Identification of Novel Bile Salt-Tolerant Genes in Lactobacillus Using Comparative Genomics and Its Application in the Rapid Screening of Tolerant Strains
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, and Growth Conditions
2.2. Bile Salt Tolerance Evaluation
2.3. Genome Sequencing and Annotation
2.4. Comparative Genomics
2.4.1. Pan-Genome Analysis, Homologous Gene Analysis, and Phylogenomic Tree Inference
2.4.2. Tolerant and Non-Tolerant Strains Comparative Genomic Analysis
2.5. Quantitative Real-Time PCR Analysis
2.6. Plasmid Construction and Expression
2.7. Development of a Method for the Rapid Screening of Bile Salt-Tolerant Lactobacilli
2.8. Statistical Analysis
3. Results
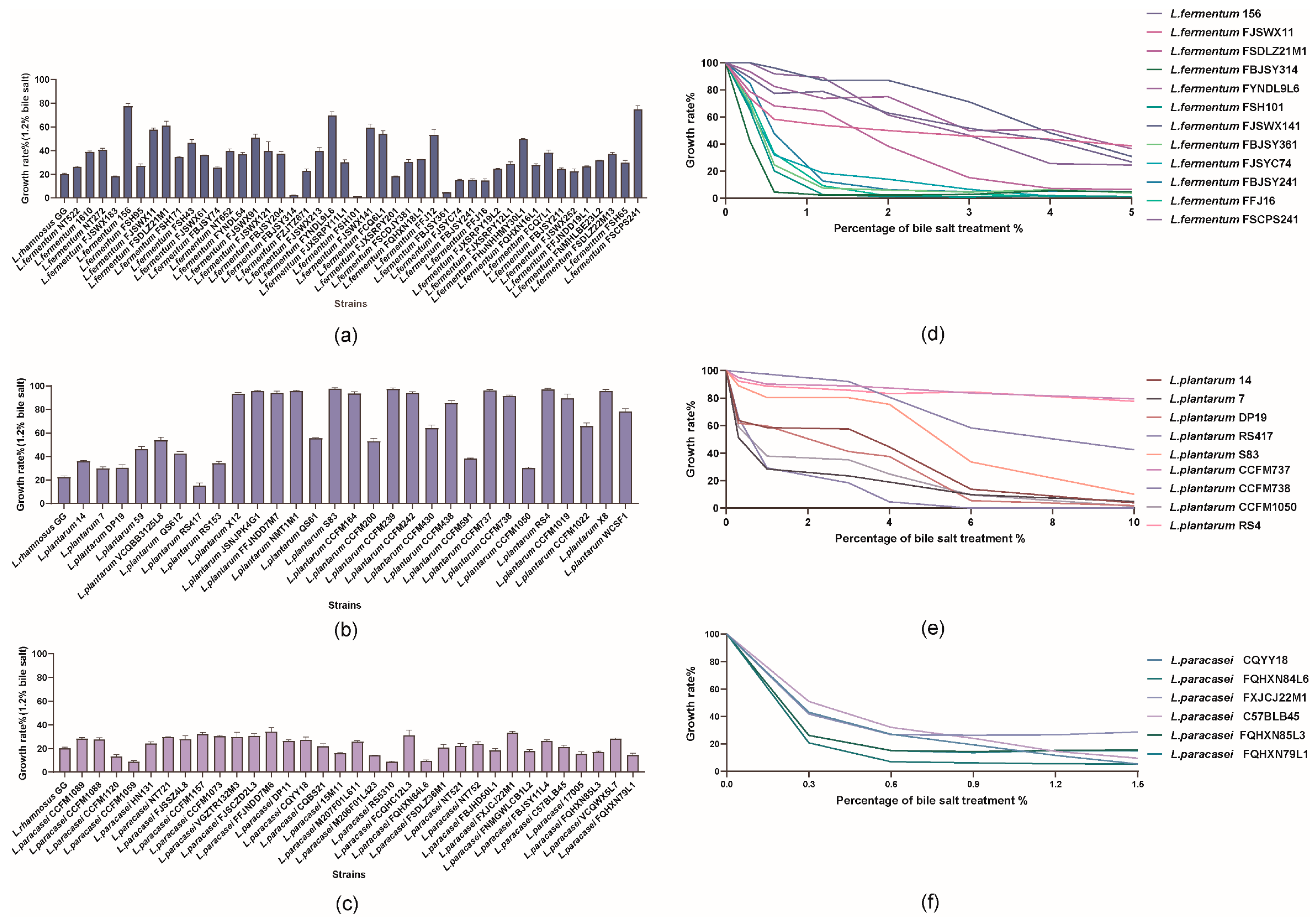
3.1. Effect of Bile Salt on the Growth of 107 Lactobacilli
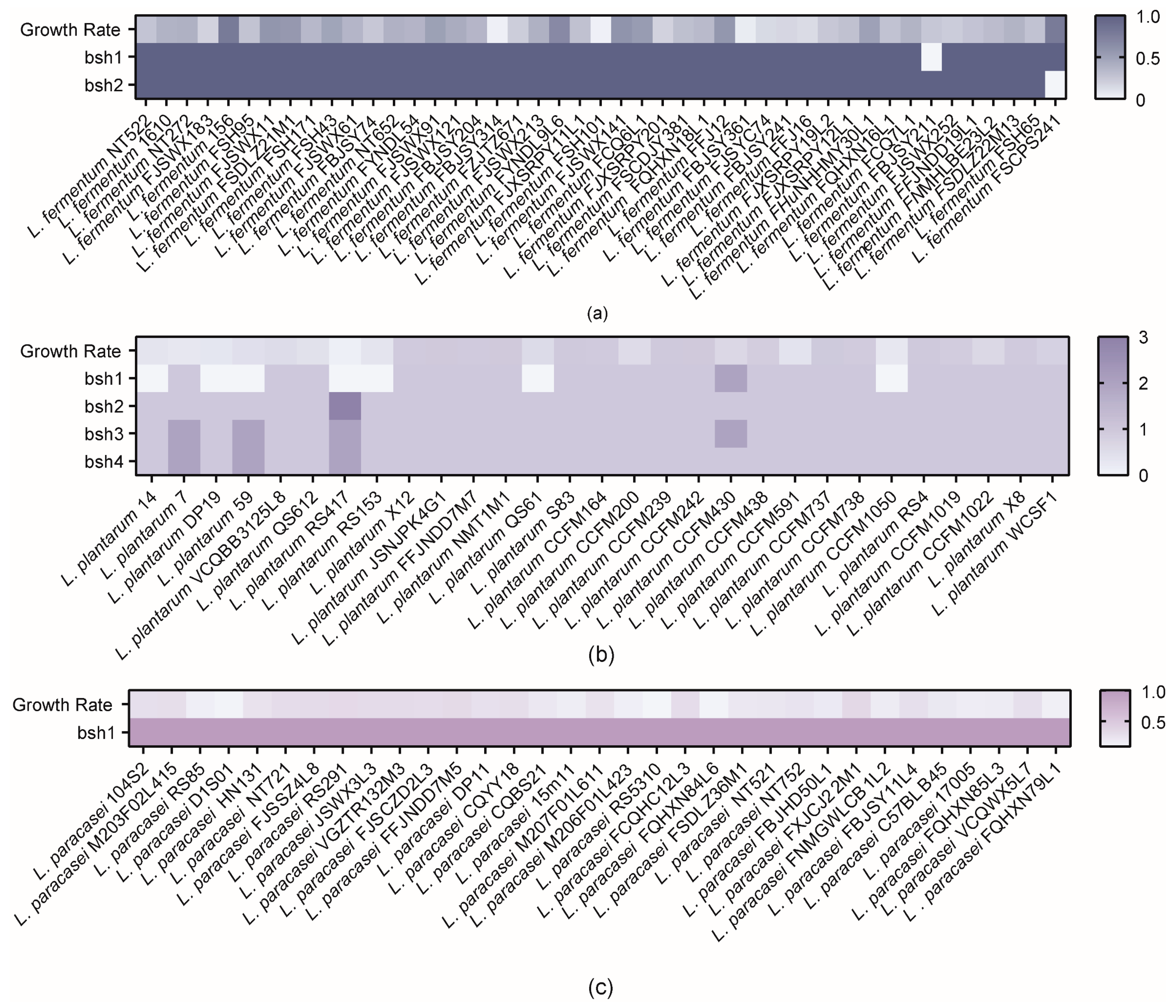
3.2. Correlation Analysis of BSHs with Bile Salt Tolerance Phenotypes
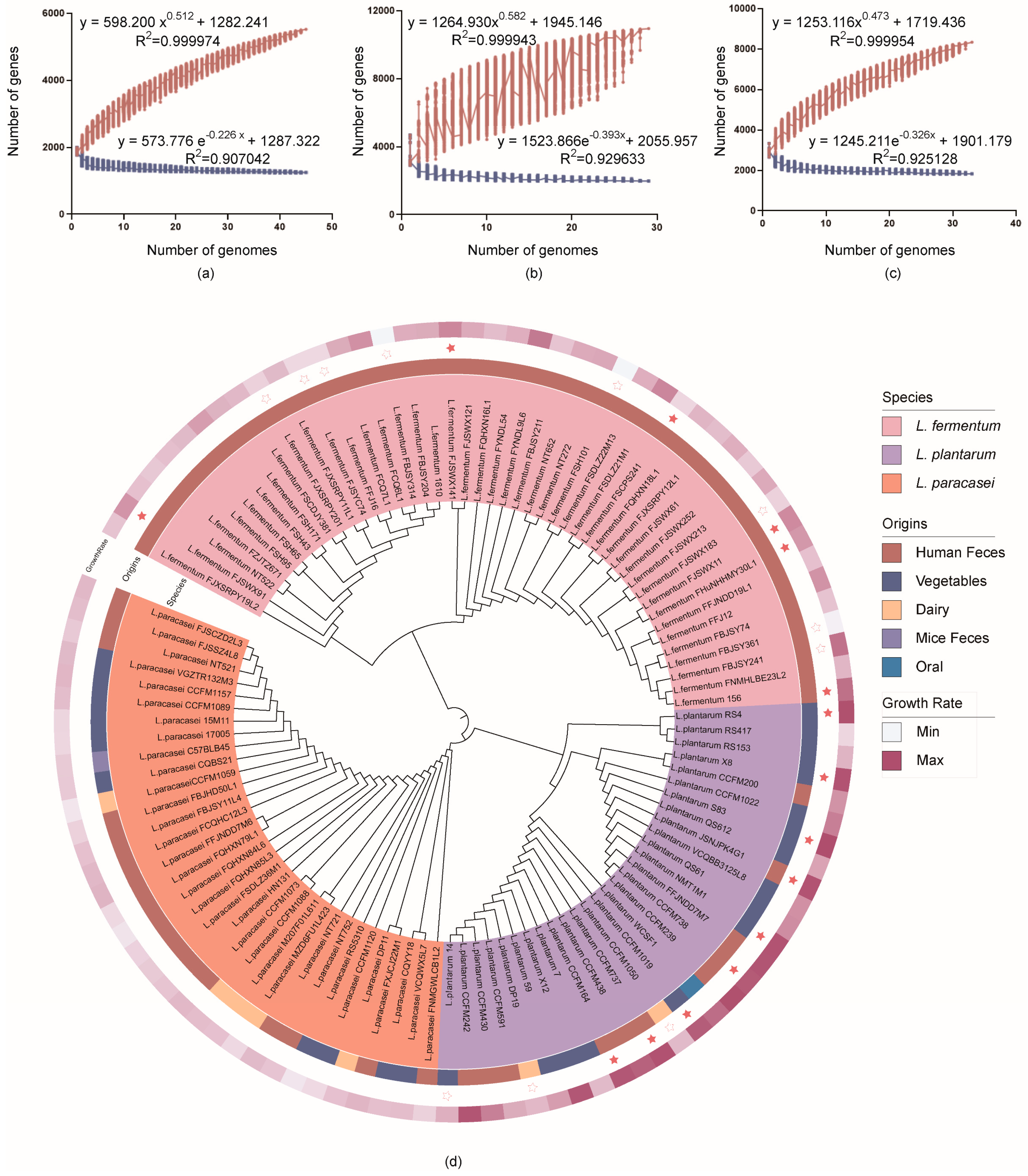
3.3. In Silico Analysis of Genome Features and Phylogenetic Analysis
3.4. Comparative Genomics Analysis
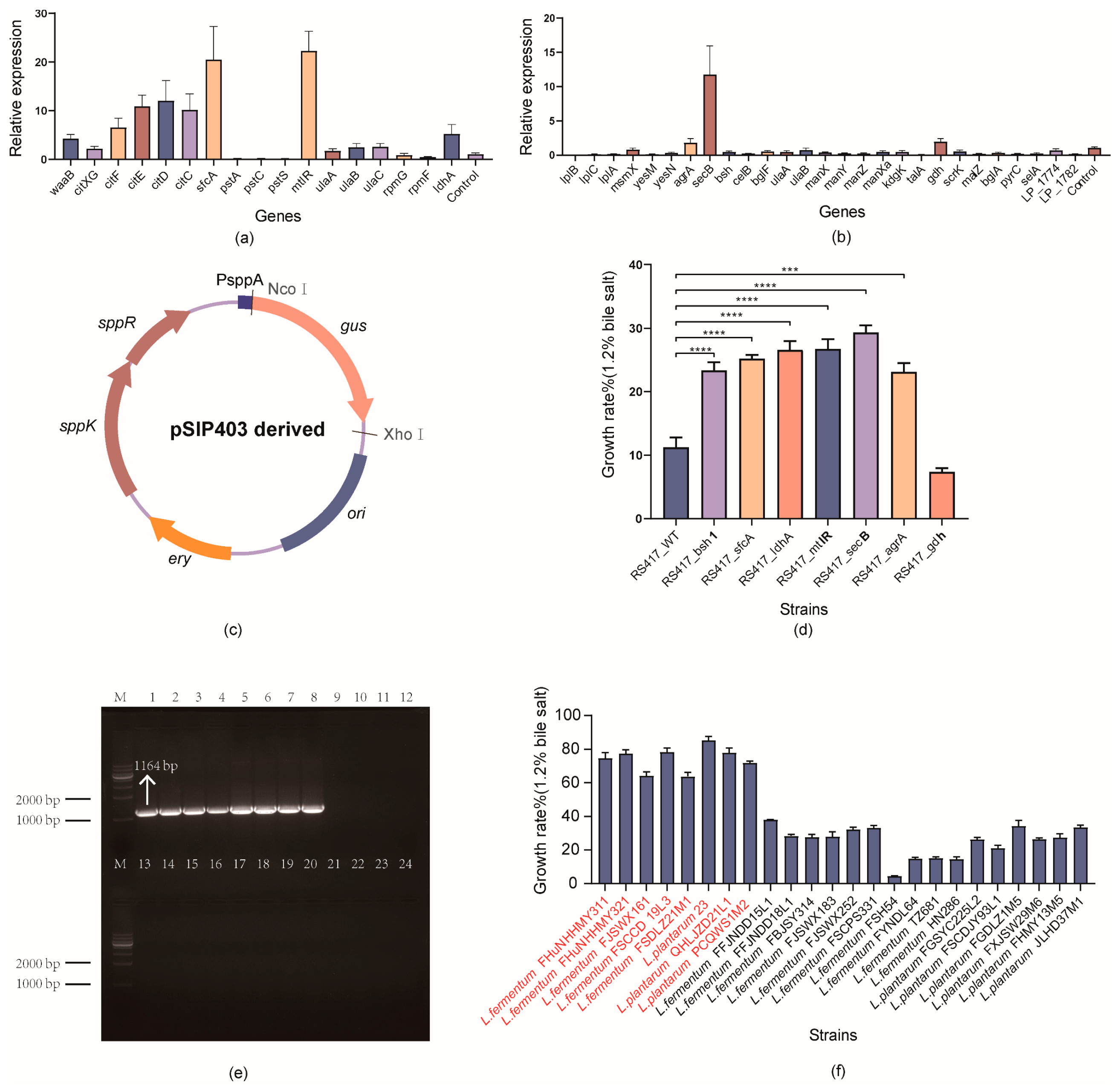
3.4.1. Potential Functional Genes Responsible for Bile Salt Tolerance
3.4.2. Potential Redundant Genes Responsible for Bile Salt Tolerance
3.5. Quantitative Real-Time PCR (qPCR) Assay for Detecting the Transcriptional Changes of Functional Genes
3.6. Validation of the Functional Genes via Overexpression in Lactobacillus
3.7. Sensitivity and Specificity of PCR Amplification Using the Developed Primer
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wauters, L.; Van Oudenhove, L.; Accarie, A.; Geboers, K.; Geysen, H.; Toth, J.; Luypaerts, A.; Verbeke, K.; Smokvina, T.; Raes, J.; et al. Lactobacillus Rhamnosus CNCM I-3690 Decreases Subjective Academic Stress in Healthy Adults: A Randomized Placebo-Controlled Trial. Gut Microbes 2022, 14, 2031695. [Google Scholar] [CrossRef]
