Molecular Profiles of Multiple Antimalarial Drug Resistance Markers in Plasmodium falciparum and Plasmodium vivax in the Mandalay Region, Myanmar
Abstract
:1. Introduction
2. Materials and Methods
2.1. Blood Samples
2.2. Amplification of Antimalarial Drug Resistance Genes
2.3. Sequence Polymorphism Analysis
2.4. Statistical Analysis
3. Results
3.1. Molecular Profiles of Antimalarial Drug Resistance Genes in Myanmar P. falciparum
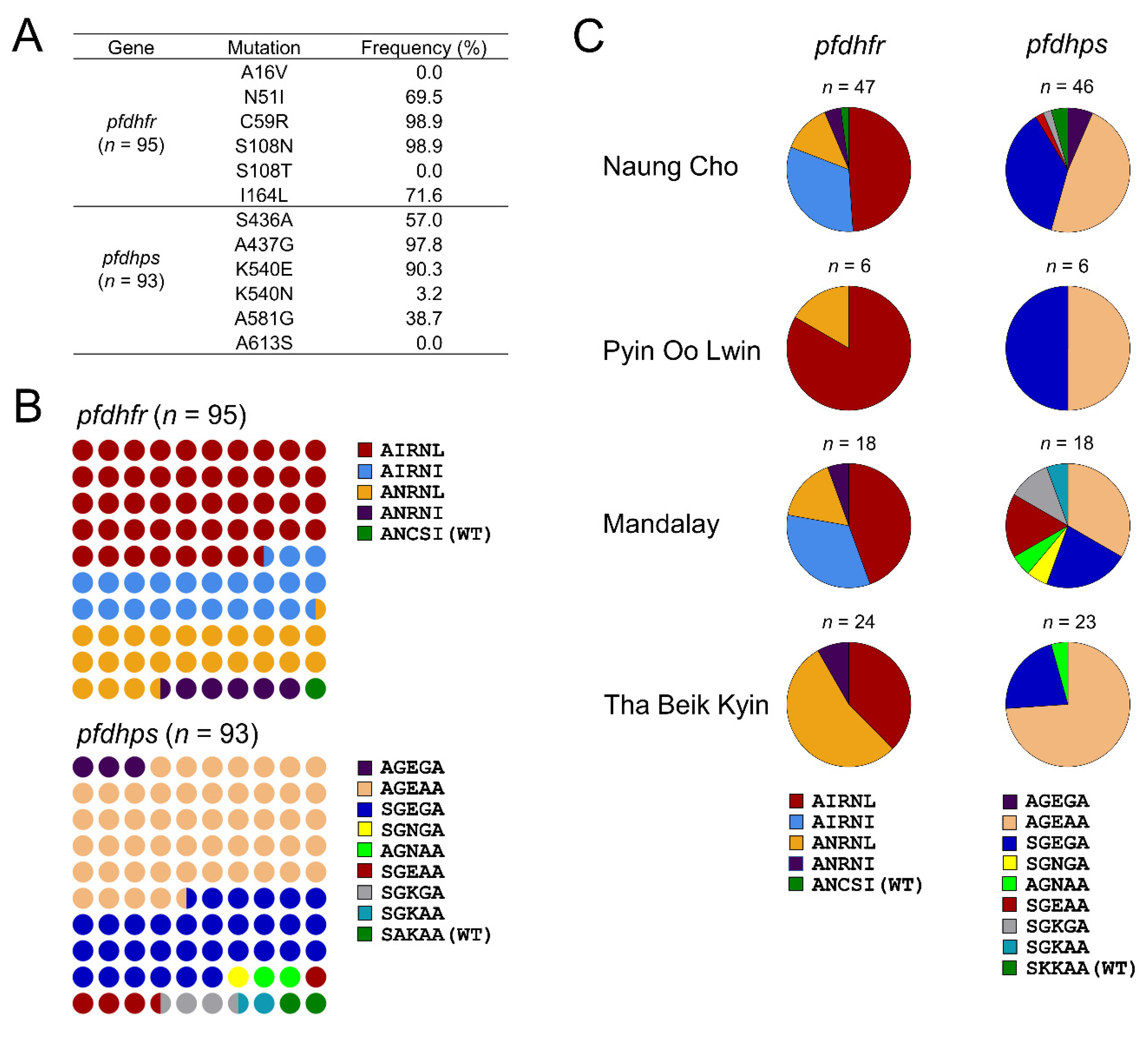
3.1.1. Molecular Profiles of pfdhfr and pfdhps
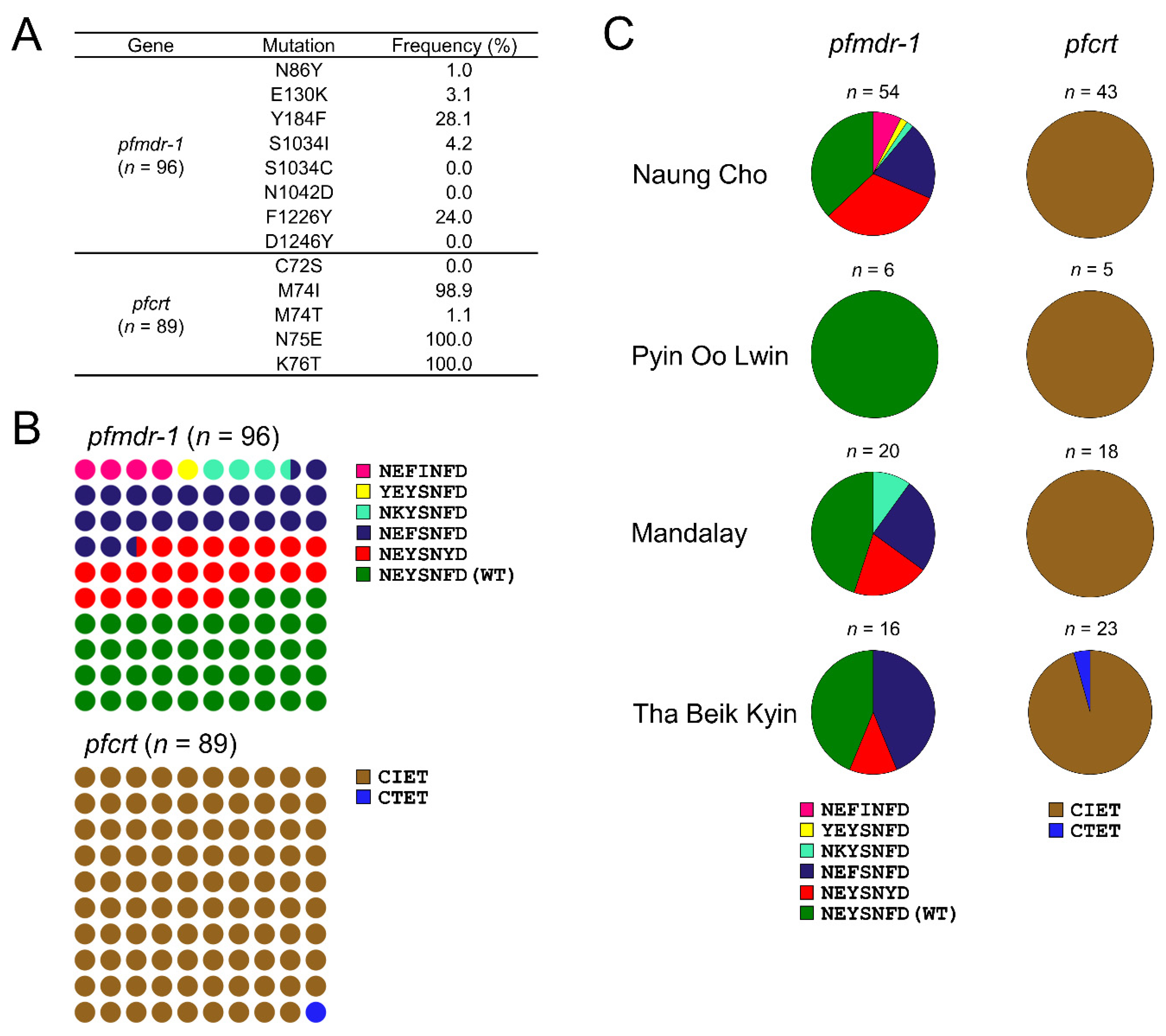
3.1.2. Molecular Profiles of pfmdr-1 and pfcrt
3.1.3. Molecular Profiles of pfk13, pfubp-1, and pfcytb
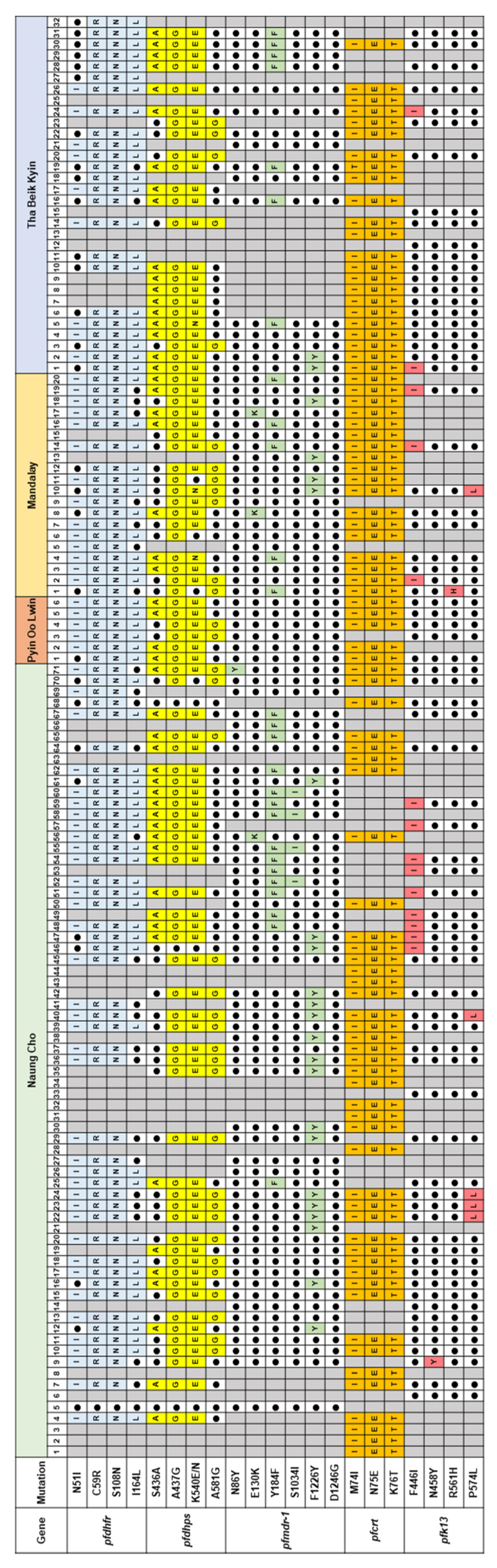
3.1.4. Summary of Mutations in Multiple Antimalarial Drug Resistance Genes
3.2. Molecular Profiles of Antimalarial Drug Resistance Genes in Myanmar P. vivax
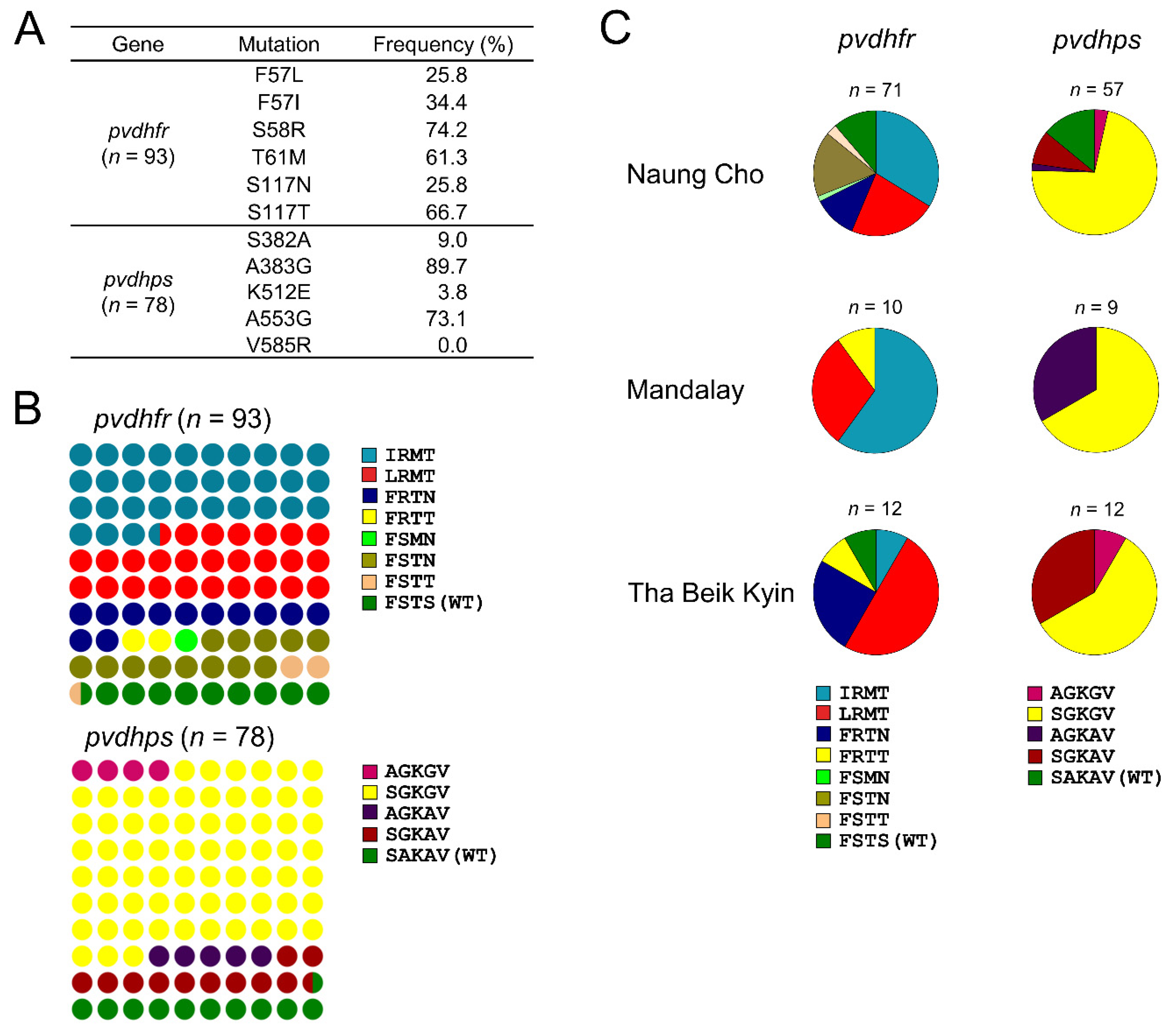
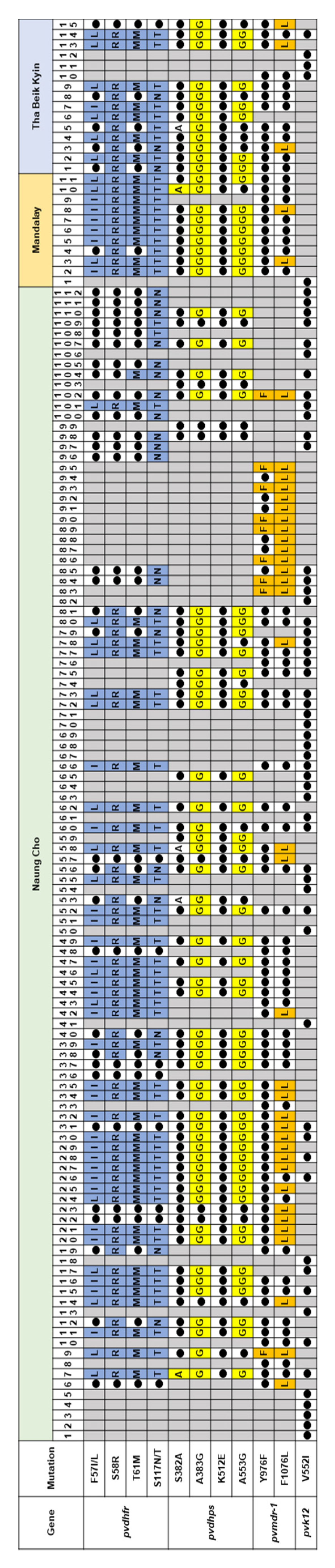
3.2.1. Molecular Profiles of pvdhfr and pvdhps
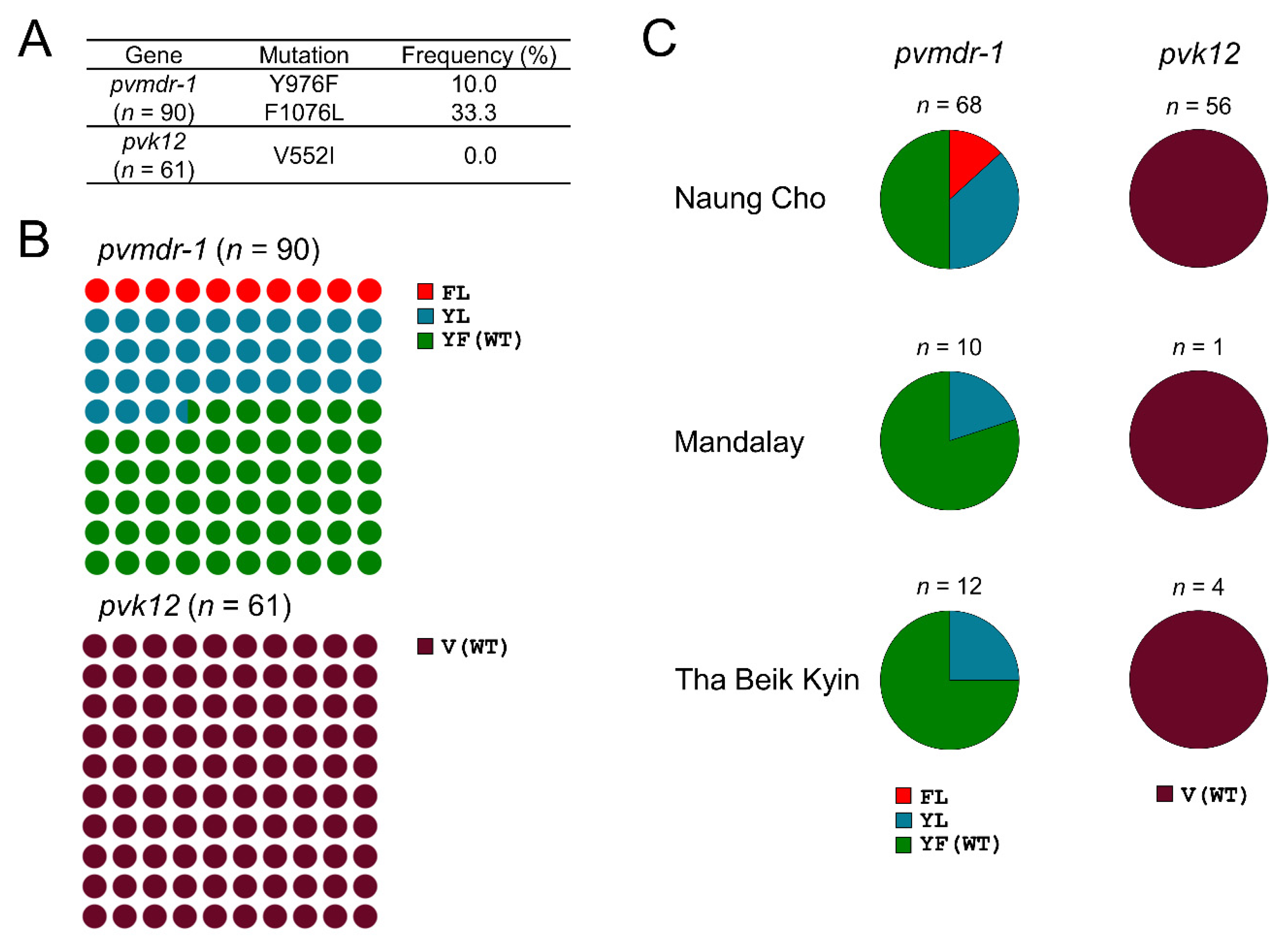
3.2.2. Molecular Profiles of pvmdr-1 and pvk12
3.2.3. Summary of Mutations in Multiple Antimalarial Drug Resistance Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. World Malaria Report; WHO: Geneva, Switzerland, 2021. [Google Scholar]
- Mayxay, M.; Pukrittayakamee, S.; Newton, P.N.; White, N.J. Mixed-species malaria infections in humans. Trends Parasitol. 2004, 20, 233–240. [Google Scholar] [CrossRef]
- Menard, D.; Dondorp, A. Antimalarial drug resistance: A threat to malaria elimination. Cold Spring Harb. Perspect. Med. 2017, 7, a025619. [Google Scholar] [CrossRef] [Green Version]
- Rocamora, F.; Winzeler, E.A. Genomic approaches to drug resistance in malaria. Annu. Rev. Microbiol. 2020, 74, 761–786. [Google Scholar] [CrossRef]
- Ippolito, M.M.; Moser, K.A.; Kabuya, J.-B.B.; Cunningham, C.; Juliano, J.J. Antimalarial drug resistance and implications for the WHO global technical strategy. Curr. Epidemiol. Reports 2021, 8, 46–62. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. The “World Malaria Report 2019” at a Glance; WHO: Geneva, Switzerland, 2019. [Google Scholar]
- Dondorp, A.M.; Fairhurst, R.M.; Slutsker, L.; MacArthur, J.R.; Breman, J.G.; Guerin, P.J.; Wellems, T.E.; Ringwald, P.; Newman, R.D.; Plowe, C.V. The threat of artemisinin-resistant malaria. N. Engl. J. Med. 2011, 365, 1073–1075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashley, E.A.; Dhorda, M.; Fairhurst, R.M.; Amaratunga, C.; Lim, P.; Suon, S.; Sreng, S.; Anderson, J.M.; Mao, S.; Sam, B.; et al. Spread of artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 2014, 371, 411–423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanboonkunupakarn, B.; White, N.J. The threat of antimalarial drug resistance. Trop. Dis. Travel Med. Vaccines 2016, 2, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woodrow, C.J.; White, N.J. The clinical impact of artemisinin resistance in Southeast Asia and the potential for future spread. FEMS Microbiol. Rev. 2017, 41, 34–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imwong, M.; Suwannasin, K.; Kunasol, C.; Sutawong, K.; Mayxay, M.; Rekol, H.; Smithuis, F.M.; Hlaing, T.M.; Tun, K.M.; van der Pluijm, R.W.; et al. The spread of artemisinin-resistant Plasmodium falciparum in the Greater Mekong Subregion: A molecular epidemiology observational study. Lancet Infect. Dis. 2017, 17, 491–497. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. World Malaria Report: 20 Years of Global Progress and Challenges; WHO: Geneva, Switzerland, 2020. [Google Scholar]
- Ejov, M.N.; Tun, T.; Aung, S.; Sein, K. Response of falciparum malaria to different antimalarials in Myanmar. Bull. World Health Organ. 1999, 77, 244–249. [Google Scholar]
- Smithuis, F.M.; Monti, F.; Grundl, M.; Zaw Oo, A.; Kyaw, T.T.; Phe, O.; White, N.J. Plasmodium falciparum: Sensitivity in vivo to chloroquine, pyrimethamine/sulfadoxine and mefloquine in Western Myanmar. Trans. R. Soc. Trop. Med. Hyg. 1997, 91, 468–472. [Google Scholar] [CrossRef]
- Tun, K.M.; Imwong, M.; Lwin, K.M.; Win, A.A.; Hlaing, T.M.; Hlaing, T.; Lin, K.; Kyaw, M.P.; Plewes, K.; Faiz, M.A.; et al. Spread of artemisinin-resistant Plasmodium falciparum in Myanmar: A cross-sectional survey of the k13 molecular marker. Lancet Infect. Dis. 2015, 15, 415–421. [Google Scholar] [CrossRef] [Green Version]
- Gething, P.W.; Patil, A.P.; Smith, D.L.; Guerra, C.A.; Elyazar, I.R.F.; Johnston, G.L.; Tatem, A.J.; Hay, S.I. A new world malaria map: Plasmodium falciparum endemicity in 2010. Malar. J. 2011, 10, 378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haldar, K.; Bhattacharjee, S.; Safeukui, I. Drug resistance in Plasmodium. Nat. Rev. Microbiol. 2018, 16, 156–170. [Google Scholar] [CrossRef]
- Na, B.K.; Lee, H.W.; Moon, S.U.; In, T.S.; Lin, K.; Maung, M.; Chung, G.T.; Lee, J.K.; Kim, T.S.; Kong, Y. Genetic Variations of the dihydrofolate reductase gene of Plasmodium vivax in Mandalay Division, Myanmar. Parasitol. Res. 2005, 96, 321–325. [Google Scholar] [CrossRef]
