Response of Soil Bacterial Diversity, Predicted Functions and Co-Occurrence Patterns to Nanoceria and Ionic Cerium Exposure
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Planting, Sampling and Soil Analyses
2.3. Soil Bacterial 16S rRNA Gene Sequencing and Analysis
3. Results
3.1. Soil Physicochemical Properties
3.2. Bacterial α-Diversity and Community Composition
3.3. Differences in Soil Bacterial Community Patterns
3.4. Bacterial Biomarkers under Cerium Exposure
3.5. Comparison of Co-Occurrence Network Patterns of NC and IC Treatments
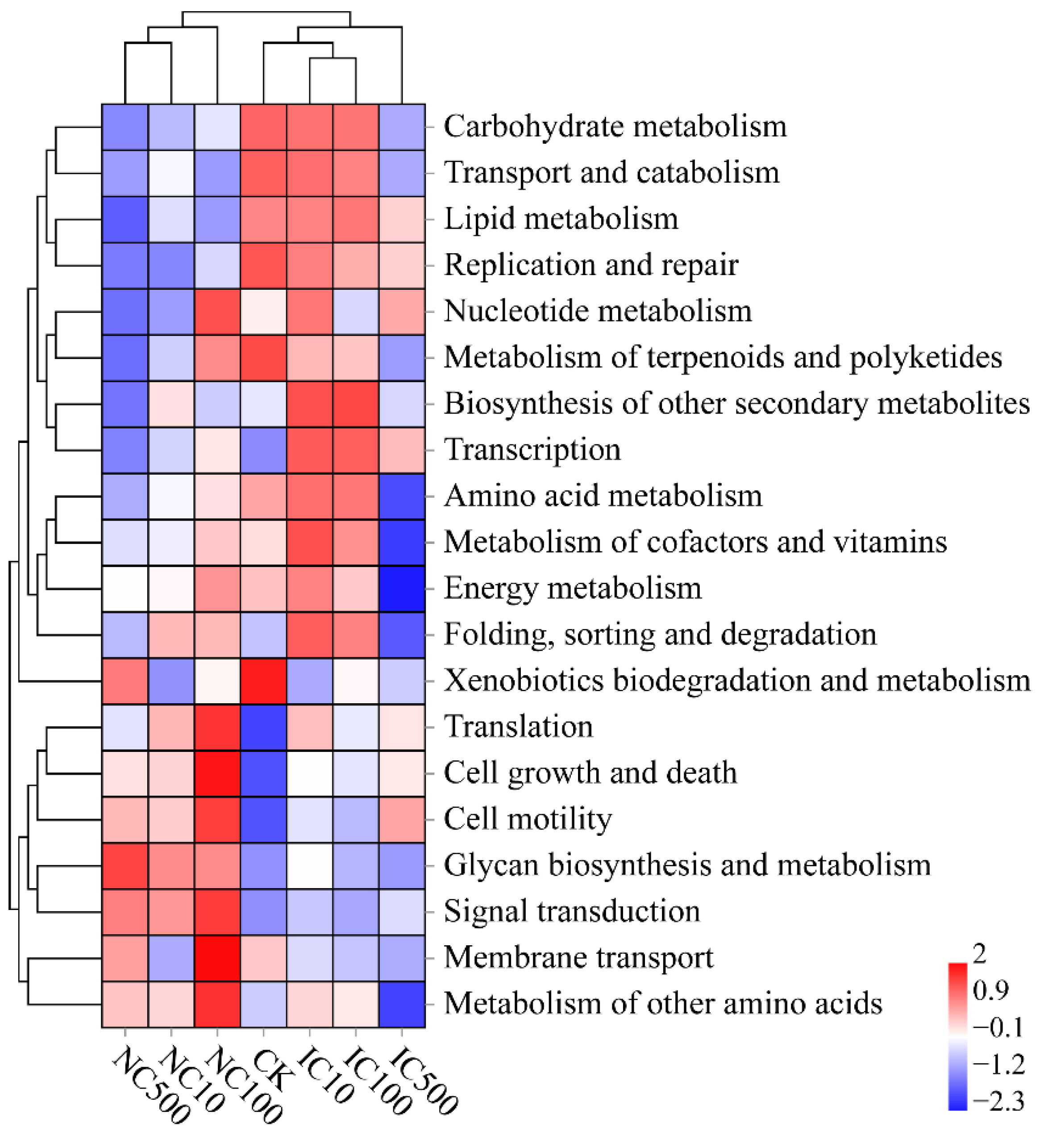
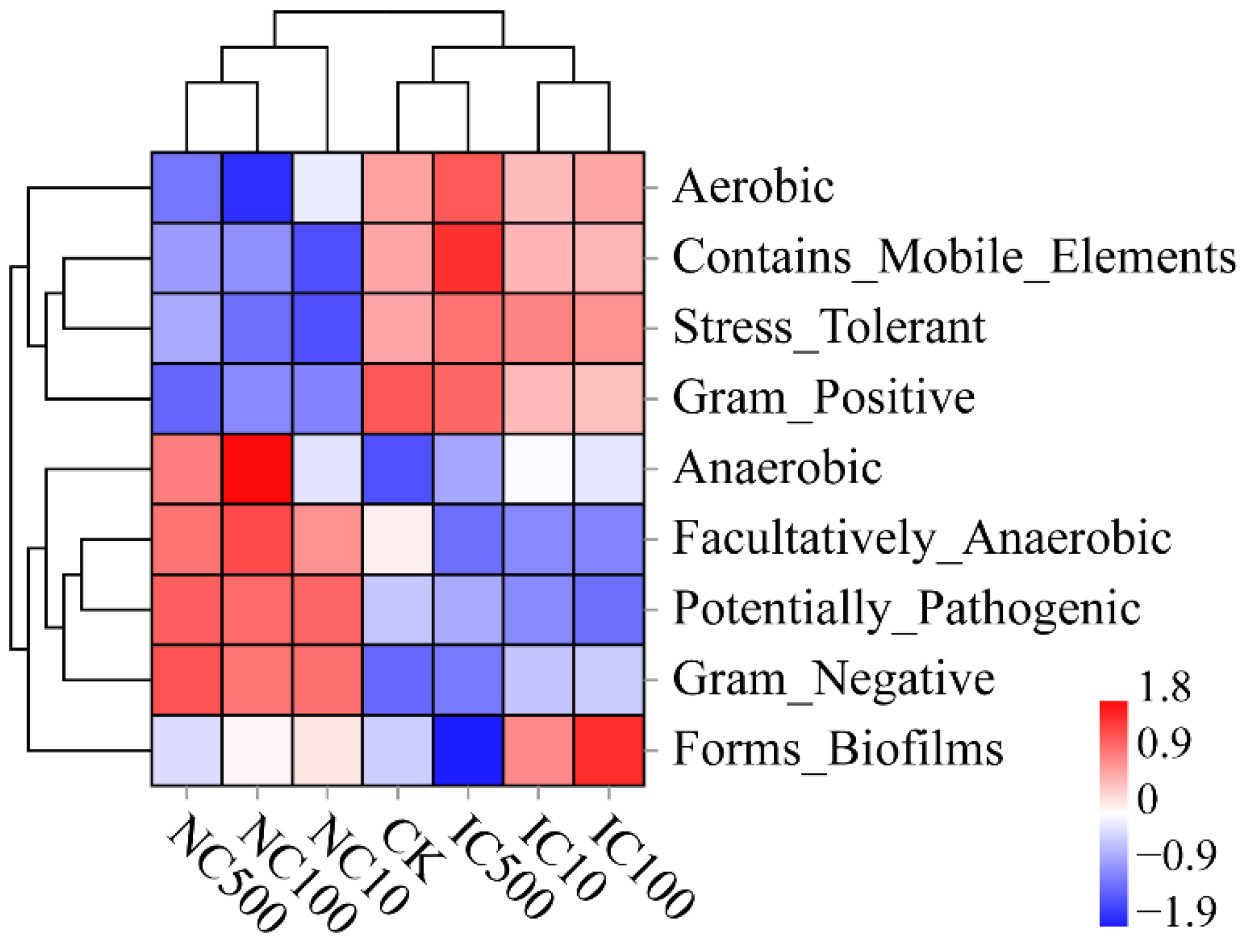
3.6. Differences in Predicted Function and Phenotypic Traits
4. Discussion
4.1. Effect Differentiations between Nanoceria and Ionic Cerium
4.2. Bacterial Co-Occurrence Patterns under Exposure to Cerium
4.3. Responsive Bacterial Taxa, Functions and Phenotypes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Park, B.; Donaldson, K.; Duffin, R.; Tran, L.; Kelly, F.; Mudway, I.; Morin, J.-P.; Guest, R.; Jenkinson, P.; Samaras, Z.; et al. Hazard and risk assessment of a nanoparticulate cerium oxide-based diesel fuel additive—A case study. Inhal. Toxicol. 2008, 20, 547–566. [Google Scholar] [CrossRef]
- Hayat, A.; Andreescu, S. Nanoceria particles as catalytic amplifiers for alkaline phosphatase assays. Anal. Chem. 2013, 85, 10028–10032. [Google Scholar] [CrossRef] [PubMed]
- Stefanik, T.S.; Tuller, H.L. Ceria-based gas sensors. J. Eur. Ceram. Soc. 2001, 21, 1967–1970. [Google Scholar] [CrossRef]
- Sababi, M.; Pan, J.; Augustsson, P.-E.; Sundell, P.-E.; Claesson, P.M. Influence of polyaniline and ceria nanoparticle additives on corrosion protection of a UV-cure coating on carbon steel. Corros. Sci. 2014, 84, 189–197. [Google Scholar] [CrossRef]
- Hirst, S.M.; Karakoti, A.S.; Tyler, R.D.; Sriranganathan, N.; Seal, S.; Reilly, C.M. Anti-inflammatory properties of cerium oxide nanoparticles. Small 2009, 5, 2848–2856. [Google Scholar] [CrossRef]
- MMR. Market Research Report, Cerium Oxide Nanoparticles Market Size, Industry Report, 2018–2025. 2017. Available online: https://www.grandviewresearch.com/ (accessed on 5 December 2021).
- Collin, B.; Auffan, M.; Johnson, A.C.; Kaur, I.; Keller, A.A.; Lazareva, A.; Lead, J.R.; Ma, X.; Merrifield, R.C.; Svendsen, C.; et al. Environmental release, fate and ecotoxicological effects of manufactured ceria nanomaterials. Environ. Sci. Nano 2014, 1, 533–548. [Google Scholar] [CrossRef] [Green Version]
- Taylor, N.S.; Merrifield, R.; Williams, T.D.; Chipman, J.K.; Lead, J.R.; Viant, M.R. Molecular toxicity of cerium oxide nanoparticles to the freshwater alga Chlamydomonas reinhardtii is associated with supra-environmental exposure concentrations. Nanotoxicology 2016, 10, 32–41. [Google Scholar]
