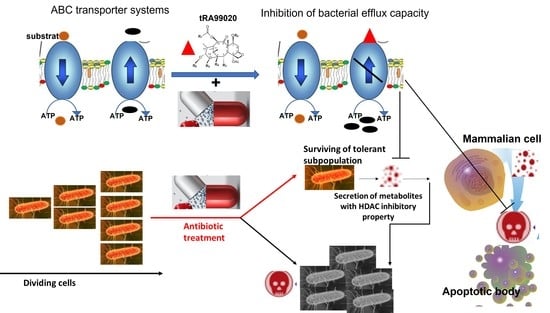
Targeting the Bet-Hedging Strategy with an Inhibitor of Bacterial Efflux Capacity Enhances Antibiotic Efficiency and Ameliorates Bacterial Persistence In Vitro
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Reagents
2.2. Enumeration of Persister Populations
2.3. Evaluation of Bacterial Export Capacity
2.4. Treatment of Human PBMCs and NHBEC with Bacterial Natural Products
2.5. Histone H3 Acetylation and Caspase 3 Activation Assays
2.6. smFISH Image Analysis
2.7. Statistical Analysis
3. Results
3.1. Taxane-Based Reversal Agent, tRA99020, Acts as an Inhibitor of B. thailandensis Efflux Capacity and Potentiates Antibiotic-Mediated Killing of Bacterial Persister Populations
3.2. At 1 µM Concentration tRA99020 Stimulates Expression of Bacterial Persistence pks Gene in B. thailandensis Populations
3.3. Natural Products of Burkholderia PKS Function Modulate HDAC Activity in Live Human Peripheral Blood Mononuclear Cells
3.4. Natural Products of B. thailandensis PKS Function Inhibit Pro-Survival Mechanisms in Host Cells
3.5. Supplementation of Antibiotic Treatment with tRA99020 Agent Ameliorates the Cytotoxic Effect of Burkholderia spp. Natural Products on Host Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Rocha-Granados, M.C.; Zenick, B.; Englander, H.E.; Mok, W.W. The social network: Impact of host and microbial interactions on bacterial antibiotic tolerance and persistence. Cell Signal. 2020, 75, 109750. [Google Scholar] [CrossRef]
- Van den Bergh, B.; Fauvart, M.; Michiels, J. Formation, physiology, ecology, evolution and clinical importance of bacterial persisters. FEMS Microbiol. Rev. 2017, 41, 219–251. [Google Scholar] [CrossRef] [PubMed]
- Windels, E.M.; Bergh, B.V.D.; Michiels, J. Bacteria under antibiotic attack: Different strategies for evolutionary adaptation. PLoS Pathog. 2020, 16, e1008431. [Google Scholar] [CrossRef]
- Fridman, O.; Goldberg, A.; Ronin, I.; Shoresh, N.; Balaban, N. Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations. Nature 2014, 513, 418–421. [Google Scholar] [CrossRef]
- Micheva-Viteva, S.N.; Ross, B.N.; Gao, J.; Adikari, S.; Zhang, P.; Mourant, J.R.; Wu, T.H.; Werner, J.H.; Torres, A.G.; Hong-Geller, E. Increased Mortality in Mice following Immunoprophylaxis Therapy with High Dosage of Nicotinamide in Burkholderia Persistent Infections. Infect. Immun. 2019, 87, e00592-18. [Google Scholar] [CrossRef] [PubMed]
- Harms, A.; Maisonneuve, E.; Gerdes, K. Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 2016, 354, aaf4268. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.H.; Bhargava, P.; McCloskey, D.; Mao, N.; Palsson, B.O.; Collins, J.J. Antibiotic-Induced Changes to the Host Metabolic Environment Inhibit Drug Efficacy and Alter Immune Function. Cell Host Microbe 2017, 22, 757–765.e3. [Google Scholar] [CrossRef] [PubMed]
- Gollan, B.; Grabe, G.; Michaux, C.; Helaine, S. Bacterial Persisters and Infection: Past, Present, and Progressing. Annu. Rev. Microbiol. 2019, 73, 359–385. [Google Scholar] [CrossRef] [PubMed]