- Yang, Z.; Li, C.; Miao, M.; Zhang, Z.; Sun, X.; Meng, H.; Li, J.; Fan, Q.; Cui, L. Multidrug- resistant genotypes of Plasmodium falciparum, Myanmar. Emerg. Infect. Dis. 2011, 17, 498–501. [Google Scholar] [CrossRef]
- Nyunt, M.H.; Han, J.H.; Wang, B.; Aye, K.M.; Aye, K.H.; Lee, S.K.; Htut, Y.; Kyaw, M.P.; Han, K.T.; Han, E.T. Clinical and molecular surveillance of drug resistant vivax malaria in Myanmar (2009–2016). Malar. J. 2017, 16, 117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Wei, C.; Pan, Y.; Wang, Z.; Ji, X.; Chen, Q.; Zhang, L.; Wang, H. Polymorphisms of potential drug resistant molecular markers in Plasmodium vivax from China-Myanmar border during 2008–2017. Infect Dis Poverty 2022, 11, 43. [Google Scholar] [CrossRef]
- Nyunt, M.H.; Hlaing, T.; Oo, H.W.; Tin-Oo, L.L.K.; Phway, H.P.; Wang, B.; Zaw, N.N.; Han, S.S.; Tun, T.; San, K.K.; et al. Molecular assessment of artemisinin resistance markers, polymorphisms in the k13 propeller, and a multidrug-resistance gene in the Eastern and Western border areas of Myanmar. Clin. Infect. Dis. 2015, 60, 1208–1215. [Google Scholar] [CrossRef] [Green Version]
- Nyunt, M.H.; Shein, T.; Zaw, N.N.; Han, S.S.; Muh, F.; Lee, S.K.; Han, J.H.; Thant, K.Z.; Han, E.T.; Kyaw, M.P. Molecular evidence of drug resistance in asymptomatic malaria infections, Myanmar, 2015. Emerg. Infect. Dis. 2017, 23, 517–520. [Google Scholar] [CrossRef] [Green Version]
- Bai, Y.; Zhang, J.; Geng, J.; Xu, S.; Deng, S.; Zeng, W.; Wang, Z.; Ngassa Mbenda, H.G.; Zhang, J.; Li, N.; et al. Longitudinal surveillance of drug resistance in Plasmodium falciparum isolates from the China-Myanmar border reveals persistent circulation of multidrug resistant parasites. Int. J. Parasitol. Drugs Drug Resist. 2018, 8, 320–328. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, Z.; Soe, M.T.; Wang, L.; Soe, T.N.; Wei, H.; Than, A.; Aung, P.L.; Li, Y.; Zhang, X.; et al. Genetic Variations associated with drug resistance markers in asymptomatic Plasmodium falciparum infections in Myanmar. Genes 2019, 10, 692. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Wang, L.; Soe, M.T.; Aung, P.L.; Wei, H.; Liu, Z.; Ma, T.; Huang, Y.; Menezes, L.J.; Wang, Q.; et al. Molecular surveillance for drug resistance markers in Plasmodium vivax isolates from symptomatic and asymptomatic infections at the China-Myanmar border. Malar. J. 2020, 19, 281. [Google Scholar] [CrossRef]
- Wang, S.; Xu, S.; Geng, J.; Si, Y.; Zhao, H.; Li, X.; Yang, Q.; Zeng, W.; Xiang, Z.; Chen, X.; et al. Molecular Surveillance and in vitro drug sensitivity study of Plasmodium falciparum isolates from the China–Myanmar border. Am. J. Trop. Med. Hyg. 2020, 103, 1100–1106. [Google Scholar] [CrossRef]