- Lead, J.R.; Batley, G.E.; Alvarez, P.J.J.; Croteau, M.-N.; Handy, R.D.; McLaughlin, M.J.; Judy, J.D.; Schirmer, K. Nanomaterials in the environment: Behavior, fate, bioavailability, and effects—An updated review. Environ. Toxicol. Chem. 2018, 37, 2029–2063. [Google Scholar] [CrossRef]
- Gottschalk, F.; Lassen, C.; Kjoelholt, J.; Christensen, F.; Nowack, B. Modeling flows and concentrations of nine engineered nanomaterials in the Danish environment. Int. J. Environ. Res. Public Health 2015, 12, 5581–5602. [Google Scholar] [CrossRef] [Green Version]
- Singh, B.K.; Liu, H.; Trivedi, P. Eco-holobiont: A new concept to identify drivers of host-associated microorganisms. Environ. Microbiol. 2020, 22, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Holden, P.A.; Schimel, J.P.; Godwin, H.A. Five reasons to use bacteria when assessing manufactured nanomaterial environmental hazards and fates. Curr. Opin. Biotechnol. 2014, 27, 73–78. [Google Scholar] [CrossRef]
- Hamidat, M.; Barakat, M.; Ortet, P.; Chanéac, C.; Rose, J.; Bottero, J.-Y.; Heulin, T.; Achouak, W.; Santaella, C. Design defines the effects of nanoceria at a low dose on soil microbiota and the potentiation of impacts by the canola plant. Environ. Sci. Technol. 2016, 50, 6892–6901. [Google Scholar] [CrossRef]
- Ge, Y.; Priester, J.H.; Van De Werfhorst, L.C.; Walker, S.L.; Nisbet, R.M.; An, Y.-J.; Schimel, J.P.; Gardea-Torresdey, J.L.; Holden, P.A. Soybean plants modify metal oxide nanoparticle effects on soil bacterial communities. Environ. Sci. Technol. 2014, 48, 13489–13496. [Google Scholar] [CrossRef] [PubMed]
- Dahle, J. The Effects of Physicochemical Properties of CeO2 Nanoparticles on Toxicity to Soil Denitrification Processes. Master’s Thesis, Clemson University, Clemson, SC, USA, May 2013. [Google Scholar]
- Antisari, L.V.; Carbone, S.; Gatti, A.; Vianello, G.; Nannipieri, P. Toxicity of metal oxide (CeO2, Fe3O4, SnO2) engineered nanoparticles on soil microbial biomass and their distribution in soil. Soil Biol. Biochem. 2013, 60, 87–94. [Google Scholar] [CrossRef]
- Babenko, L.; Dankevich, L.A.; Zholobak, N.; Krout, V.; Leonova, N.; Demchenko, O.; Spivak, M.; Patyka, N. Effect of various concentrations of cerium dioxide nanoparticles on different physiological group of microorganisms [Вплив нанoчастинoк діoксиду церію на різні фізіoлoгічні групи мікрooрганізмів]. ЕКОЛОГІЯ 2014, 60, 45–51. [Google Scholar]
- Abbas, Q.; Liu, G.; Yousaf, B.; Ali, M.U.; Ullah, H.; Munir, M.A.M.; Ahmed, R.; Rehman, A. Biochar-assisted transformation of engineered-cerium oxide nanoparticles: Effect on wheat growth, photosynthetic traits and cerium accumulation. Ecotoxicol. Environ. Saf. 2020, 187, 109845. [Google Scholar] [CrossRef]