- Rodríguez-Sánchez, S.; Ramos, I.M.; Seseña, S.; Poveda, J.M.; Palop, M.L. Potential of Lactobacillus Strains for Health-Promotion and Flavouring of Fermented Dairy Foods. LWT 2021, 143, 111102. [Google Scholar] [CrossRef]
- Begley, M.; Gahan, C.G.M.; Hill, C. The Interaction between Bacteria and Bile. FEMS Microbiol. Rev. 2005, 29, 625–651. [Google Scholar] [CrossRef] [PubMed]
- van Zyl, W.F.; Deane, S.M.; Dicks, L.M.T. Molecular Insights into Probiotic Mechanisms of Action Employed against Intestinal Pathogenic Bacteria. Gut Microbes 2020, 12, 1831339. [Google Scholar] [CrossRef] [PubMed]
- Kenfack, C.; Kaktcham, P.; Ngoufack, F.; Wang, Y.; Yin, L.; Zhu, T. Screening and Characterization of Putative Probiotic Lactobacillus Strains from Honey Bee Gut (Apis Mellifera). J. Adv. Microbiol. 2018, 10, 1–18. [Google Scholar] [CrossRef]
- Reale, A.; Di Renzo, T.; Rossi, F.; Zotta, T.; Iacumin, L.; Preziuso, M.; Parente, E.; Sorrentino, E.; Coppola, R. Tolerance of Lactobacillus Casei, Lactobacillus Paracasei and Lactobacillus Rhamnosus Strains to Stress Factors Encountered in Food Processing and in the Gastro-Intestinal Tract. LWT Food Sci. Technol. 2015, 60, 721–728. [Google Scholar] [CrossRef]
- Merritt, M.E.; Donaldson, J.R. Effect of Bile Salts on the DNA and Membrane Integrity of Enteric Bacteria. J. Med. Microbiol. 2009, 58, 1533–1541. [Google Scholar] [CrossRef]
- Ruiz, L.; Margolles, A.; Sánchez, B. Bile Resistance Mechanisms in Lactobacillus and Bifidobacterium. Front. Microbiol. 2013, 4, 396. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.-X.; Yan, R.; Shi, H.-Y.; Shi, D.; Fang, D.-Q.; Jiang, H.-Y.; Wu, W.-R.; Guo, F.-F.; Jiang, X.-W.; Gu, S.-L.; et al. Integrated Transcriptomic and Proteomic Analysis of the Bile Stress Response in Probiotic Lactobacillus Salivarius LI01. J. Proteomics 2017, 150, 216–229. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Wang, G.; Zhai, Z.; Zhou, P.; Hao, Y. Global Transcriptomic Analysis and Function Identification of Malolactic Enzyme Pathway of Lactobacillus Paracasei L9 in Response to Bile Stress. Front. Microbiol. 2018, 9, 1978. [Google Scholar] [CrossRef] [PubMed]