- Tang, T.; Xu, Y.; Cao, L.; Tian, P.; Shao, J.; Deng, Y.; Zhou, H.; Xiao, B. Ten-Year molecular surveillance of drug-resistant Plasmodium spp. isolated from the China–Myanmar border. Front. Cell. Infect. Microbiol. 2021, 11, 733788. [Google Scholar] [CrossRef]
- Kang, J.M.; Cho, P.Y.; Moe, M.; Lee, J.; Jun, H.; Lee, H.W.; Ahn, S.K.; Kim, T.I.; Pak, J.H.; Myint, M.K.; et al. Comparison of the diagnostic performance of microscopic examination with nested polymerase chain reaction for optimum malaria diagnosis in Upper Myanmar. Malar. J. 2017, 16, 119. [Google Scholar] [CrossRef] [Green Version]
- Snounou, G.; Viriyakosol, S.; Jarra, W.; Thaithong, S.; Brown, K.N. Identification of the four human malaria parasite species in field samples by the polymerase chain reaction and detection of a high prevalence of mixed infections. Mol. Biochem. Parasitol. 1993, 58, 283–292. [Google Scholar] [CrossRef]
- Naidoo, I.; Roper, C. Mapping “partially resistant”, “fully resistant”, and “super resistant” malaria. Trends Parasitol. 2013, 29, 505–515. [Google Scholar] [CrossRef]
- Eyase, F.L.; Akala, H.M.; Ingasia, L.; Cheruiyot, A.; Omondi, A.; Okudo, C.; Juma, D.; Yeda, R.; Andagalu, B.; Wanja, E.; et al. The role of pfmdr1 and pfcrt in changing chloroquine, amodiaquine, mefloquine and lumefantrine susceptibility in Western-Kenya P. falciparum samples during 2008–2011. PLoS ONE 2013, 8, e64299. [Google Scholar] [CrossRef] [Green Version]
- Imwong, M.; Pukrittayakamee, S.; Cheng, Q.; Moore, C.; Looareesuwan, S.; Snounou, G.; White, N.J.; Day, N.P.J. Limited polymorphism in the dihydropteroate synthetase gene (dhps) of Plasmodium vivax isolates from Thailand. Antimicrob. Agents Chemother. 2005, 49, 4393–4395. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Soe, M.T.; Aung, P.L.; Zhao, L.; Zeng, W.; Menezes, L.; Yang, Z.; Kyaw, M.P.; Cui, L. Efficacy of artemether-lumefantrine for treating uncomplicated Plasmodium falciparum cases and molecular surveillance of drug resistance genes in Western Myanmar. Malar. J. 2020, 19, 304. [Google Scholar] [CrossRef] [PubMed]
- Kublin, J.G.; Dzinjalamala, F.K.; Kamwendo, D.D.; Malkin, E.M.; Cortese, J.F.; Martino, L.M.; Mukadam, R.A.G.; Rogerson, S.J.; Lescano, A.G.; Molyneux, M.E.; et al. Molecular markers for failure of sulfadoxine-pyrimethamine and chlorproguanil-dapsone treatment of Plasmodium falciparum malaria. J. Infect. Dis. 2002, 185, 380–388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turkiewicz, A.; Manko, E.; Sutherland, C.J.; Benavente, E.D.; Campino, S.; Clark, T.G. Genetic diversity of the Plasmodium falciparum GTP-Cyclohydrolase 1, dihydrofolate reductase and dihydropteroate synthetase genes reveals new insights into sulfadoxine-pyrimethamine antimalarial drug resistance. PLoS Genet. 2020, 16, e1009268. [Google Scholar] [CrossRef]