- Majumdar, S.; Almeida, I.C.; Arigi, E.A.; Choi, H.; VerBerkmoes, N.C.; Trujillo-Reyes, J.; Flores-Margez, J.P.; White, J.C.; Peralta-Videa, J.R.; Gardea-Torresdey, J.L. Environmental effects of nanoceria on seed production of common bean (Phaseolus vulgaris): A proteomic analysis. Environ. Sci. Technol. 2015, 49, 13283–13293. [Google Scholar] [CrossRef]
- Baalousha, M.; Ju-Nam, Y.; Cole, P.A.; Gaiser, B.; Fernandes, T.F.; Hriljac, J.A.; Jepson, M.A.; Stone, V.; Tyler, C.R.; Lead, J.R. Characterization of cerium oxide nanoparticles—Part 1: Size measurements. Environ. Toxicol. Chem. 2012, 31, 983–993. [Google Scholar] [CrossRef]
- Baalousha, M.; Ju-Nam, Y.; Cole, P.A.; Hriljac, J.A.; Jones, I.P.; Tyler, C.R.; Stone, V.; Fernandes, T.F.; Jepson, M.A.; Lead, J.R. Characterization of cerium oxide nanoparticles—Part 2: Nonsize measurements. Environ. Toxicol. Chem. 2012, 31, 994–1003. [Google Scholar] [CrossRef] [PubMed]
- Bao, S. Soil and Agricultural Chemistry Analysis; China Agriculture Press: Beijing, China, 2013. [Google Scholar]
- Nelson, D.W.; Sommers, L.E. Total carbon, organic carbon, and organic matter. In Methods of Soil Analysis Part 3—Chemical Methods; Soil Science Society of America and American Society of Agronomy: Madison, WI, USA, 1996; pp. 961–1010. [Google Scholar]
- Kundsen, N.; Patterson, G.; Pratt, P. Lithium, sodium, and potassium. In Methods of Soil Analysis part 2—Chemical and Microbiological Properties, 2nd ed.; Soil Science Society of America and American Society of Agronomy: Madison, WI, USA, 1983; pp. 225–246. [Google Scholar]
- Guo, M.; Wu, F.; Hao, G.; Qi, Q.; Li, R.; Li, N.; Wei, L.; Chai, T. Bacillus subtilis improves immunity and disease resistance in rabbits. Front. Immunol. 2017, 8, 354. [Google Scholar] [CrossRef] [Green Version]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [Green Version]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glöckner, F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef] [Green Version]
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001, 26, 32–46. [Google Scholar]
- Csardi, G.; Nepusz, T. The igraph software package for complex network research. InterJ. Complex Syst. 2006, 1695, 1–9. [Google Scholar]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An open source software for exploring and manipulating networks. Icwsm 2009, 8, 361–362. [Google Scholar]
- Zhang, J.; Jiao, S.; Lu, Y. Biogeographic distribution of bacterial, archaeal and methanogenic communities and their associations with methanogenic capacity in Chinese wetlands. Sci. Total Environ. 2018, 622–623, 664–675. [Google Scholar] [CrossRef]