- Hamon, E.; Horvatovich, P.; Izquierdo, E.; Bringel, F.; Marchioni, E.; Aoudé-Werner, D.; Ennahar, S. Comparative Proteomic Analysis of Lactobacillus Plantarum for the Identification of Key Proteins in Bile Tolerance. BMC Microbiol. 2011, 11, 63. [Google Scholar] [CrossRef] [PubMed]
- Koskenniemi, K.; Laakso, K.; Koponen, J.; Kankainen, M.; Greco, D.; Auvinen, P.; Savijoki, K.; Nyman, T.A.; Surakka, A. Proteomics and Transcriptomics Characterization of Bile Stress Response in Probiotic Lactobacillus Rhamnosus GG. Mol. Cell. Proteomics 2011, 10, 18. [Google Scholar] [CrossRef] [PubMed]
- Hamon, E.; Horvatovich, P.; Bisch, M.; Bringel, F.; Marchioni, E.; Aoudé-Werner, D.; Ennahar, S. Investigation of Biomarkers of Bile Tolerance in Lactobacillus Casei Using Comparative Proteomics. J. Proteome Res. 2012, 11, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Kwon, E.-A.; Lee, J.-I.; Park, J.W.; Kim, S.-S. Application of Comparative Genomics in the Development of DNA Probes to Detect Bacillus Cereus and Bacillus Subtilis. LWT 2021, 142, 110996. [Google Scholar] [CrossRef]
- Lee, J.-I.; Kim, S.-S.; Kang, D.-H. Development of DNA Probes to Detect Cronobacter Sakazakii Based on Comparative Genomics and Its Application in Food Samples. Food Control 2022, 137, 108853. [Google Scholar] [CrossRef]
- Sun, Z.; Liu, W.; Song, Y.; Xu, H.; Yu, J.; Bilige, M.; Zhang, H.; Chen, Y. Population Structure of Lactobacillus Helveticus Isolates from Naturally Fermented Dairy Products Based on Multilocus Sequence Typing. J. Dairy Sci. 2015, 98, 2962–2972. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Pajarillo, E.A.B.; Kim, M.J.; Chae, J.P.; Kang, D.-K. Proteomic and Transcriptional Analysis of Lactobacillus Johnsonii PF01 during Bile Salt Exposure by ITRAQ Shotgun Proteomics and Quantitative RT-PCR. J. Proteome Res. 2013, 12, 432–443. [Google Scholar] [CrossRef] [PubMed]
