Marine Bioprospecting, Biocatalysis and Process Development
Abstract
1. Introduction
2. Marine Biocatalysts to Improve Industrial Processes
3. Assessing Not-Yet-Cultured Biocatalysts
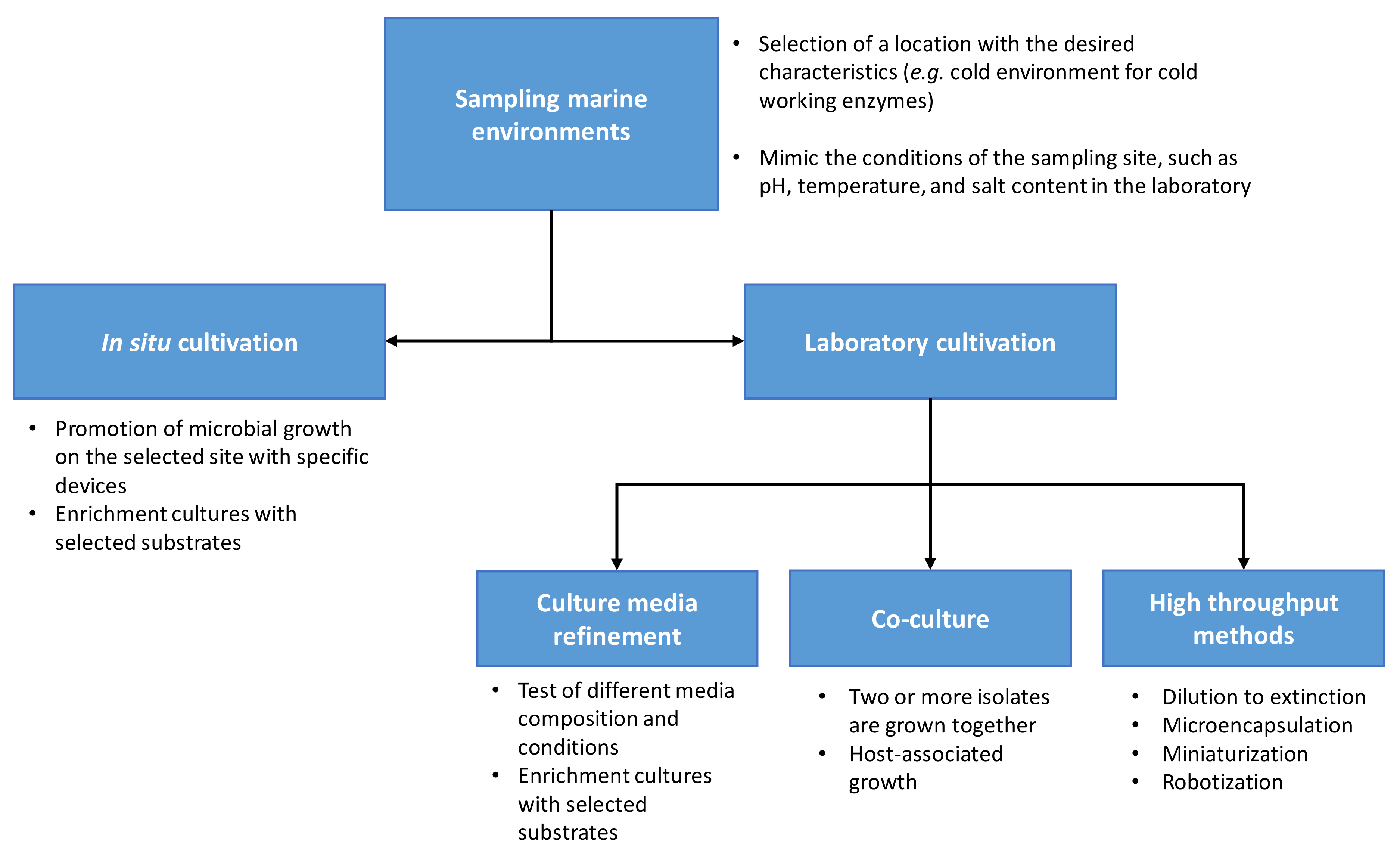
Techniques for Cultivation of Marine Bacteria
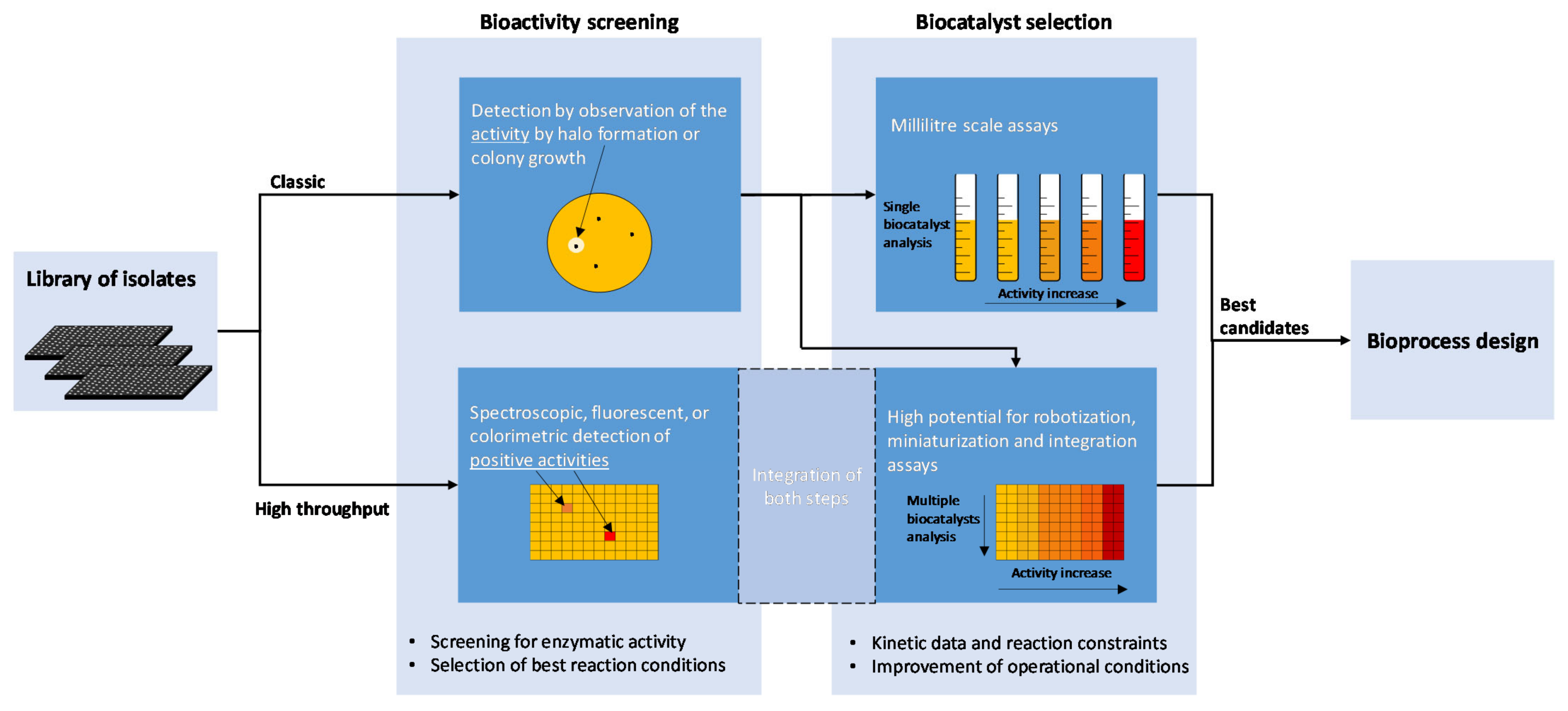
4. Detection of Biocatalysts from Cultured Bacteria
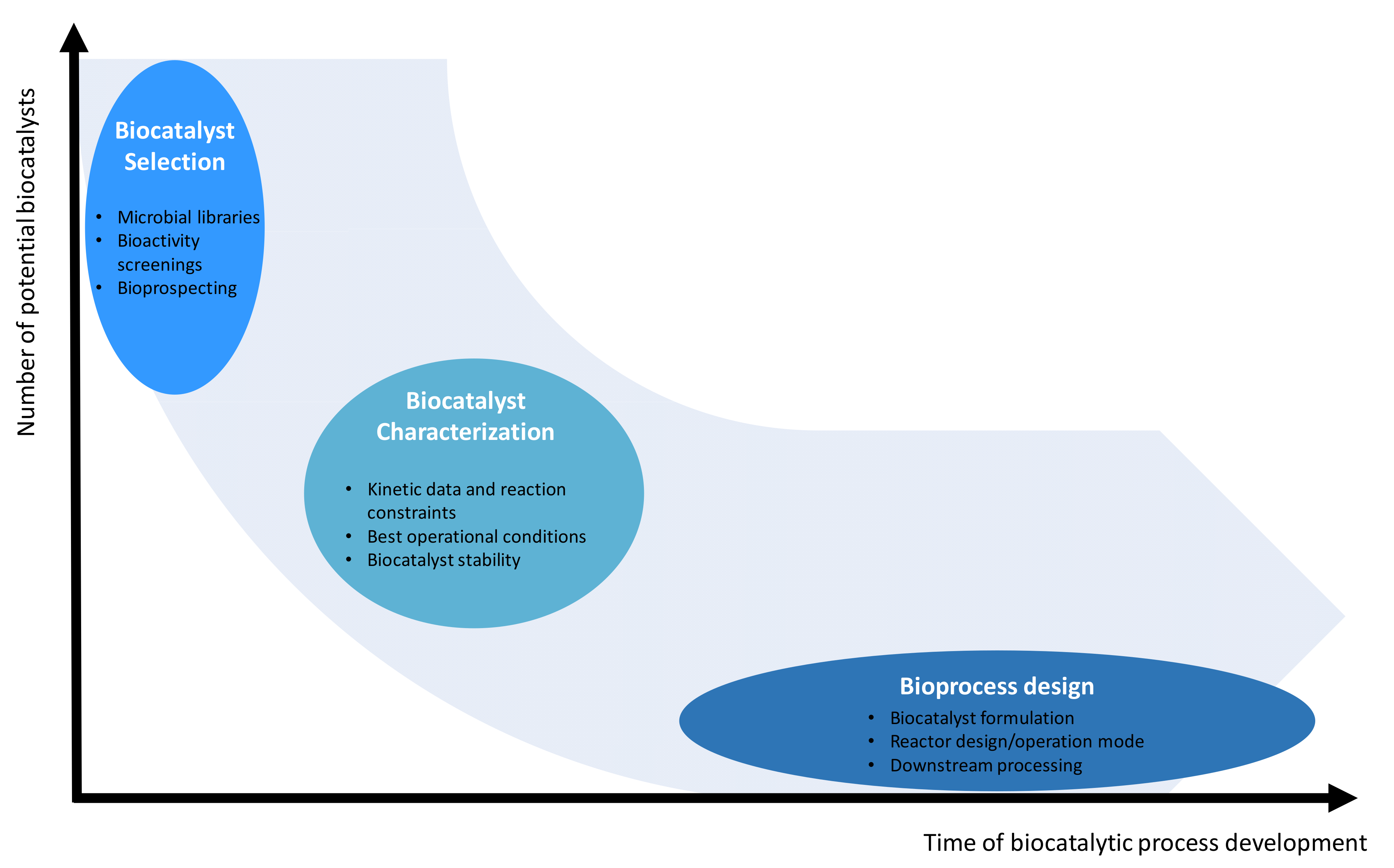
5. Bioprocess Development
6. Final Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dodd, M.S.; Papineau, D.; Grenne, T.; Slack, J.F.; Rittner, M.; Pirajno, F.; O’Neil, J.; Little, C.T.S. Evidence for early life in Earth’s oldest hydrothermal vent precipitates. Nature 2017, 543, 60–64. [Google Scholar] [CrossRef]
- Locey, K.J.; Lennon, J.T. Scaling laws predict global microbial diversity. Proc. Natl. Acad. Sci. USA 2016, 113, 5970. [Google Scholar] [CrossRef] [PubMed]
- Nikapitiya, C. Bioactive secondary metabolites from marine microbes for drug discovery. Adv. Food Nutr. Res. 2012, 65, 363–387. [Google Scholar] [CrossRef] [PubMed]
- De Carvalho, C.C.C.R.; Fernandes, P. Production of metabolites as bacterial responses to the marine environment. Mar. Drugs 2010, 8, 705–727. [Google Scholar] [CrossRef] [PubMed]
- Blevins, S.M.; Bronze, M.S. Robert Koch and the ‘golden age’ of bacteriology. Int. J. Infect. Dis. 2010, 14, e744–e751. [Google Scholar] [CrossRef] [PubMed]
- Zobell, C.E. Studies on marine bacteria. I. The cultural requirements of heterotrophic aerobes. J. Mar. Res. 2019, 4, 41–75. [Google Scholar]
- Razumov, A.S. The direct method of calculation of bacteria in water: Comparison with the Koch method. Mikrobiologija 1932, 1, 131–146. [Google Scholar]
- Staley, J.T.; Konopka, A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu. Rev. Microbiol. 1985, 39, 321–346. [Google Scholar] [CrossRef]