- Myint, -O.; Myat, -P.-K.; Myint, -L.; Kyin, -H.-A.; Thaw, -Z.; Nwe, -N.-Y. Emergence of chloroquine-resistant Plasmodium vivax in Myanmar (Burma). Trans. R. Soc. Trop. Med. Hyg. 1993, 87, 687. [Google Scholar] [CrossRef]
- Warhurst, D.C. A molecular marker for chloroquine-resistant falciparum malaria. N. Engl. J. Med. 2001, 344, 299–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Djimde, A.A.; Barger, B.; Kone, A.; Beavogui, A.H.; Tekete, M.; Fofana, B.; Dara, A.; Maiga, H.; Dembele, D.; Toure, S.; et al. A molecular map of chloroquine resistance in Mali. FEMS. Immunol. Med. Microbiol. 2010, 58, 113–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duraisingh, M.T.; Jones, P.; Sambou, I.; Von Seidlein, L.; Pinder, M.; Warhurst, D.C. The tyrosine-86 allele of the pfmdr1 gene of Plasmodium falciparum is associated with increased sensitivity to the anti-malarials mefloquine and artemisinin. Mol. Biochem. Parasitol. 2000, 108, 13–23. [Google Scholar] [CrossRef]
- Babiker, H.A.; Pringle, S.J.; Abdel-Muhsin, A.; Mackinnon, M.; Hunt, P.; Walliker, D. High-level chloroquine resistance in Sudanese isolates of Plasmodium falciparum is associated with mutations in the chloroquine resistance transporter gene pfcrt and the multidrug resistance gene pfmdr1. J. Infect. Dis. 2001, 183, 1535–1538. [Google Scholar] [CrossRef] [Green Version]
- Calçada, C.; Silva, M.; Baptista, V.; Thathy, V.; Silva-Pedrosa, R.; Granja, D.; Ferreira, P.E.; Gil, J.P.; Fidock, D.A.; Veiga, M.I. Expansion of a specific Plasmodium falciparum pfmdr1 haplotype in southeast asia with increased substrate transport. MBio 2020, 11, e02093-20. [Google Scholar] [CrossRef]
- Yuan, L.; Wang, Y.; Parker, D.M.; Gupta, B.; Yang, Z.; Liu, H.; Fan, Q.; Cao, Y.; Xiao, Y.; Lee, M.C.; et al. Therapeutic responses of Plasmodium vivax malaria to chloroquine and primaquine treatment in Northeastern Myanmar. Antimicrob. Agents Chemother. 2015, 59, 1230–1235. [Google Scholar] [CrossRef] [Green Version]
- Htun, M.W.; Mon, N.C.N.; Aye, K.M.; Hlaing, C.M.; Kyaw, M.P.; Handayuni, I.; Trimarsanto, H.; Bustos, D.; Ringwald, P.; Price, R.N.; et al. Chloroquine efficacy for Plasmodium vivax in Myanmar in populations with high genetic diversity and moderate parasite gene flow. Malar. J. 2017, 16, 281. [Google Scholar] [CrossRef]