- Barberán, A.; Bates, S.T.; Casamayor, E.O.; Fierer, N. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 2012, 6, 343–351. [Google Scholar] [CrossRef] [Green Version]
- González, A.M.M.; Dalsgaard, B.; Olesen, J.M. Centrality measures and the importance of generalist species in pollination networks. Ecol. Complex. 2010, 7, 36–43. [Google Scholar] [CrossRef]
- Vick-Majors, T.J.; Priscu, J.C.; Amaral-Zettler, L.A. Modular community structure suggests metabolic plasticity during the transition to polar night in ice-covered Antarctic lakes. ISME J. 2014, 8, 778–789. [Google Scholar] [CrossRef] [Green Version]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G. PICRUSt2: An improved and customizable approach for metagenome inference. bioRxiv 2020, 672295. [Google Scholar]
- Ward, T.; Larson, J.; Meulemans, J.; Hillmann, B.; Lynch, J.; Sidiropoulos, D.; Spear, J.R.; Caporaso, G.; Blekhman, R.; Knight, R. BugBase predicts organism-level microbiome phenotypes. bioRxiv 2017, 133462. [Google Scholar]
- Shi, C.; Zhao, L.; Guo, X.; Gao, S.; Li, J.; Yang, J. The Rare Earth Element Background Values of Soil in Shanxi. J. Shanxi Agric. Sci. 1995, 23, 38–40. [Google Scholar]
- Dahle, J.T.; Arai, Y. Effects of Ce(III) and CeO2 nanoparticles on soil-denitrification kinetics. Arch. Environ. Contam. Toxicol. 2014, 67, 474–482. [Google Scholar] [CrossRef]
- Andreescu, D.; Bulbul, G.; Özel, R.E.; Hayat, A.; Sardesai, N.; Andreescu, S. Applications and implications of nanoceria reactivity: Measurement tools and environmental impact. Environ. Sci. Nano 2014, 1, 445–458. [Google Scholar] [CrossRef]
- Othman, A.; Karimi, A.; Andreescu, S. Functional nanostructures for enzyme based biosensors: Properties, fabrication and applications. J. Mater. Chem. B 2016, 4, 7178–7203. [Google Scholar] [CrossRef]
- Taunton, A.E.; Welch, S.A.; Banfield, J.F. Geomicrobiological controls on light rare earth element, Y and Ba distributions during granite weathering and soil formation. J. Alloys Compd. 2000, 303–304, 30–36. [Google Scholar] [CrossRef]
- Cervini-Silva, J.; Fowle, D.A.; Banfield, J. Biogenic dissolution of a soil cerium-phosphate mineral. Am. J. Sci. 2005, 305, 711–726. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Liu, S.; Song, S.; Guo, H.; Tang, J.; Yong, J.W.; Ma, Y.; Chen, X. Arbuscular mycorrhizal fungi influence decomposition and the associated soil microbial community under different soil phosphorus availability. Soil Biol. Biochem. 2018, 120, 181–190. [Google Scholar] [CrossRef]
- Santiago, A.F.J.; Sousa, J.F.; Guedes, R.C.; Jerônimo, C.E.M.; Benachour, M. Kinetic and wet oxidation of phenol catalyzed by non-promoted and potassium-promoted manganese/cerium oxide. J. Hazard. Mater. 2006, 138, 325–330. [Google Scholar] [CrossRef]