- Staley, J.T.; Lehmicke, L.G.; Palmer, F.E.; Peet, R.W.; Wissmar, R.C. Impact of Mount St. Helens eruption on bacteriology of lakes in the blast zone. Appl. Environ. Microbiol. 1982, 43, 664–670. [Google Scholar] [CrossRef]
- Giovannoni, S.J.; Britschgi, T.B.; Moyer, C.L.; Field, K.G. Genetic diversity in Sargasso Sea bacterioplankton. Nature 1990, 345, 60–63. [Google Scholar] [CrossRef]
- National Research Council (US). Committee on Metagenomics: Challenges and Functional Applications. The New Science of Metagenomics: Revealing the Secrets of Our Microbial Planet; National Academies Press: Washington, DC, USA, 2007. [Google Scholar]
- Stein, J.L.; Marsh, T.L.; Wu, K.-Y.; Shizuya, H.; Delong, E.F. Characterization of uncultivated prokaryotes: Isolation and analysis of a 40-kilobase-pair genome fragment from a planktonic marine archaeon. J. Bacteriol. 1996, 178, 591–599. [Google Scholar] [CrossRef]
- Ngara, T.R.; Zhang, H. Recent Advances in Function-based Metagenomic Screening. Genom. Proteom. Bioinform. 2018, 16, 405–415. [Google Scholar] [CrossRef]
- Lagier, J.C.; Dubourg, G.; Million, M.; Cadoret, F.; Bilen, M.; Fenollar, F.; Levasseur, A.; Rolain, J.M.; Fournier, P.E.; Raoult, D. Culturing the human microbiota and culturomics. Nat. Rev. Microbiol. 2018, 16, 540–550. [Google Scholar] [CrossRef] [PubMed]
- Wooley, J.C.; Ye, Y. Metagenomics: Facts and artifacts, and computational challenges. J. Comput. Syst. Sci. 2010, 25, 71–81. [Google Scholar] [CrossRef]
- Sun, Z.; Huang, S.; Zhang, M.; Zhu, Q.; Haiminen, N.; Carrieri, A.P.; Vázquez-Baeza, Y.; Parida, L.; Kim, H.-C.; Knight, R.; et al. Challenges in benchmarking metagenomic profilers. Nat. Methods 2021, 18, 618–626. [Google Scholar] [CrossRef]
- Keller, M.; Zengler, K. Tapping into microbial diversity. Nat. Rev. Microbiol. 2004, 2, 141–150. [Google Scholar] [CrossRef]