- Kaur, G.; Ali, S.A.; Kumar, S.; Mohanty, A.K.; Behare, P. Label-Free Quantitative Proteomic Analysis of Lactobacillus Fermentum NCDC 400 during Bile Salt Exposure. J. Proteomics 2017, 167, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Turpin, W.; Humblot, C.; Guyot, J.-P. Genetic Screening of Functional Properties of Lactic Acid Bacteria in a Fermented Pearl Millet Slurry and in the Metagenome of Fermented Starchy Foods. Appl. Environ. Microbiol. 2011, 77, 8722–8734. [Google Scholar] [CrossRef]
- Huang, Z.; Zhou, X.; Stanton, C.; Ross, R.P.; Zhao, J.; Zhang, H.; Yang, B.; Chen, W. Comparative Genomics and Specific Functional Characteristics Analysis of Lactobacillus Acidophilus. Microorganisms 2021, 9, 1992. [Google Scholar] [CrossRef] [PubMed]
- García-Ruiz, A.; González de Llano, D.; Esteban-Fernández, A.; Requena, T.; Bartolomé, B.; Moreno-Arribas, M.V. Assessment of Probiotic Properties in Lactic Acid Bacteria Isolated from Wine. Food Microbiol. 2014, 44, 220–225. [Google Scholar] [CrossRef]
- Besemer, J. GeneMarkS: A Self-Training Method for Prediction of Gene Starts in Microbial Genomes. Implications for Finding Sequence Motifs in Regulatory Regions. Nucleic Acids Res. 2001, 29, 2607–2618. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG Database: A Tool for Genome-Scale Analysis of Protein Functions and Evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef]
- Li, L.; Stoeckert, C.J., Jr.; Roos, D.S. OrthoMCL: Identification of Ortholog Groups for Eukaryotic Genomes. Genome Res. 2003, 13, 2178–2189. [Google Scholar] [CrossRef]