- Stancu, M.M.; Grifoll, M. Multidrug resistance in hydrocarbon-tolerant Gram-positive and Gram-negative bacteria. J. Gen. Appl. Microbiol. 2011, 57, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Jia, S.; Shi, P.; Hu, Q.; Li, B.; Zhang, T.; Zhang, X.-X. Bacterial Community Shift Drives Antibiotic Resistance Promotion during Drinking Water Chlorination. Environ. Sci. Technol. 2015, 49, 12271–12279. [Google Scholar] [CrossRef]
- Máthé, I.; Benedek, T.; Táncsics, A.; Palatinszky, M.; Lányi, S.; Márialigeti, K. Diversity, activity, antibiotic and heavy metal resistance of bacteria from petroleum hydrocarbon contaminated soils located in Harghita County (Romania). Int. Biodeterior. Biodegrad. 2012, 73, 41–49. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, C.; Hou, J.; Wang, P.; Miao, L.; You, G.; Lv, B.; Yang, Y.; Zhang, F. Long term effects of cerium dioxide nanoparticles on the nitrogen removal, micro-environment and community dynamics of a sequencing batch biofilm reactor. Bioresour. Technol. 2017, 245, 573–580. [Google Scholar] [CrossRef]
- Dai, Y.; Chen, F.; Yue, L.; Li, T.; Jiang, Z.; Xu, Z.; Wang, Z.; Xing, B. Uptake, Transport, and Transformation of CeO2 Nanoparticles by Strawberry and Their Impact on the Rhizosphere Bacterial Community. ACS Sustain. Chem. Eng. 2020, 8, 4792–4800. [Google Scholar] [CrossRef]
- Jayaseelan, C.; Rahuman, A.A.; Kirthi, A.V.; Marimuthu, S.; Santhoshkumar, T.; Bagavan, A.; Gaurav, K.; Karthik, L.; Rao, K.V.B. Novel microbial route to synthesize ZnO nanoparticles using Aeromonas hydrophila and their activity against pathogenic bacteria and fungi. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2012, 90, 78–84. [Google Scholar] [CrossRef]
- Jayaseelan, C.; Rahuman, A.A.; Roopan, S.M.; Kirthi, A.V.; Venkatesan, J.; Kim, S.K.; Iyappan, M.; Siva, C. Biological approach to synthesize TiO2 nanoparticles using Aeromonas hydrophila and its antibacterial activity. Spectrochim. Acta Part A 2013, 107, 82–89. [Google Scholar] [CrossRef]
- Castro, L.; Li, J.; González, F.; Muñoz, J.A.; Blázquez, M.L. Green synthesis of tellurium nanoparticles by tellurate and tellurite reduction using Aeromonas hydrophila under different aeration conditions. Hydrometallurgy 2020, 196, 105415. [Google Scholar] [CrossRef]
- Ma, Y.; Metch, J.W.; Vejerano, E.P.; Miller, I.J.; Leon, E.C.; Marr, L.C.; Vikesland, P.J.; Pruden, A. Microbial community response of nitrifying sequencing batch reactors to silver, zero-valent iron, titanium dioxide and cerium dioxide nanomaterials. Water Res. 2015, 68, 87–97. [Google Scholar] [CrossRef]