- Vartoukian, S.R.; Palmer, R.M.; Wade, W.G. Strategies for culture of ‘unculturable’ bacteria. FEMS Microbiol. Lett. 2010, 309, 1–7. [Google Scholar] [CrossRef]
- Marx, V. Microbiology: The return of culture. Nat. Meth. 2017, 14, 37–40. [Google Scholar] [CrossRef]
- Marques, M.P.C.; Walshe, K.; Doyle, S.; Fernandes, P.; de Carvalho, C.C.C.R. Anchoring high-throughput screening methods to scale-up bioproduction of siderophores. Process Biochem. 2012, 47, 416–421. [Google Scholar] [CrossRef][Green Version]
- Lagier, J.C.; Hugon, P.; Khelaifia, S.; Fournier, P.E.; La Scola, B.; Raoult, D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin. Microbiol. Rev. 2015, 28, 237–264. [Google Scholar] [CrossRef]
- Gest, H. The Modern Myth of “Unculturable” Bacteria/Scotoma of contemporary microbiology. Available online: http://hdl.handle.net/2022/3149 (accessed on 1 July 2022).
- Puspita, I.D.; Kamagata, Y.; Tanaka, M.; Asano, K.; Nakatsu, C.H. Are uncultivated bacteria really uncultivable? Microbes Environ. 2012, 27, 356–366. [Google Scholar] [CrossRef]
- Rodrigues, C.J.C.; de Carvalho, C.C.C.R. Cultivating marine bacteria under laboratory conditions: Overcoming the “unculturable” dogma. Front. Bioeng. Biotechnol. 2022, 10, 964589. [Google Scholar] [CrossRef]
- Rodrigues, C.J.C.; Pereira, R.F.S.; Fernandes, P.; Cabral, J.M.S.; de Carvalho, C.C.C.R. Cultivation-based strategies to find efficient marine biocatalysts. Biotechnol. J. 2017, 12, 1700036. [Google Scholar] [CrossRef]
- Imachi, H.; Nobu, M.K.; Miyazaki, M.; Tasumi, E.; Saito, Y.; Sakai, S.; Ogawara, M.; Ohashi, A.; Takai, K. Cultivation of previously uncultured microorganisms with a continuous-flow down-flow hanging sponge (DHS) bioreactor, using a syntrophic archaeon culture obtained from deep marine sediment as a case study. Nat. Protoc. 2022. online ahead of print. [Google Scholar] [CrossRef]