- Katoh, K. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Aukrust, T.; Blom, H. Transformation of Lactobacillus Strains Used in Meat and Vegetable Fermentations. Food Res. Int. 1992, 25, 253–261. [Google Scholar] [CrossRef]
- Nguyen, T.-T.; Mathiesen, G.; Fredriksen, L.; Kittl, R.; Nguyen, T.-H.; Eijsink, V.G.H.; Haltrich, D.; Peterbauer, C.K. A Food-Grade System for Inducible Gene Expression in Lactobacillus Plantarum Using an Alanine Racemase-Encoding Selection Marker. J. Agric. Food Chem. 2011, 59, 5617–5624. [Google Scholar] [CrossRef] [PubMed]
- Foley, M.H.; O’Flaherty, S.; Allen, G.; Rivera, A.J.; Stewart, A.K.; Barrangou, R.; Theriot, C.M. Lactobacillus Bile Salt Hydrolase Substrate Specificity Governs Bacterial Fitness and Host Colonization. Proc. Natl. Acad. Sci. USA 2021, 118, e2017709118. [Google Scholar] [CrossRef] [PubMed]
- Jayashree, S.; Pooja, S.; Pushpanathan, M.; Rajendhran, J.; Gunasekaran, P. Identification and Characterization of Bile Salt Hydrolase Genes from the Genome of Lactobacillus Fermentum MTCC 8711. Appl. Biochem. Biotechnol. 2014, 174, 855–866. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Zhang, Y.; Zhang, Y.; Liu, Y.; Wang, S.; Dong, X.; Wang, Y.; Zhang, H. Screening of Potential Probiotic Properties of Lactobacillus Fermentum Isolated from Traditional Dairy Products. Food Control 2010, 21, 695–701. [Google Scholar] [CrossRef]
- Leite, A.M.O.; Miguel, M.A.L.; Peixoto, R.S.; Ruas-Madiedo, P.; Paschoalin, V.M.F.; Mayo, B.; Delgado, S. Probiotic Potential of Selected Lactic Acid Bacteria Strains Isolated from Brazilian Kefir Grains. J. Dairy Sci. 2015, 98, 3622–3632. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.-L.; Yuan, Y.-H.; Yue, T.-L.; Guo, C.-F. A New Method for the in Vitro Determination of the Bile Tolerance of Potentially Probiotic Lactobacilli. Appl. Microbiol. Biotechnol. 2018, 102, 1903–1910. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.-H.; Yu, B.; Jang, S.-H.; Tsen, H.-Y. Different Probiotic Properties for Lactobacillus Fermentum Strains Isolated from Swine and Poultry. Anaerobe 2007, 13, 107–113. [Google Scholar] [CrossRef]
