critically regulates mitochondrial biogenesis through the transcriptional co-activator Hap4p in Saccharomyces cerevisiae. J. Biol. Chem. 2020, 295, 5095–5109. [Google Scholar] [CrossRef]
- Yoboue, E.D.; Augier, E.; Galinier, A.; Blancard, C.; Pinson, B.; Casteilla, L.; Rigoulet, M.; Devin, A. cAMP-induced mitochondrial compartment biogenesis: Role of glutathione redox state. J. Biol. Chem. 2012, 287, 14569–14578. [Google Scholar] [CrossRef]
- Amberg, D.C.; Burke, D.J.; Strathern, J.N. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual; Cold Spring Harbor Laboratory: New York, NY, USA, 2005. [Google Scholar]
- Morris, M.C.; Kaiser, P.; Rudyak, S.; Baskerville, C.; Watson, M.H.; Reed, S.I. Cks1-dependent proteasome recruitment and activation of CDC20 transcription in budding yeast. Nature 2003, 423, 1009–1013. [Google Scholar] [CrossRef]
- Kaiser, P.; Sia, R.A.; Bardes, E.G.; Lew, D.J.; Reed, S.I. Cdc34 and the F-box protein Met30 are required for degradation of the Cdk-inhibitory kinase Swe1. Genes Dev. 1998, 12, 2587–2597. [Google Scholar] [CrossRef]
- Lipford, J.R.; Smith, G.T.; Chi, Y.; Deshaies, R.J. A putative stimulatory role for activator turnover in gene expression. Nature 2005, 438, 113–116. [Google Scholar] [CrossRef]
- Seufert, W.; McGrath, J.P.; Jentsch, S. UBC1 encodes a novel member of an essential subfamily of yeast ubiquitin-conjugating enzymes involved in protein degradation. EMBO J. 1990, 9, 4535–4541. [Google Scholar] [CrossRef] [PubMed]
- Yaffe, M.P.; Schatz, G. Two nuclear mutations that block mitochondrial protein import in yeast. Proc. Natl. Acad. Sci. USA 1984, 81, 4819–4823. [Google Scholar] [CrossRef]
- Chelstowska, A.; Liu, Z.; Jia, Y.; Amberg, D.; Butow, R.A. Signalling between mitochondria and the nucleus regulates the expression of a new D-lactate dehydrogenase activity in yeast. Yeast 1999, 15, 1377–1391. [Google Scholar] [CrossRef]
- Liao, X.S.; Small, W.C.; Srere, P.A.; Butow, R.A. Intramitochondrial functions regulate nonmitochondrial citrate synthase (CIT2) expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 1991, 11, 38–46. [Google Scholar] [PubMed]
- Repetto, B.; Tzagoloff, A. Structure and regulation of KGD1, the structural gene for yeast alpha-ketoglutarate dehydrogenase. Mol. Cell. Biol. 1989, 9, 2695–2705. [Google Scholar] [PubMed]
- Fontanesi, F.; Jin, C.; Tzagoloff, A.; Barrientos, A. Transcriptional activators HAP/NF-Y rescue a cytochrome c oxidase defect in yeast and human cells. Hum. Mol. Genet. 2008, 17, 775–788. [Google Scholar] [CrossRef]
- Mumberg, D.; Muller, R.; Funk, M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 1995, 156, 119–122. [Google Scholar] [CrossRef]
- Epstein, C.B.; Waddle, J.A.; Hale, W.t.; Dave, V.; Thornton, J.; Macatee, T.L.; Garner, H.R.; Butow, R.A. Genome-wide responses to mitochondrial dysfunction. Mol. Biol. Cell 2001, 12, 297–308. [Google Scholar] [CrossRef]
- Mayor, T.; Graumann, J.; Bryan, J.; MacCoss, M.J.; Deshaies, R.J. Quantitative profiling of ubiquitylated proteins reveals proteasome substrates and the substrate repertoire influenced by the Rpn10 receptor pathway. Mol. Cell. Proteom. 2007, 6, 1885–1895. [Google Scholar] [CrossRef]
- Lee, D.H.; Goldberg, A.L. Selective inhibitors of the proteasome-dependent and vacuolar pathways of protein degradation in Saccharomyces cerevisiae. J. Biol. Chem. 1996, 271, 27280–27284. [Google Scholar] [CrossRef]
- Hochstrasser, M. Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 1996, 30, 405–439. [Google Scholar] [CrossRef] [PubMed]
- Devaux, F.; Carvajal, E.; Moye-Rowley, S.; Jacq, C. Genome-wide studies on the nuclear PDR3-controlled response to mitochondrial dysfunction in yeast. FEBS Lett. 2002, 515, 25–28. [Google Scholar] [CrossRef] [PubMed]
- Fleming, J.A.; Lightcap, E.S.; Sadis, S.; Thoroddsen, V.; Bulawa, C.E.; Blackman, R.K. Complementary whole-genome technologies reveal the cellular response to proteasome inhibition by PS-341. Proc. Natl. Acad. Sci. USA 2002, 99, 1461–1466. [Google Scholar] [CrossRef] [PubMed]
- Hershko, A.; Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef]