- Hauer, B. Embracing Nature’s Catalysts: A Viewpoint on the Future of Biocatalysis. ACS Catal. 2020, 10, 8418–8427. [Google Scholar] [CrossRef]
- Wu, S.; Snajdrova, R.; Moore, J.C.; Baldenius, K.; Bornscheuer, U.T. Biocatalysis: Enzymatic Synthesis for Industrial Applications. Angew. Chem. Int. Ed. 2021, 60, 88–119. [Google Scholar] [CrossRef]
- De Carvalho, C.C.C.R. Whole cell biocatalysts: Essential workers from Nature to the industry. Microb. Biotechnol. 2017, 10, 250–263. [Google Scholar] [CrossRef]
- Bornscheuer, U.T. The fourth wave of biocatalysis is approaching. Phil. Trans. R. Soc. A 2018, 376, 20170063. [Google Scholar] [CrossRef]
- Trincone, A. Marine biocatalysts: Enzymatic features and applications. Mar. Drugs 2011, 9, 478–499. [Google Scholar] [CrossRef]
- De Santi, C.; Altermark, B.; de Pascale, D.; Willassen, N.-P. Bioprospecting around Arctic islands: Marine bacteria as rich source of biocatalysts. J. Basic Microbiol. 2016, 56, 238–253. [Google Scholar] [CrossRef]
- Krühne, U.; Heintz, S.; Ringborg, R.; Rosinha, I.P.; Tufvesson, P.; Gernaey, K.V.; Woodley, J.M. Biocatalytic process development using microfluidic miniaturized systems. Green Process. Synth. 2014, 3, 23–31. [Google Scholar] [CrossRef]
- Marques, M.P.C.; Szita, N. Bioprocess microfluidics: Applying microfluidic devices for bioprocessing. Curr. Opin. Chem. Eng. 2017, 18, 61–68. [Google Scholar] [CrossRef]
- Fernandes, P.; de Carvalho, C.C.C.R. Multi-enzyme systems in flow chemistry. Processes 2021, 9, 225. [Google Scholar] [CrossRef]
- Manjrekar, S.; Wadekar, T.; Sumant, O. Global Enzymes Market. 2021. Available online: https://www.alliedmarketresearch.com/ (accessed on 1 July 2022).