- Chu, C.S.; Phyo, A.P.; Lwin, K.M.; Win, H.H.; San, T.; Aung, A.A.; Raksapraidee, R.; Carrara, V.I.; Bancone, G.; Watson, J.; et al. Comparison of the cumulative efficacy and safety of chloroquine, artesunate, and chloroquine-primaquine in Plasmodium vivax malaria. Clin. Infect. Dis. 2018, 67, 1543–1549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, S.; Zeng, W.; Ngassa Mbenda, H.G.; Liu, H.; Chen, X.; Xiang, Z.; Li, C.; Zhang, Y.; Baird, J.K.; Yang, Z.; et al. Efficacy of directly-observed chloroquine-primaquine treatment for uncomplicated acute Plasmodium Vivax malaria in Northeast Myanmar: A Prospective Open-Label Efficacy Trial. Travel Med. Infect. Dis. 2020, 36, 101499. [Google Scholar] [CrossRef] [PubMed]
- Djimdé, A.; Doumbo, O.K.; Cortese, J.F.; Kayentao, K.; Doumbo, S.; Diourté, Y.; Coulibaly, D.; Dicko, A.; Su, X.; Nomura, T.; et al. A molecular marker for chloroquine-resistant falciparum malaria. N. Engl. J. Med. 2001, 344, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Brega, S.; Meslin, B.; De Monbrison, F.; Severini, C.; Gradoni, L.; Udomsangpetch, R.; Sutanto, I.; Peyron, F.; Picot, S. Identification of the Plasmodium vivax mdr-like gene (pvmdr1) and analysis of single-nucleotide polymorphisms among isolates from different areas of endemicity. J. Infect. Dis. 2005, 191, 272–277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suwanarusk, R.; Russell, B.; Chavchich, M.; Chalfein, F.; Kenangalem, E.; Kosaisavee, V.; Prasetyorini, B.; Piera, K.A.; Barends, M.; Brockman, A.; et al. Chloroquine resistant Plasmodium vivax: In vitro characterisation and association with molecular polymorphisms. PLoS ONE 2007, 2, e1089. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, J.; Li, Q.; Hu, Y.; Ruan, Y.; Tao, Z.; Xia, H.; Qiao, J.; Meng, L.; Zeng, W.; et al. Ex vivo susceptibilities of Plasmodium vivax isolates from the China-Myanmar border to antimalarial drugs and association with polymorphisms in pvmdr1 and pvcrt-o genes. PLoS Negl. Trop. Dis. 2020, 14, e0008255. [Google Scholar] [CrossRef] [PubMed]
- Nwe, T.W.; Oo, T.; Wai, K.T.; Zhou, S.; van Griensven, J.; Chinnakali, P.; Shah, S.; Thi, A. Malaria profiles and challenges in artemisinin resistance containment in Myanmar. Infect. Dis. Poverty 2017, 6, 76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Organization. World Malaria Report 2015; WHO: Geneva, Switzerland. Available online: https://Apps.Who.Int/Iris/Handle/10665/200018rldMalariaReport2015 (accessed on 23 March 2022).