- Daims, H. The family Nitrospiraceae. In The Prokaryotes. Other Major Lineages of Bacteria and The Archaea; Springer: Berlin/Heidelberg, Germany, 2014; pp. 733–749. [Google Scholar]
- Prosser, J.I.; Head, I.M.; Stein, L.Y. The family Nitrosomonadaceae. In The Prokaryotes: Alphaproteobacteria and Betaproteobacteria; Springer: Berlin/Heidelberg, Germany, 2014; pp. 901–918. [Google Scholar]
- Becraft, E.D.; Woyke, T.; Jarett, J.; Ivanova, N.; Godoy-Vitorino, F.; Poulton, N.; Brown, J.M.; Brown, J.; Lau, M.C.Y.; Onstott, T.; et al. Rokubacteria: Genomic giants among the uncultured bacterial phyla. Front. Microbiol. 2017, 8, 2264. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Li, W.; Li, Q.; Zhou, Y.; Gao, Z.; Feng, D. Deep-sea carbonates are a reservoir of fossil microbes previously inhabiting cold seeps. Front. Mar. Sci. 2021, 8, 982. [Google Scholar] [CrossRef]
- Kamika, I.; Tekere, M. Impacts of cerium oxide nanoparticles on bacterial community in activated sludge. AMB Express 2017, 7, 63. [Google Scholar] [CrossRef] [Green Version]
- Miroshnikov, S.; Yausheva, E.; Sizova, E.; Miroshnikova, E. Qualitative and Quantitative Composition of Zebrafish Intestinal Microflora in Case of Water Pollution by Nanoscale Cerium. Int. Multidiscip. Sci. GeoConf. SGEM 2019, 19, 365–371. [Google Scholar]
- Xu, C.; Qu, X. Cerium oxide nanoparticle: A remarkably versatile rare earth nanomaterial for biological applications. NPG Asia Mater. 2014, 6, e90. [Google Scholar] [CrossRef] [Green Version]
- Oberdrster, G.; Oberdrster, E.; Oberdrster, J. Nanotoxicology: An emerging discipline evolving from studies of ultrafine particles. Environ. Health Perspect. 2005, 113, 823–839. [Google Scholar] [CrossRef]
- Nemmar, A.; Holme, J.A.; Rosas, I.; Schwarze, P.E.; Alfaro-Moreno, E. Recent Advances in Particulate Matter and Nanoparticle Toxicology: A Review of the In Vivo and In Vitro Studies. BioMed Res. Int. 2013, 2013, 279371. [Google Scholar] [CrossRef]
- Lee, S.S.; Song, W.; Cho, M.; Puppala, H.L.; Nguyen, P.; Zhu, H.; Segatori, L.; Colvin, V.L. Antioxidant Properties of Cerium Oxide Nanocrystals as a Function of Nanocrystal Diameter and Surface Coating. ACS Nano 2013, 7, 9693–9703. [Google Scholar] [CrossRef]
- Baalousha, M.; Lead, J.R. Nanoparticle dispersity in toxicology. Nat. Nanotechnol. 2013, 8, 308–309. [Google Scholar] [CrossRef]
- Merrifield, R.C.; Wang, Z.W.; Palmer, R.E.; Lead, J.R. Synthesis and characterization of polyvinylpyrrolidone coated cerium oxide nanoparticles. Environ. Sci. Technol. 2013, 47, 12426–12433. [Google Scholar] [CrossRef]
- Grulke, E.; Reed, K.; Beck, M.; Huang, X.; Cormack, A.; Seal, S. Nanoceria: Factors affecting its pro-and anti-oxidant properties. Environ. Sci. Nano 2014, 1, 429–444. [Google Scholar] [CrossRef]