- Singh, R.; Kumar, M.; Mittal, A.; Mehta, P.K. Microbial enzymes: Industrial progress in 21st century. 3 Biotech. 2016, 6, 174. [Google Scholar] [CrossRef]
- Fuchs, M.; Farnberger, J.E.; Kroutil, W. The Industrial Age of Biocatalytic Transamination. Eur. J. Org. Chem. 2015, 2015, 6965–6982. [Google Scholar] [CrossRef]
- Porter, J.L.; Rusli, R.A.; Ollis, D.L. Directed Evolution of Enzymes for Industrial Biocatalysis. ChemBioChem 2016, 17, 197–203. [Google Scholar] [CrossRef]
- Demain, A.L.; Adrio, J.L. Contributions of Microorganisms to Industrial Biology. Mol. Biotechnol. 2007, 38, 41–55. [Google Scholar] [CrossRef]
- Cira, L.; Guevara-Luna, J.; Soto Padilla, M.; Román-Ponce, B.; Vasquez Murrieta, M.; Estrada Alvarado, M. Kinetics of Halophilic Enzymes; IntechOpen: London, UK, 2018. [Google Scholar]
- DasSarma, S.; DasSarma, P. Halophiles and their enzymes: Negativity put to good use. Curr. Opin. Microbiol. 2015, 25, 120–126. [Google Scholar] [CrossRef]
- Salameh, M.d.; Wiegel, J. Lipases from extremophiles and potential for industrial applications. Adv. Appl. Microbiol. 2007, 61, 253–283. [Google Scholar] [CrossRef]
- Karan, R.; Capes, M.D.; DasSarma, S. Function and biotechnology of extremophilic enzymes in low water activity. Aquat. Biosyst. 2012, 8, 4. [Google Scholar] [CrossRef]
- Yin, J.; Chen, J.-C.; Wu, Q.; Chen, G.-Q. Halophiles, coming stars for industrial biotechnology. Biotechnol. Adv. 2015, 33, 1433–1442. [Google Scholar] [CrossRef] [PubMed]
- Shafiei, M.; Ziaee, A.-A.; Amoozegar, M.A. Purification and characterization of an organic-solvent-tolerant halophilic α-amylase from the moderately halophilic Nesterenkonia sp. strain F. J. Ind. Microbiol. Biotechnol. 2011, 38, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, M.; Yokouchi, H.; Suzuki, N.; Ohata, H.; Matsunaga, T. Saccharification of marine microalgae using marine bacteria for ethanol production. Appl. Biochem. Biotechnol. 2003, 10–108, 247–254. [Google Scholar] [CrossRef]
- Chakraborty, S.; Khopade, A.; Kokare, C.; Mahadik, K.; Chopade, B. Isolation and characterization of novel α-amylase from marine Streptomyces sp. D1. J. Mol. Catal. B: Enzym. 2009, 58, 17–23. [Google Scholar] [CrossRef]
- Lailaja, V.P.; Chandrasekaran, M. Detergent compatible alkaline lipase produced by marine Bacillus smithii BTMS 11. World J. Microbiol. Biotechnol. 2013, 29, 1349–1360. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Malik, H.T.; Hua, L. Asymmetric ketone reduction by a hyperthermophilic alcohol dehydrogenase. The substrate specificity, enantioselectivity and tolerance of organic solvents. Tetrahedron: Asymmetry 2006, 17, 3010–3014. [Google Scholar] [CrossRef]
- Vieille, C.; Zeikus, G.J. Hyperthermophilic enzymes: Sources, uses, and molecular mechanisms for thermostability. Microbiol. Mol. Biol. Rev. MMBR 2001, 65, 1–43. [Google Scholar] [CrossRef]
- Santiago, M.; Ramírez-Sarmiento, C.A.; Zamora, R.A.; Parra, L.P. Discovery, molecular mechanisms, and industrial applications of cold-active enzymes. Front. Microbiol. 2016, 7, 1408. [Google Scholar] [CrossRef]
- Lo Giudice, A.; Michaud, L.; de Pascale, D.; De Domenico, M.; di Prisco, G.; Fani, R.; Bruni, V. Lipolytic activity of Antarctic cold-adapted marine bacteria (Terra Nova Bay, Ross Sea). J. Appl. Microbiol. 2006, 101, 1039–1048. [Google Scholar] [CrossRef]
- Plisova, Y.E.; Balabanova, L.A.; Ivanova, E.P.; Kozhemyako, V.B.; Mikhailov, V.V.; Agafonova, E.V.; Rasskazov, V.A. A highly active alkaline phosphatase from the marine bacterium Cobetia. Mar. Biotechnol. 2005, 7, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Wang, L.; Yu, Y. Production and characterization of alkaline protease from a high yielding and moderately halophilic strain of SD11 marine bacteria. J. Chem. 2015, 2015, 798304. [Google Scholar] [CrossRef]
- Wang, X.; Kan, G.; Ren, X.; Yu, G.; Shi, C.; Xie, Q.; Wen, H.; Betenbaugh, M. Molecular cloning and characterization of a novel α-amylase from Antarctic Sea ice bacterium Pseudoalteromonas sp. M175 and its primary application in detergent. BioMed Res. Int. 2018, 2018, 3258383. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.-L.; Li, Y.; Chi, Z.; Chi, Z.-M. Purification and characterization of κ-carrageenase from the marine bacterium Pseudoalteromonas porphyrae for hydrolysis of κ-carrageenan. Process Biochem. 2011, 46, 265–271. [Google Scholar] [CrossRef]