- Levin-Reisman, I.; Ronin, I.; Gefen, O.; Braniss, I.; Shoresh, N.; Balaban, N.Q. Antibiotic tolerance facilitates the evolution of resistance. Science 2017, 355, 826–830. [Google Scholar] [CrossRef] [PubMed]
- Micheva-Viteva, S.N.; Shakya, M.; Adikari, S.H.; Gleasner, C.D.; Velappan, N.; Mourant, J.R.; Chain, P.S.G.; Hong-Geller, E. A Gene Cluster That Encodes Histone Deacetylase Inhibitors Contributes to Bacterial Persistence and Antibiotic Tolerance in Burkholderia thailandensis. mSystems 2020, 5, e00609-19. [Google Scholar] [CrossRef]
- Marques, C.N.H.; Morozov, A.; Planzos, P.; Zelaya, H.M. The Fatty Acid Signaling Molecule cis-2-Decenoic Acid Increases Metabolic Activity and Reverts Persister Cells to an Antimicrobial-Susceptible State. Appl. Environ. Microbiol. 2014, 80, 6976–6991. [Google Scholar] [CrossRef] [PubMed]
- Pu, Y.; Zhao, Z.; Li, Y.; Zou, J.; Ma, Q.; Zhao, Y.; Ke, Y.; Zhu, Y.; Chen, H.; Baker, M.A.; et al. Enhanced Efflux Activity Facilitates Drug Tolerance in Dormant Bacterial Cells. Mol. Cell 2016, 62, 284–294. [Google Scholar] [CrossRef] [PubMed]
- Alcalde-Rico, M.; Hernando-Amado, S.; Blanco, P.; Martínez, J.L. Multidrug Efflux Pumps at the Crossroad between Antibiotic Resistance and Bacterial Virulence. Front. Microbiol. 2016, 7, 1483. [Google Scholar] [CrossRef] [PubMed]
- Rosen, B.C.; Dillon, N.; Peterson, N.D.; Minato, Y.; Baughn, A.D. Long-Chain Fatty Acyl Coenzyme A Ligase FadD2 Mediates Intrinsic Pyrazinamide Resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2017, 61, 753. [Google Scholar] [CrossRef]
- Pandey, A.K.; Sassetti, C.M. Mycobacterial persistence requires the utilization of host cholesterol. Proc. Natl. Acad. Sci. USA 2008, 105, 4376–4380. [Google Scholar] [CrossRef]
- Lomovskaya, O.; Warren, M.S.; Lee, A.; Galazzo, J.; Fronko, R.; Lee, M.; Blais, J.; Cho, D.; Chamberland, S.; Renau, T.; et al. Identification and Characterization of Inhibitors of Multidrug Resistance Efflux Pumps in Pseudomonas aeruginosa: Novel Agents for Combination Therapy. Antimicrob. Agents Chemother. 2001, 45, 105–116. [Google Scholar] [CrossRef]
- Li, X.-Z.; Plésiat, P.; Nikaido, H. The Challenge of Efflux-Mediated Antibiotic Resistance in Gram-Negative Bacteria. Clin. Microbiol. Rev. 2015, 28, 337–418. [Google Scholar] [CrossRef]
- Brooks, T.A.; Minderman, H.; O’Loughlin, K.L.; Pera, P.; Ojima, I.; Baer, M.R.; Bernacki, R.J. Taxane-based reversal agents modulate drug resistance mediated by P-glycoprotein, multidrug resistance protein, and breast cancer resistance protein. Mol. Cancer Ther. 2003, 2, 1195–1205. [Google Scholar]
- Ojima, I.; Bounaud, P.-Y.; Takeuchi, C.; Pera, P.; Bernacki, R.J. New taxanes as highly efficient reversal agents for multi-drug resistance in cancer cells. Bioorg. Med. Chem. Lett. 1998, 8, 189–194. [Google Scholar] [CrossRef]