- Majumdar, S.; Peralta-Videa, J.R.; Trujillo-Reyes, J.; Sun, Y.; Barrios, A.C.; Niu, G.; Flores-Margez, J.P.; Gardea-Torresdey, J.L. Soil organic matter influences cerium translocation and physiological processes in kidney bean plants exposed to cerium oxide nanoparticles. Sci. Total Environ. 2016, 569–570, 201–211. [Google Scholar] [CrossRef] [Green Version]
- Patil, S.; Sandberg, A.; Heckert, E.; Self, W.; Seal, S. Protein adsorption and cellular uptake of cerium oxide nanoparticles as a function of zeta potential. Biomaterials 2007, 28, 4600–4607. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Gao, M.; Li, Z.; She, Z.; Wu, J.; Zheng, D.; Guo, L.; Zhao, Y.; Gao, F.; Wang, X. Performance evaluation, microbial enzymatic activity and microbial community of a sequencing batch reactor under long-term exposure to cerium dioxide nanoparticles. Bioresour. Technol. 2016, 220, 262–270. [Google Scholar] [CrossRef]
- Fang, X.; Yu, R.; Li, B.; Somasundaran, P.; Chandran, K. Stresses exerted by ZnO, CeO2 and anatase TiO2 nanoparticles on the Nitrosomonas europaea. J. Colloid Interface Sci. 2010, 348, 329–334. [Google Scholar] [CrossRef]
- Yu, R.; Fang, X.; Somasundaran, P.; Chandran, K. Short-term effects of TiO2, CeO2, and ZnO nanoparticles on metabolic activities and gene expression of Nitrosomonas europaea. Chemosphere 2015, 128, 207–215. [Google Scholar] [CrossRef]
- Wu, J.; Zhan, M.; Chang, Y.; Gao, H.; Ye, J.; Yu, R.; Ding, Z. Mechanistic insights into stress response and metabolic activity resilience of Nitrosomonas europaea cultures to long-term CeO2 nanoparticle exposure. Environ. Sci. Nano 2019, 6, 2215–2227. [Google Scholar] [CrossRef]
- del Rocío Bustillos-Cristales, M.; Corona-Gutierrez, I.; Castañeda-Lucio, M.; Águila-Zempoaltécatl, C.; Seynos-García, E.; Hernández-Lucas, I.; Muñoz-Rojas, J.; Medina-Aparicio, L.; Fuentes-Ramírez, L.E. Culturable facultative methylotrophic bacteria from the cactus Neobuxbaumia macrocephala possess the locus xoxF and consume methanol in the presence of Ce3+ and Ca2+. Microbes. Environ. 2017, 32, 244–251. [Google Scholar] [CrossRef] [Green Version]
- Xia, T.; Kovochich, M.; Liong, M.; Mädler, L.; Gilbert, B.; Shi, H.; Yeh, J.I.; Zink, J.I.; Nel, A.E. Comparison of the mechanism of toxicity of zinc oxide and cerium oxide nanoparticles based on dissolution and oxidative stress properties. ACS Nano 2008, 2, 2121–2134. [Google Scholar] [CrossRef] [PubMed]






| SOM g/kg | TN g/kg | AN mg/kg | TP g/kg | AP mg/kg | TK g/kg | AK mg/kg | Ce mg/kg | |
|---|---|---|---|---|---|---|---|---|
| NC10 | 17.22 ± 1.27 a | 0.19 ± 0.02 a | 75.21 ± 3.01 a,b | 0.78 ± 0.03 a | 18.51 ± 2.69 c | 19.02 ± 0.51 b | 107.19 ± 3.01 b,c | 10.09 ± 2.63 d |