- Stefanidi, E.; Vorgias, C.E. Molecular analysis of the gene encoding a new chitinase from the marine psychrophilic bacterium Moritella marina and biochemical characterization of the recombinant enzyme. Extremophiles 2008, 12, 541–552. [Google Scholar] [CrossRef] [PubMed]
- Cordas, C.M.; Nguyen, G.-S.; Valério, G.N.; Jønsson, M.; Söllner, K.; Aune, I.H.; Wentzel, A.; Moura, J.J.G. Discovery and characterization of a novel Dyp-type peroxidase from a marine actinobacterium isolated from Trondheim fjord, Norway. J. Inorg. Biochem. 2022, 226, 111651. [Google Scholar] [CrossRef] [PubMed]
- De Santi, C.; Leiros, H.-K.S.; Di Scala, A.; de Pascale, D.; Altermark, B.; Willassen, N.-P. Biochemical characterization and structural analysis of a new cold-active and salt-tolerant esterase from the marine bacterium Thalassospira sp. Extremophiles 2016, 20, 323–336. [Google Scholar] [CrossRef]
- Mahajan, P.M.; Nayak, S.; Lele, S.S. Fibrinolytic enzyme from newly isolated marine bacterium Bacillus subtilis ICTF-1: Media optimization, purification and characterization. J. Biosci. Bioeng. 2012, 113, 307–314. [Google Scholar] [CrossRef]
- Nam, E.; Ahn, J. Antarctic marine bacterium Pseudoalteromonas sp. KNOUC808 as a source of cold-adapted lactose hydrolyzing enzyme. Braz. J. Microbiol. 2011, 42, 927–936. [Google Scholar] [CrossRef]
- Salwoom, L.; Raja Abd Rahman, R.N.Z.; Salleh, A.B.; Mohd Shariff, F.; Convey, P.; Pearce, D.; Mohamad Ali, M.S. Isolation, characterisation, and lipase production of a cold-adapted bacterial strain Pseudomonas sp. LSK25 isolated from Signy Island, Antarctica. Molecules 2019, 24, 715. [Google Scholar] [CrossRef] [PubMed]
- Mikkelsen, M.D.; Cao, H.T.T.; Roret, T.; Rhein-Knudsen, N.; Holck, J.; Tran, V.T.T.; Nguyen, T.T.; Tran, V.H.N.; Lezyk, M.J.; Muschiol, J.; et al. A novel thermostable prokaryotic fucoidan active sulfatase PsFucS1 with an unusual quaternary hexameric structure. Sci. Rep. 2021, 11, 19523. [Google Scholar] [CrossRef]
- Lajoie, C.A.; Kitner, J.B.; Potochnik, S.J.; Townsend, J.M.; Beatty, C.C.; Kelly, C.J. Cloning, expression and characterization of xylose isomerase from the marine bacterium Fulvimarina pelagi in Escherichia coli. Biotechnol. Progr. 2016, 32, 1230–1237. [Google Scholar] [CrossRef] [PubMed]
- Hugenholtz, P.; Goebel, B.M.; Pace, N.R. Impact of Culture-Independent Studies on the Emerging Phylogenetic View of Bacterial Diversity. J. Bacteriol. 1998, 180, 4765–4774. [Google Scholar] [CrossRef]
- Amann, R.I.; Ludwig, W.; Schleifer, K.H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 1995, 59, 143–169. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Handelsman, J. Metagenomics for studying unculturable microorganisms: Cutting the Gordian knot. Genome Biol. 2005, 6, 229. [Google Scholar] [CrossRef] [PubMed]
- Solden, L.; Lloyd, K.; Wrighton, K. The bright side of microbial dark matter: Lessons learned from the uncultivated majority. Curr. Opin. Microbiol. 2016, 31, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Zha, Y.; Chong, H.; Yang, P.; Ning, K. Microbial dark matter: From discovery to applications. Genom. Proteom. Bioinform. 2022, in press. [CrossRef]
- Cheng, M.; Cao, L.; Ning, K. Microbiome Big-Data mining and applications using single-cell technologies and metagenomics approaches toward precision medicine. Front. Genet. 2019, 10, 972. [Google Scholar] [CrossRef]
- Bordoloi, N.K.; Bhagowati, P.; Chaudhuri, M.K.; Mukherjee, A.K. Proteomics and metabolomics analyses to elucidate the desulfurization pathway of Chelatococcus sp. PLoS ONE 2016, 11, e0153547. [Google Scholar] [CrossRef]
- Betancur, L.A.; Naranjo-Gaybor, S.J.; Vinchira-Villarraga, D.M.; Moreno-Sarmiento, N.C.; Maldonado, L.A.; Suarez-Moreno, Z.R.; Acosta-González, A.; Padilla-Gonzalez, G.F.; Puyana, M.; Castellanos, L.; et al. Marine Actinobacteria as a source of compounds for phytopathogen control: An integrative metabolic-profiling/bioactivity and taxonomical approach. PLoS ONE 2017, 12, e0170148. [Google Scholar] [CrossRef]