- Ayyash, M.M.; Abdalla, A.K.; AlKalbani, N.S.; Baig, M.A.; Turner, M.S.; Liu, S.-Q.; Shah, N.P. Invited Review: Characterization of New Probiotics from Dairy and Nondairy Products—Insights into Acid Tolerance, Bile Metabolism and Tolerance, and Adhesion Capability. J. Dairy Sci. 2021, 104, 8363–8379. [Google Scholar] [CrossRef] [PubMed]
- Haghshenas, B.; Nami, Y.; Haghshenas, M.; Abdullah, N.; Rosli, R.; Radiah, D.; Yari Khosroushahi, A. Bioactivity Characterization of Lactobacillus Strains Isolated from Dairy Products. MicrobiologyOpen 2015, 4, 803–813. [Google Scholar] [CrossRef]
- Lambert, J.M.; Bongers, R.S.; de Vos, W.M.; Kleerebezem, M. Functional Analysis of Four Bile Salt Hydrolase and Penicillin Acylase Family Members in Lactobacillus Plantarum WCFS1. Appl. Environ. Microbiol. 2008, 74, 4719–4726. [Google Scholar] [CrossRef] [PubMed]
- De Boever, P.; Wouters, R.; Verschaeve, L.; Berckmans, P.; Schoeters, G.; Verstraete, W. Protective Effect of the Bile Salt Hydrolase-Active Lactobacillus Reuteri against Bile Salt Cytotoxicity. Appl. Microbiol. Biotechnol. 2000, 53, 709–714. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, J.; Zhang, X.; Tong, Y.; Yang, R. Genomic and Transcriptomic Analysis of Bacillus Subtilis JNFE1126 with Higher Nattokinase Production through Ultraviolet Combined 60Co-γ Ray Mutagenesis. LWT 2021, 147, 111652. [Google Scholar] [CrossRef]
- Su, M.S.-W.; Oh, P.L.; Walter, J.; Gänzle, M.G. Intestinal Origin of Sourdough Lactobacillus Reuteri Isolates as Revealed by Phylogenetic, Genetic, and Physiological Analysis. Appl. Environ. Microbiol. 2012, 78, 6777–6780. [Google Scholar] [CrossRef]