- Schweizer, H.P. Mechanisms of antibiotic resistance in Burkholderia pseudomallei: Implications for treatment of melioidosis. Futur. Microbiol. 2012, 7, 1389–1399. [Google Scholar] [CrossRef]
- Rhodes, K.A.; Schweizer, H.P. Antibiotic resistance in Burkholderia species. Drug Resist. Updates Rev. Comment. Antimicrob. Anticancer. Chemother. 2016, 28, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Nierman, W.C.; Yu, Y.; Losada, L. The In vitro Antibiotic Tolerant Persister Population in Burkholderia pseudomallei is Altered by Environmental Factors. Front. Microbiol. 2015, 6, 1338. [Google Scholar] [CrossRef] [PubMed]
- Scoffone, V.C.; Chiarelli, L.R.; Trespidi, G.; Mentasti, M.; Riccardi, G.; Buroni, S. Burkholderia cenocepacia Infections in Cystic Fibrosis Patients: Drug Resistance and Therapeutic Approaches. Front. Microbiol. 2017, 8, 1592. [Google Scholar] [CrossRef] [PubMed]
- Kovacs-Simon, A.; Hemsley, C.M.; Scott, A.E.; Prior, J.L.; Titball, R.W. Burkholderia thailandensis strain E555 is a surrogate for the investigation of Burkholderia pseudomallei replication and survival in macrophages. BMC Microbiol. 2019, 19, 97. [Google Scholar] [CrossRef]
- Barker, S.; Harding, S.V.; Gray, D.; Richards, M.I.; Atkins, H.S.; Harmer, N.J. Drug screening to identify compounds to act as co-therapies for the treatment of Burkholderia species. PLoS ONE 2021, 16, e0248119. [Google Scholar] [CrossRef]
- Biggins, J.B.; Gleber, C.D.; Brady, S.F. Acyldepsipeptide HDAC Inhibitor Production Induced in Burkholderia thailandensis. Org. Lett. 2011, 13, 1536–1539. [Google Scholar] [CrossRef]
- Ge, X.; Cai, Y.; Chen, Z.; Gao, S.; Geng, X.; Li, Y.; Jia, J.; Sun, Y. Bifunctional Enzyme SpoT Is Involved in Biofilm Formation of Helicobacter pylori with Multidrug Resistance by Upregulating Efflux Pump Hp1174 (gluP). Antimicrob. Agents Chemother. 2018, 62, e00957-18. [Google Scholar] [CrossRef]
- Reuter, G.; Janvilisri, T.; Venter, H.; Shahi, S.; Balakrishnan, L.; van Veen, H.W. The ATP Binding Cassette Multidrug Transporter LmrA and Lipid Transporter MsbA Have Overlapping Substrate Specificities*. J. Biol. Chem. 2003, 278, 35193–35198. [Google Scholar] [CrossRef]
- Qu, Q.; Sharom, F. Proximity of Bound Hoechst 33342 to the ATPase Catalytic Sites Places the Drug Binding Site of P-glycoprotein within the Cytoplasmic Membrane Leaflet. Biochemistry 2002, 41, 4744–4752. [Google Scholar] [CrossRef]
- Robey, R.; Honjo, Y.; van de Laar, A.; Miyake, K.; Regis, J.T.; Litman, T.; Bates, S.E. A functional assay for detection of the mitoxantrone resistance protein, MXR (ABCG2). Biochim. Biophys. Acta 2001, 1512, 171–182. [Google Scholar] [CrossRef]
- Schotterl, S.; Brennenstuhl, H.; Naumann, U. Modulation of immune responses by histone deacetylase inhibitors. Crit. Rev. Oncog. 2015, 20, 139–154. [Google Scholar] [CrossRef] [PubMed]
- Edelstein, L.; Micheva-Viteva, S.; Phelan, B.D.; Dougherty, J.P. Short Communication: Activation of Latent HIV Type 1 Gene Expression by Suberoylanilide Hydroxamic Acid (SAHA), an HDAC Inhibitor Approved for Use to Treat Cutaneous T Cell Lymphoma. AIDS Res. Hum. Retrovir. 2009, 25, 883–887. [Google Scholar] [CrossRef] [PubMed]