- Lin, k.; Myint, M.K.; Thu, A.; Win, P.P.; Moe, M.; Hlaing, T.; Myint, Y.Y. A study on drug resistant malaria in sentinel site of Upper Myanmar. In Proceedings of the 43rd Myanmar Health Research Congress, Yangon, Myanmar, 5–9 January 2015. [Google Scholar]
- Tun, K.M.; Jeeyapant, A.; Imwong, M.; Thein, M.; Aung, S.S.M.; Hlaing, T.M.; Yuentrakul, P.; Promnarate, C.; Dhorda, M.; Woodrow, C.J.; et al. Parasite Clearance rates in Upper Myanmar indicate a distinctive artemisinin resistance phenotype: A therapeutic efficacy study. Malar. J. 2016, 15, 185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Myint, M.K.; Rasmussen, C.; Thi, A.; Bustos, D.; Ringwald, P.; Lin, K. Therapeutic efficacy and artemisinin resistance in Northern Myanmar: Evidence from in vivo and molecular marker studies. Malar. J. 2017, 16, 143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, K.T.; Lin, K.; Han, Z.Y.; Myint, M.K.; Aye, K.H.; Thi, A.; Thapa, B.; Bustos, M.D.; Borghini-Fuhrer, I.; Ringwald, P.; et al. Efficacy and safety of pyronaridine–artesunate for the treatment of uncomplicated Plasmodium falciparum and Plasmodium vivax malaria in Myanmar. Am. J. Trop. Med. Hyg. 2020, 103, 1088–1093. [Google Scholar] [CrossRef] [PubMed]
- Popovici, J.; Kao, S.; Eal, L.; Bin, S.; Kim, S.; Ménard, D. Reduced polymorphism in the kelch propeller domain in Plasmodium vivax isolates from Cambodia. Antimicrob. Agents Chemother. 2015, 59, 730–733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imwong, M.; Nakeesathit, S.; Day, N.P.J.; White, N.J. A Review of mixed malaria species infections in Anopheline Mosquitoes. Malar. J. 2011, 10, 253. [Google Scholar] [CrossRef] [Green Version]
- Pearson, R.D.; Amato, R.; Auburn, S.; Miotto, O.; Almagro-Garcia, J.; Amaratunga, C.; Suon, S.; Mao, S.; Noviyanti, R.; Trimarsanto, H.; et al. Genomic analysis of local variation and recent evolution in Plasmodium vivax. Nat. Genet. 2016, 48, 959–964. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Ruan, Y.; Bai, Y.; Hu, Y.; Deng, Z.; He, Y.; Ruan, R.; Wu, Y.; Yang, Z.; Cui, L. Genetic diversity of the pvk12 gene in Plasmodium vivax from the China-Myanmar border area. Malar. J. 2016, 15, 528. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Siddiqui, F.A.; Fan, Q.; Luo, E.; Cao, Y.; Cui, L. Limited genetic diversity in the pvk12 kelch protein in Plasmodium vivax isolates from Southeast Asia. Malar. J. 2016, 15, 537. [Google Scholar] [CrossRef] [Green Version]
- Adams, T.; Ennuson, N.A.A.; Quashie, N.B.; Futagbi, G.; Matrevi, S.; Hagan, O.C.K.; Abuaku, B.; Koram, K.A.; Duah, N.O. Prevalence of Plasmodium falciparum delayed clearance associated polymorphisms in adaptor protein complex 2 mu subunit (pfap2mu) and ubiquitin specific protease 1 (pfubp1) genes in Ghanaian isolates. Parasites Vectors 2018, 11, 175. [Google Scholar] [CrossRef] [Green Version]
- Borrmann, S.; Straimer, J.; Mwai, L.; Abdi, A.; Rippert, A.; Okombo, J.; Muriithi, S.; Sasi, P.; Kortok, M.M.; Lowe, B.; et al. Genome-wide screen identifies new candidate genes associated with artemisinin susceptibility in Plasmodium falciparum in Kenya. Sci. Rep. 2013, 3, 3318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Looareesuwan, S.; Chulay, J.D.; Canfield, C.J.; Hutchinson, D.B.A.; De Alencar, F.; Anabwani, G.; Attanath, P.; Bienzle, U.; Bouchaud, O.; Bustos, D.; et al. Malarone (TM) (Atovaquone and Proguanil Hydrochloride): A review of its clinical development for treatment of malaria. Am. J. Trop. Med. Hyg. 1999, 60, 533–541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Massamba, L.; Madamet, M.; Benoit, N.; Chevalier, A.; Fonta, I.; Mondain, V.; Jeandel, P.Y.; Amalvict, R.; Delaunay, P.; Mosnier, J.; et al. Late clinical failure associated with cytochrome b codon 268 mutation during treatment of falciparum malaria with atovaquone-proguanil in traveller returning from Congo. Malar. J. 2020, 19, 37. [Google Scholar] [CrossRef]
- Naoshima-Ishibashi, Y.; Iwagami, M.; Kawazu, S.-I.; Looareesuwan, S.; Kano, S. Analyses of cytochrome b mutations in Plasmodium falciparum isolates in Thai-Myanmar border. Travel Med. Infect. Dis. 2007, 5, 132–134. [Google Scholar] [CrossRef] [PubMed]
- Lê, H.G.; Kang, J.-M.; Jun, H.; Lee, J.; Thái, T.L.; Myint, M.K.; Aye, K.S.; Sohn, W.-M.; Shin, H.-J.; Kim, T.-S.; et al. Changing pattern of the genetic diversities of Plasmodium falciparum merozoite surface protein-1 and merozoite surface protein-2 in Myanmar isolates. Malar. J. 2019, 18, 241. [Google Scholar] [CrossRef] [PubMed]
- Naw, H.; Kang, J.M.; Moe, M.; Lee, J.; Lê, H.G.; Võ, T.C.; Mya, Y.Y.; Myint, M.K.; Htun, Z.T.; Kim, T.S.; et al. Temporal changes in the genetic diversity of Plasmodium vivax merozoite surface protein-1 in Myanmar. Pathogens 2021, 10, 916. [Google Scholar] [CrossRef]
- Geng, J.; Malla, P.; Zhang, J.; Xu, S.; Li, C.; Zhao, Y.; Wang, Q.; Kyaw, M.P.; Cao, Y.; Yang, Z.; et al. Increasing trends of malaria in a border area of the Greater Mekong Subregion. Malar. J. 2019, 18, 309. [Google Scholar] [CrossRef] [Green Version]