- Verce, M.; De Vuyst, L.; Weckx, S. Comparative Genomics of Lactobacillus Fermentum Suggests a Free-Living Lifestyle of This Lactic Acid Bacterial Species. Food Microbiol. 2020, 89, 103448. [Google Scholar] [CrossRef]
- Pfeiler, E.A.; Azcarate-Peril, M.A.; Klaenhammer, T.R. Characterization of a Novel Bile-Inducible Operon Encoding a Two-Component Regulatory System in Lactobacillus Acidophilus. J. Bacteriol. 2007, 189, 4624–4634. [Google Scholar] [CrossRef]
- Whitehead, K.; Versalovic, J.; Roos, S.; Britton, R.A. Genomic and Genetic Characterization of the Bile Stress Response of Probiotic Lactobacillus Reuteri ATCC 55730. Appl. Environ. Microbiol. 2008, 74, 1812–1819. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Sun, Z.; Wu, J.; Meng, H.; Zhang, H. Effect of Bile Salts Stress on Protein Synthesis of Lactobacillus Casei Zhang Revealed by 2-Dimensional Gel Electrophoresis. J. Dairy Sci. 2010, 93, 3858–3868. [Google Scholar] [CrossRef] [PubMed]
- Cochu, A.; Vadeboncoeur, C.; Moineau, S.; Frenette, M. Genetic and Biochemical Characterization of the Phosphoenolpyruvate:Glucose/Mannose Phosphotransferase System of Streptococcus Thermophilus. Appl. Environ. Microbiol. 2003, 69, 5423–5432. [Google Scholar] [CrossRef] [PubMed]
- Pfeiler, E.A.; Klaenhammer, T.R. Role of Transporter Proteins in Bile Tolerance of Lactobacillus Acidophilus. Appl. Environ. Microbiol. 2009, 75, 6013–6016. [Google Scholar] [CrossRef]
- O’Flaherty, S.J.; Klaenhammer, T.R. Functional and Phenotypic Characterization of a Protein from Lactobacillus Acidophilus Involved in Cell Morphology, Stress Tolerance and Adherence to Intestinal Cells. Microbiology 2010, 156, 3360–3367. [Google Scholar] [CrossRef]
- Nguyen, H.A.; Nguyen, T.-H.; Nguyen, T.-T.; Peterbauer, C.K.; Mathiesen, G.; Haltrich, D. Chitinase from Bacillus Licheniformis DSM13: Expression in Lactobacillus Plantarum WCFS1 and Biochemical Characterisation. Protein Expr. Purif. 2012, 81, 166–174. [Google Scholar] [CrossRef]