- Ram, D.R.; Ilyukha, V.; Volkova, T.; Buzdin, A.; Tai, A.; Smirnova, I.; Poltorak, A. Balance between short and long isoforms of cFLIP regulates Fas-mediated apoptosis in vivo. Proc. Natl. Acad. Sci. USA 2016, 113, 1606–1611. [Google Scholar] [CrossRef] [PubMed]
- Loris, R.; Garcia-Pino, A. Disorder- and Dynamics-Based Regulatory Mechanisms in Toxin–Antitoxin Modules. Chem. Rev. 2014, 114, 6933–6947. [Google Scholar] [CrossRef] [PubMed]
- Heaton, B.E.; Herrou, J.; Blackwell, A.E.; Wysocki, V.H.; Crosson, S. Molecular Structure and Function of the Novel BrnT/BrnA Toxin-Antitoxin System of Brucella abortus. J. Biol. Chem. 2012, 287, 12098–12110. [Google Scholar] [CrossRef]
- Yang, Q.E.; Walsh, T.R. Toxin–antitoxin systems and their role in disseminating and maintaining antimicrobial resistance. FEMS Microbiol. Rev. 2017, 41, 343–353. [Google Scholar] [CrossRef]
- Chan, Y.A.; Podevels, A.M.; Kevany, B.M.; Thomas, M.G. Biosynthesis of polyketide synthase extender units. Nat. Prod. Rep. 2009, 26, 90–114. [Google Scholar] [CrossRef]
- Locher, K.P. Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat. Struct. Mol. Biol. 2016, 23, 487–493. [Google Scholar] [CrossRef]
- Cuthbertson, L.; Kos, V.; Whitfield, C. ABC transporters involved in export of cell surface glycoconjugates. Microbiol. Mol. Biol. Rev. 2010, 74, 341–362. [Google Scholar] [CrossRef]
- Wong, K.; Ma, J.; Rothnie, A.; Biggin, P.C.; Kerr, I.D. Towards understanding promiscuity in multidrug efflux pumps. Trends Biochem. Sci. 2014, 39, 8–16. [Google Scholar] [CrossRef]
- Sarkadi, B.; Homolya, L.; Szakacs, G.; Váradi, A. Human Multidrug Resistance ABCB and ABCG Transporters: Participation in a Chemoimmunity Defense System. Physiol. Rev. 2006, 86, 1179–1236. [Google Scholar] [CrossRef] [PubMed]
- Kerr, I.D.; Jones, P.M.; George, A.M. Multidrug efflux pumps: The structures of prokaryotic ATP-binding cassette transporter efflux pumps and implications for our understanding of eukaryotic P-glycoproteins and homologues. FEBS J. 2010, 277, 550–563. [Google Scholar] [CrossRef]
- Richardson, M.; Khosla, C. Structure, Function, and Engineering of Bacterial Aromatic Polyketide Synthases. In Comprehensive Natural Products Chemistry; Barton, S.D., Nakanishi, K., Meth-Cohn, O., Eds.; Pergamon: Oxford, UK, 1999; pp. 473–494. [Google Scholar] [CrossRef]
- Kastrinsky, D.B.; McBride, N.S.; Backus, K.M.; LeBlanc, J.J.; Barry, C.E. Mycolic Acid/Cyclopropane Fatty Acid/Fatty Acid Biosynthesis and Health Relations. In Comprehensive Natural Products II; Liu, H.-W., Mander, L., Eds.; Elsevier: Oxford, UK, 2010; pp. 65–145. [Google Scholar] [CrossRef]