- Varshavsky, A. The ubiquitin system. Trends Biochem. Sci 1997, 22, 383–387. [Google Scholar] [CrossRef]
- Johnson, E.S.; Blobel, G. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J. Biol. Chem. 1997, 272, 26799–26802. [Google Scholar] [CrossRef]
- Seufert, W.; Jentsch, S. Yeast ubiquitin-conjugating enzymes involved in selective protein degradation are essential for cell viability. Acta Biol. Hung. 1991, 42, 27–37. [Google Scholar]
- Stoll, K.E.; Brzovic, P.S.; Davis, T.N.; Klevit, R.E. The essential Ubc4/Ubc5 function in yeast is HECT E3-dependent, and RING E3-dependent pathways require only monoubiquitin transfer by Ubc4. J. Biol. Chem. 2011, 286, 15165–15170. [Google Scholar] [CrossRef]
- Foury, F.; Roganti, T.; Lecrenier, N.; Purnelle, B. The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. FEBS Lett. 1998, 440, 325–331. [Google Scholar] [CrossRef]
- Rodrigo-Brenni, M.C.; Morgan, D.O. Sequential E2s drive polyubiquitin chain assembly on APC targets. Cell 2007, 130, 127–139. [Google Scholar] [CrossRef]
- Schork, S.M.; Thumm, M.; Wolf, D.H. Catabolite inactivation of fructose-1,6-bisphosphatase of Saccharomyces cerevisiae. Degradation occurs via the ubiquitin pathway. J. Biol. Chem. 1995, 270, 26446–26450. [Google Scholar] [CrossRef] [PubMed]
- Seufert, W.; Jentsch, S. Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. EMBO J. 1990, 9, 543–550. [Google Scholar] [CrossRef] [PubMed]
- Michelle, C.; Vourc’h, P.; Mignon, L.; Andres, C.R. What was the set of ubiquitin and ubiquitin-like conjugating enzymes in the eukaryote common ancestor? J. Mol. Evol. 2009, 68, 616–628. [Google Scholar] [CrossRef] [PubMed]
- Ghaemmaghami, S.; Huh, W.K.; Bower, K.; Howson, R.W.; Belle, A.; Dephoure, N.; O’Shea, E.K.; Weissman, J.S. Global analysis of protein expression in yeast. Nature 2003, 425, 737–741. [Google Scholar] [CrossRef] [PubMed]
- Morano, K.A.; Grant, C.M.; Moye-Rowley, W.S. The response to heat shock and oxidative stress in Saccharomyces cerevisiae. Genetics 2012, 190, 1157–1195. [Google Scholar] [CrossRef]
- Halliwell, B. Reactive species and antioxidants. Redox biology is a fundamental theme of aerobic life. Plant Physiol. 2006, 141, 312–322. [Google Scholar] [CrossRef]
- Ozkaynak, E.; Finley, D.; Solomon, M.J.; Varshavsky, A. The yeast ubiquitin genes: A family of natural gene fusions. EMBO J. 1987, 6, 1429–1439. [Google Scholar] [CrossRef]
- Cheng, L.; Watt, R.; Piper, P.W. Polyubiquitin gene expression contributes to oxidative stress resistance in respiratory yeast (Saccharomyces cerevisiae). Mol. Gen. Genet. 1994, 243, 358–362. [Google Scholar] [CrossRef]
- Watt, R.; Piper, P.W. UBI4, the polyubiquitin gene of Saccharomyces cerevisiae, is a heat shock gene that is also subject to catabolite derepression control. Mol. Gen. Genet. 1997, 253, 439–447. [Google Scholar] [CrossRef]
- Rasmussen, A.K.; Chatterjee, A.; Rasmussen, L.J.; Singh, K.K. Mitochondria-mediated nuclear mutator phenotype in Saccharomyces cerevisiae. Nucleic Acids Res. 2003, 31, 3909–3917. [Google Scholar] [CrossRef]
- Zhang, F.; Pracheil, T.; Thornton, J.; Liu, Z. Adenosine Triphosphate (ATP) Is a Candidate Signaling Molecule in the Mitochondria-to-Nucleus Retrograde Response Pathway. Genes 2013, 4, 86–100. [Google Scholar] [CrossRef] [PubMed]
- Hershko, A.; Leshinsky, E.; Ganoth, D.; Heller, H. ATP-dependent degradation of ubiquitin-protein conjugates. Proc. Natl. Acad. Sci. USA 1984, 81, 1619–1623. [Google Scholar] [CrossRef] [PubMed]
- Ocampo, A.; Liu, J.; Schroeder, E.A.; Shadel, G.S.; Barrientos, A. Mitochondrial respiratory thresholds regulate yeast chronological life span and its extension by caloric restriction. Cell Metab. 2012, 16, 55–67. [Google Scholar] [CrossRef] [PubMed]
- Vowinckel, J.; Hartl, J.; Marx, H.; Kerick, M.; Runggatscher, K.; Keller, M.A.; Mülleder, M.; Day, J.; Weber, M.; Rinnerthaler, M.; et al. The metabolic growth limitations of petite cells lacking the mitochondrial genome. Nat. Metab. 2021, 3, 1521–1535. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).