- Ranadheera, C.S.; Evans, C.A.; Adams, M.C.; Baines, S.K. In Vitro Analysis of Gastrointestinal Tolerance and Intestinal Cell Adhesion of Probiotics in Goat’s Milk Ice Cream and Yogurt. Food Res. Int. 2012, 49, 619–625. [Google Scholar] [CrossRef]

| Bacteria and Plasmids | Relevant Characteristics |
|---|---|
| Strains | |
| Lactobacillus fermentum 156 | Source of bsh1, sfcA, ldhA, and mtlR genes |
| Lactobacillus plantarum S83 | Source of secB, agrA, and gdh genes |
| Lactobacillus plantarum RS417 | Expression host |
| Escherichia coli DH5α | Cloning host |
| Plasmids | |
| pSIP403 | spp-based expression vector with promoter PsppA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, C.; Yu, L.; Tian, F.; Zhao, J.; Zhai, Q. Identification of Novel Bile Salt-Tolerant Genes in Lactobacillus Using Comparative Genomics and Its Application in the Rapid Screening of Tolerant Strains. Microorganisms 2022, 10, 2371. https://doi.org/10.3390/microorganisms10122371
Chen C, Yu L, Tian F, Zhao J, Zhai Q. Identification of Novel Bile Salt-Tolerant Genes in Lactobacillus Using Comparative Genomics and Its Application in the Rapid Screening of Tolerant Strains. Microorganisms. 2022; 10(12):2371. https://doi.org/10.3390/microorganisms10122371
Chicago/Turabian StyleChen, Chunfei, Leilei Yu, Fengwei Tian, Jianxin Zhao, and Qixiao Zhai. 2022. "Identification of Novel Bile Salt-Tolerant Genes in Lactobacillus Using Comparative Genomics and Its Application in the Rapid Screening of Tolerant Strains" Microorganisms 10, no. 12: 2371. https://doi.org/10.3390/microorganisms10122371
APA StyleChen, C., Yu, L., Tian, F., Zhao, J., & Zhai, Q. (2022). Identification of Novel Bile Salt-Tolerant Genes in Lactobacillus Using Comparative Genomics and Its Application in the Rapid Screening of Tolerant Strains. Microorganisms, 10(12), 2371. https://doi.org/10.3390/microorganisms10122371