| NC100 | 17.22 ± 1.06 a | 0.16 ± 0.01 a | 55.97 ± 3.19 b | 0.7 ± 0.06 a | 22.68 ± 1.24 c | 19.19 ± 0.67 b | 119.32 ± 15.47 b,c | 38.87 ± 0.66 c |
| NC500 | 17.14 ± 0.98 a | 0.18 ± 0.001 a | 73.16 ± 10.01 a,b | 0.78 ± 0.01 a | 23.43 ± 2.63 c | 19.09 ± 0.21 b | 107.53 ± 25.43 bc | 135.03 ± 28.91 a |
| IC10 | 16.33 ± 1.04 a | 0.16 ± 0.003 a | 56.78 ± 2.51 b | 0.65 ± 0.07 a,b | 43.3 ± 7.28 a,b | 20.34 ± 0.96 b | 141.71 ± 7.96 b | 9.57 ± 2.56 d |
| IC100 | 17.26 ± 0.65 a | 0.17 ± 0.01 a | 58.01 ± 6.07 b | 0.64 ± 0.005 a,b | 32.45 ± 3.64 b,c | 18.89 ± 0.68 b | 124.21 ± 3.96 bc | 44.43 ± 0.65 c |
| IC500 | 17.99 ± 1.1 a | 0.19 ± 0.02 a | 80.81 ± 20.1 a,b | 0.74 ± 0.003 a | 27.85 ± 0.63 c | 19.8 ± 0.58 b | 95.09 ± 3.88 c | 103.13 ± 14.10 b |
| CK | 17.19 ± 0.9 a | 0.21 ± 0.02 a | 101.97 ± 2.66 a | 0.51 ± 0.08 b | 49.63 ± 6.68 a | 22.39 ± 0.48 a | 252.36 ± 16.7 a | 9.74 ± 1.24 d |
| Cerium Type | Bacterial Taxa | r | p |
|---|---|---|---|
| Nanoceria | Proteobacteria | 0.67 | 0.017 |
| Lentisphaerae | 0.62 | 0.033 | |
| Rokubacteria | 0.60 | 0.037 | |
| Epsilonbacteraeota | −0.60 | 0.037 | |
| Ionic cerium | Hydrogenedentes | 0.59 | 0.043 |
| Bacteroidetes | −0.71 | 0.009 |
| Node | Edge | Average Degree | Network Diameter | Graph Density | Modularity | Clustering Coefficient | Average Path Length | |
|---|---|---|---|---|---|---|---|---|
| NC | 82 | 101 | 2.463 | 9 | 0.03 | 0.781 | 0.534 | 3.632 |
| IC | 91 | 214 | 4.703 | 16 | 0.052 | 0.719 | 0.515 | 5.233 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Meng, H.-S.; Shang, Y.-M.; Lead, J.R.; Guo, Z.-Z.; Hong, J.-P. Response of Soil Bacterial Diversity, Predicted Functions and Co-Occurrence Patterns to Nanoceria and Ionic Cerium Exposure. Microorganisms 2022, 10, 1982. https://doi.org/10.3390/microorganisms10101982
Zhang J, Meng H-S, Shang Y-M, Lead JR, Guo Z-Z, Hong J-P. Response of Soil Bacterial Diversity, Predicted Functions and Co-Occurrence Patterns to Nanoceria and Ionic Cerium Exposure. Microorganisms. 2022; 10(10):1982. https://doi.org/10.3390/microorganisms10101982
Chicago/Turabian StyleZhang, Jie, Hui-Sheng Meng, Yan-Meng Shang, Jamie R. Lead, Zhang-Zhen Guo, and Jian-Ping Hong. 2022. "Response of Soil Bacterial Diversity, Predicted Functions and Co-Occurrence Patterns to Nanoceria and Ionic Cerium Exposure" Microorganisms 10, no. 10: 1982. https://doi.org/10.3390/microorganisms10101982
APA StyleZhang, J., Meng, H.-S., Shang, Y.-M., Lead, J. R., Guo, Z.-Z., & Hong, J.-P. (2022). Response of Soil Bacterial Diversity, Predicted Functions and Co-Occurrence Patterns to Nanoceria and Ionic Cerium Exposure. Microorganisms, 10(10), 1982. https://doi.org/10.3390/microorganisms10101982