- Guazzaroni, M.E.; Silva-Rocha, R.; Ward, R.J. Synthetic biology approaches to improve biocatalyst identification in metagenomic library screening. Microb. Biotechnol. 2015, 8, 52–64. [Google Scholar] [CrossRef]
- Terrón-González, L.; Medina, C.; Limón-Mortés, M.C.; Santero, E. Heterologous viral expression systems in fosmid vectors increase the functional analysis potential of metagenomic libraries. Sci. Rep. 2013, 3, 1107. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Lopez, O.; Cerdan, M.E.; Gonzalez Siso, M.I. New extremophilic lipases and esterases from metagenomics. Curr. Protein Pept. Sci. 2014, 15, 445–455. [Google Scholar] [CrossRef] [PubMed]
- Liaw, R.B.; Cheng, M.P.; Wu, M.C.; Lee, C.Y. Use of metagenomic approaches to isolate lipolytic genes from activated sludge. Bioresour. Technol. 2010, 101, 8323–8329. [Google Scholar] [CrossRef]
- Grosse, S.; Bergeron, H.; Imura, A.; Boyd, J.; Wang, S.; Kubota, K.; Miyadera, A.; Sulea, T.; Lau, P.C.K. Nature versus nurture in two highly enantioselective esterases from Bacillus cereus and Thermoanaerobacter tengcongensis. Microb. Biotechnol. 2010, 3, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, T.; Miyazaki, K. Product-Induced Gene Expression, a Product-Responsive Reporter Assay Used To Screen Metagenomic Libraries for Enzyme-Encoding Genes. Appl. Environ. Microbiol. 2010, 76, 7029–7035. [Google Scholar] [CrossRef]
- Ferrer, M.; Martinez-Abarca, F.; Golyshin, P.N. Mining genomes and ‘metagenomes’ for novel catalysts. Curr. Opin. Biotechnol. 2005, 16, 588–593. [Google Scholar] [CrossRef]
- Lorenz, P.; Eck, J. Metagenomics and industrial applications. Nat. Rev. Micro. 2005, 3, 510–516. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Arrojo, L.; Guazzaroni, M.E.; Lopez-Cortes, N.; Beloqui, A.; Ferrer, M. Metagenomic era for biocatalyst identification. Curr. Opin. Biotechnol. 2010, 21, 725–733. [Google Scholar] [CrossRef] [PubMed]
- Berini, F.; Casciello, C.; Marcone, G.L.; Marinelli, F. Metagenomics: Novel enzymes from non-culturable microbes. FEMS Microbiol. Lett. 2017, 364, 19. [Google Scholar] [CrossRef]
- Li, L.; Li, G.; Cao, L.-c.; Ren, G.-h.; Kong, W.; Wang, S.-d.; Guo, G.-s.; Liu, Y.-H. Characterization of the cross-linked enzyme aggregates of a novel β-galactosidase, a potential catalyst for the synthesis of galacto-oligosaccharides. J. Agric. Food Chem. 2015, 63, 894–901. [Google Scholar] [CrossRef]
- Tchigvintsev, A.; Tran, H.; Popovic, A.; Kovacic, F.; Brown, G.; Flick, R.; Hajighasemi, M.; Egorova, O.; Somody, J.C.; Tchigvintsev, D.; et al. The environment shapes microbial enzymes: Five cold-active and salt-resistant carboxylesterases from marine metagenomes. Appl. Microbiol. Biotechnol. 2015, 99, 2165–2178. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, M.; Bargiela, R.; Martínez-Martínez, M.; Mir, J.; Koch, R.; Golyshina, O.V.; Golyshin, P.N. Biodiversity for biocatalysis: A review of the α/β-hydrolase fold superfamily of esterases-lipases discovered in metagenomes. Biocatal. Biotransformation 2015, 33, 235–249. [Google Scholar] [CrossRef]
- Greub, G. Culturomics: A new approach to study the human microbiome. Clin. Microbiol. Infect. 2012, 18, 1157–1159. [Google Scholar] [CrossRef] [PubMed]
- Gabor, E.M.; Alkema, W.B.; Janssen, D.B. Quantifying the accessibility of the metagenome by random expression cloning techniques. Environ. Microbiol. 2004, 6, 879–886. [Google Scholar] [CrossRef]
- Galperin, M.Y.; Koonin, E.V. From complete genome sequence to “complete“ understanding? Trends. Biotechnol. 2010, 28, 398–406. [Google Scholar] [CrossRef]
- Prakash, T.; Taylor, T.D. Functional assignment of metagenomic data: Challenges and applications. Brief Bioinform. 2012, 13, 711–727. [Google Scholar] [CrossRef]