- Chung, D.-I.; Jeong, S.; Dinzouna-Boutamba, S.-D.; Yang, H.-W.; Yeo, S.-G.; Hong, Y.; Goo, Y.-K. Evaluation of single nucleotide polymorphisms of pvmdr1 and microsatellite genotype in Plasmodium vivax isolates from Republic of Korea military personnel. Malar. J. 2015, 14, 336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, B.; Deng, C.; Yang, T.; Xue, L.; Wang, Q.; Huang, S.; Su, X.-Z.; Liu, Y.; Zheng, S.; Guan, Y.; et al. Polymorphisms of the artemisinin resistant marker (K13) in Plasmodium falciparum parasite populations of Grande Comore Island 10 years after artemisinin combination therapy. Parasites Vectors 2015, 8, 634. [Google Scholar] [CrossRef] [Green Version]
- Deida, J.M.; Khalef, Y.O.; Semane, E.M.; Salem, M.S.O.A.; Bogreau, H.; Basco, L.; Boukhary, A.O.M.S.; Tahar, R. Assessment of drug resistance associated genetic diversity in Mauritanian isolates of Plasmodium vivax reveals limited polymorphism. Malar. J. 2018, 17, 416. [Google Scholar] [CrossRef] [PubMed]
- Pearce, R.J.; Drakeley, C.; Chandramohan, D.; Mosha, F.; Roper, C. Molecular Determination of Point Mutation Haplotypes in the Dihydrofolate Reductase and Dihydropteroate Synthase of Plasmodium falciparum in Three Districts of Northern Tanzania. Antimicrob. Agents Chemother. 2003, 47, 1347–1354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwöbel, B.; Alifrangis, M.; Salanti, A.; Jelinek, T. Different mutation patterns of atovaquone resistance to Plasmodium falciparum in vitro and in vivo: rapid detection of codon 268 polymorphisms in the cytochrome b as potential in vivo resistance marker. Malar. J. 2003, 2, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, H.; Kong, X.; Zhang, T.; Xiao, H.; Feng, X.; Tu, H.; Feng, J. Prevalence of Plasmodium falciparum Kelch 13 ( PfK13 ) and Ubiquitin-Specific Protease 1 ( pfubp1 ) Gene Polymorphisms in Returning Travelers from Africa Reported in Eastern China. Antimicrob. Agents Chemother. 2020, 64, e00981-20. [Google Scholar] [CrossRef] [PubMed]

| Gene Combination | Mutations | Naung Cho (n = 38) | Mandalay (n = 17) | Pyin Oo Lwin (n = 6) | Tha Beik Kyin (n = 19) |
| pfdhfr + pfdhps | N51I/C59R/S108N + A437G | 0 | 0 | 0 | 0 |
| N51I/C59R/S108N + A437G/K540E | 1 (2.6%) | 2 (11.8%) | 0 | 0 | |
| N51I/C59R/S108N + A437G/K540E/A581G | 8 (21.1%) | 1 (5.9%) | 0 | 0 | |
| Gene combination | Mutations | Naung Cho (n = 16) | Mandalay (n = 11) | Pyin Oo Lwin (n = 0) | Tha Beik Kyin (n = 6) |
| pfmdr-1 + pfcrt | N86Y + K76T | 1 (6.3%) | 0 | 0 | 0 |
| Gene Combination | Mutations | Naung Cho (n = 43) | Mandalay (n = 9) | Tha Beik Kyin (n = 12) |
|---|---|---|---|---|
| pvdhfr + pvdhps | F57I/S58R/T61M/S117T + S382A/A383G/A553G | 1 (2.3%) | 0 | 0 |
| F57I/S58R/T61M/S117T + A383G/A553G | 19 (41.1%) | 5 (55.6%) | 1 (8.3%) | |
| F57L/S58R/T61M/S117T + A383G/A553G | 7 (16.3%) | 2 (22.2%) | 5 (41.7%) | |
| F57I/S58R/T61M/S117T + S382A/A383G | 0 | 0 | 0 | |
| F57L/S58R/T61M/S117T + S382A/A383G | 0 | 1 (11.1%) | 0 | |
| F57L/S58R/T61M/S117T + A383G | 2 (4.7%) | 0 | 1 (8.3%) | |
| F57I/S58R/T61M/S117T + A383G | 1 (2.3%) | 0 | 0 | |
| S58R/S117N + A383G/A553G | 6 (13.9%) | 0 | 1 (8.3%) | |
| S58R/S117T + S382A/A383G/A553G | 0 | 0 | 1 (8.3%) | |
| S58R/S117N + S382A/A553G | 1 (2.3%) | 0 | 0 | |
| T61M/S117N + A383G/A553G | 1 (2.3%) | 0 | 0 | |
| S58R/S117T + A383G/A553G | 0 | 1 (11.1%) | 0 | |
| S117N + A383G/A553G | 3 (7.0%) | 0 | 0 | |
| S58R/S117N + A553G | 0 | 0 | 2 (16.7%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lê, H.G.; Naw, H.; Kang, J.-M.; Võ, T.C.; Myint, M.K.; Htun, Z.T.; Lee, J.; Yoo, W.G.; Kim, T.-S.; Shin, H.-J.; et al. Molecular Profiles of Multiple Antimalarial Drug Resistance Markers in Plasmodium falciparum and Plasmodium vivax in the Mandalay Region, Myanmar. Microorganisms 2022, 10, 2021. https://doi.org/10.3390/microorganisms10102021
Lê HG, Naw H, Kang J-M, Võ TC, Myint MK, Htun ZT, Lee J, Yoo WG, Kim T-S, Shin H-J, et al. Molecular Profiles of Multiple Antimalarial Drug Resistance Markers in Plasmodium falciparum and Plasmodium vivax in the Mandalay Region, Myanmar. Microorganisms. 2022; 10(10):2021. https://doi.org/10.3390/microorganisms10102021
Chicago/Turabian StyleLê, Hương Giang, Haung Naw, Jung-Mi Kang, Tuấn Cường Võ, Moe Kyaw Myint, Zaw Than Htun, Jinyoung Lee, Won Gi Yoo, Tong-Soo Kim, Ho-Joon Shin, and et al. 2022. "Molecular Profiles of Multiple Antimalarial Drug Resistance Markers in Plasmodium falciparum and Plasmodium vivax in the Mandalay Region, Myanmar" Microorganisms 10, no. 10: 2021. https://doi.org/10.3390/microorganisms10102021
APA StyleLê, H. G., Naw, H., Kang, J.-M., Võ, T. C., Myint, M. K., Htun, Z. T., Lee, J., Yoo, W. G., Kim, T.-S., Shin, H.-J., & Na, B.-K. (2022). Molecular Profiles of Multiple Antimalarial Drug Resistance Markers in Plasmodium falciparum and Plasmodium vivax in the Mandalay Region, Myanmar. Microorganisms, 10(10), 2021. https://doi.org/10.3390/microorganisms10102021