- Minderman, H.; Brooks, T.A.; O’Loughlin, K.L.; Ojima, I.; Bernacki, R.J.; Baer, M.R. Broad-spectrum modulation of ATP-binding cassette transport proteins by the taxane derivatives ortataxel (IDN-5109, BAY 59-8862) and tRA96023. Cancer Chemother. Pharmacol. 2004, 53, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Roger, T.; Lugrin, J.; LE Roy, D.; Goy, G.; Mombelli, M.; Koessler, T.; Ding, X.C.; Chanson, A.-L.; Reymond, M.K.; Miconnet, I.; et al. Histone deacetylase inhibitors impair innate immune responses to Toll-like receptor agonists and to infection. Blood 2011, 117, 1205–1217. [Google Scholar] [CrossRef] [PubMed]
- Fortuin, S.; Soares, N.C. The Integration of Proteomics and Metabolomics Data Paving the Way for a Better Understanding of the Mechanisms Underlying Microbial Acquired Drug Resistance. Front. Med. 2022, 9, 849838. [Google Scholar] [CrossRef] [PubMed]
- Peng, B.; Li, H.; Peng, X. Proteomics approach to understand bacterial antibiotic resistance strategies. Expert Rev. Proteom. 2019, 16, 829–839. [Google Scholar] [CrossRef] [PubMed]
- Sulaiman, J.E.; Lam, H. Application of proteomics in studying bacterial persistence. Expert Rev. Proteom. 2019, 16, 227–239. [Google Scholar] [CrossRef]
- Zhang, D.; Hu, Y.; Zhu, Q.; Huang, J.; Chen, Y. Proteomic interrogation of antibiotic resistance and persistence in Escherichia coli—Progress and potential for medical research. Expert Rev. Proteom. 2020, 17, 393–409. [Google Scholar] [CrossRef]
- Wang, L.; Abdulla, A.; Wang, A.; Warden, A.R.; Ahmad, K.Z.; Xin, Y.; Ding, X. Sickle-like Inertial Microfluidic System for Online Rare Cell Separation and Tandem Label-Free Quantitative Proteomics (Orcs-Proteomics). Anal. Chem. 2022, 94, 6026–6035. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morales, D.; Micheva-Viteva, S.; Adikari, S.; Werner, J.; Wolinsky, M.; Hong-Geller, E.; Kim, J.; Ojima, I. Targeting the Bet-Hedging Strategy with an Inhibitor of Bacterial Efflux Capacity Enhances Antibiotic Efficiency and Ameliorates Bacterial Persistence In Vitro. Microorganisms 2022, 10, 1966. https://doi.org/10.3390/microorganisms10101966
Morales D, Micheva-Viteva S, Adikari S, Werner J, Wolinsky M, Hong-Geller E, Kim J, Ojima I. Targeting the Bet-Hedging Strategy with an Inhibitor of Bacterial Efflux Capacity Enhances Antibiotic Efficiency and Ameliorates Bacterial Persistence In Vitro. Microorganisms. 2022; 10(10):1966. https://doi.org/10.3390/microorganisms10101966
Chicago/Turabian StyleMorales, Demosthenes, Sofiya Micheva-Viteva, Samantha Adikari, James Werner, Murray Wolinsky, Elizabeth Hong-Geller, Jinwoo Kim, and Iwao Ojima. 2022. "Targeting the Bet-Hedging Strategy with an Inhibitor of Bacterial Efflux Capacity Enhances Antibiotic Efficiency and Ameliorates Bacterial Persistence In Vitro" Microorganisms 10, no. 10: 1966. https://doi.org/10.3390/microorganisms10101966
APA StyleMorales, D., Micheva-Viteva, S., Adikari, S., Werner, J., Wolinsky, M., Hong-Geller, E., Kim, J., & Ojima, I. (2022). Targeting the Bet-Hedging Strategy with an Inhibitor of Bacterial Efflux Capacity Enhances Antibiotic Efficiency and Ameliorates Bacterial Persistence In Vitro. Microorganisms, 10(10), 1966. https://doi.org/10.3390/microorganisms10101966