- Ferrer, M.; Méndez-García, C.; Bargiela, R.; Chow, J.; Alonso, S.; García-Moyano, A.; Bjerga, G.E.K.; Steen, I.H.; Schwabe, T.; Blom, C.; et al. Decoding the ocean’s microbiological secrets for marine enzyme biodiscovery. FEMS Microbiol. Lett. 2019, 366, fny285. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, T.; Miyazaki, K. Functional metagenomics for enzyme discovery: Challenges to efficient screening. Curr. Opin. Biotechnol. 2009, 20, 616–622. [Google Scholar] [CrossRef] [PubMed]
- Grealey, J.; Lannelongue, L.; Saw, W.-Y.; Marten, J.; Méric, G.; Ruiz-Carmona, S.; Inouye, M. The carbon footprint of bioinformatics. Mol. Biol. Evol. 2022, 39, msac034. [Google Scholar] [CrossRef]
- Donachie, S.P.; Foster, J.S.; Brown, M.V. Culture clash: Challenging the dogma of microbial diversity. Isme J. 2007, 1, 97. [Google Scholar] [CrossRef]
- Stewart, E.J. Growing unculturable bacteria. J. Bacteriol. 2012, 194, 4151–4160. [Google Scholar] [CrossRef]
- Angelakis, E. Microbial culturomics. Int. J. Infect. Dis. 2016, 45, 27. [Google Scholar] [CrossRef][Green Version]
- Pham, V.H.T.; Kim, J. Cultivation of unculturable soil bacteria. Trends Biotechnol. 2012, 30, 475–484. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Li, M.; Huang, L.; Zhang, X.-H. Cultivation of uncultured marine microorganisms. Mar. Life Sci. Technol. 2021, 3, 117–120. [Google Scholar] [CrossRef]
- Tidjani Alou, M.; Naud, S.; Khelaifia, S.; Bonnet, M.; Lagier, J.C.; Raoult, D. State of the Art in the Culture of the Human Microbiota: New Interests and Strategies. Clin. Microbiol. Rev. 2020, 34, e00129-19. [Google Scholar] [CrossRef] [PubMed]
- Lagier, J.C.; Armougom, F.; Million, M.; Hugon, P.; Pagnier, I.; Robert, C.; Bittar, F.; Fournous, G.; Gimenez, G.; Maraninchi, M.; et al. Microbial culturomics: Paradigm shift in the human gut microbiome study. Clin. Microbiol. Infect. 2012, 18, 1185–1193. [Google Scholar] [CrossRef]
- Dubourg, G.; Lagier, J.C.; Armougom, F.; Robert, C.; Hamad, I.; Brouqui, P.; Raoult, D. The gut microbiota of a patient with resistant tuberculosis is more comprehensively studied by culturomics than by metagenomics. Eur. J. Clin. Microbiol. Infect. Dis. 2013, 32, 637–645. [Google Scholar] [CrossRef]
- Pfleiderer, A.; Lagier, J.C.; Armougom, F.; Robert, C.; Vialettes, B.; Raoult, D. Culturomics identified 11 new bacterial species from a single anorexia nervosa stool sample. Eur. J. Clin. Microbiol. Infect. Dis. 2013, 32, 1471–1481. [Google Scholar] [CrossRef]
- Alain, K.; Querellou, J. Cultivating the uncultured: Limits, advances and future challenges. Extremophiles 2009, 13, 583–594. [Google Scholar] [CrossRef] [PubMed]
- Overmann, J.; Abt, B.; Sikorski, J. Present and future of culturing bacteria. Annu. Rev. Microbiol. 2017, 71, 711–730. [Google Scholar] [CrossRef]
- Koch, A.L. Oligotrophs versus copiotrophs. Bioessays 2001, 23, 657–661. [Google Scholar] [CrossRef] [PubMed]
- Song, H.-K.; Song, W.; Kim, M.; Tripathi, B.M.; Kim, H.; Jablonski, P.; Adams, J.M. Bacterial strategies along nutrient and time gradients, revealed by metagenomic analysis of laboratory microcosms. FEMS Microbiol. Ecol. 2017, 93, fix114. [Google Scholar] [CrossRef]
- Kim, H.; Jeon, J.; Lee, K.K.; Lee, Y.-H. Compositional shift of Bacterial, Archaeal, and Fungal communities is dependent on trophic lifestyles in Rice Paddy Soil. Front. Microbiol. 2021, 12, 719486. [Google Scholar] [CrossRef] [PubMed]
- Jung, D.; Liu, B.; He, X.; Owen, J.S.; Liu, L.; Yuan, Y.; Zhang, W.; He, S. Accessing previously uncultured marine microbial resources by a combination of alternative cultivation methods. Microb. Biotechnol. 2021, 14, 1148–1158. [Google Scholar] [CrossRef] [PubMed]
- Bruns, A.; Nübel, U.; Cypionka, H.; Overmann, J. Effect of Signal Compounds and Incubation Conditions on the Culturability of Freshwater Bacterioplankton. Appl. Environ. Microbiol. 2003, 69, 1980–1989. [Google Scholar] [CrossRef] [PubMed]
- Nai, C.; Meyer, V. From axenic to mixed cultures: Technological advances accelerating a paradigm shift in microbiology. Trends Microbiol. 2018, 26, 538–554. [Google Scholar] [CrossRef]
- Song, J.; Oh, H.M.; Cho, J.C. Improved culturability of SAR11 strains in dilution-to-extinction culturing from the East Sea, West Pacific Ocean. FEMS Microbiol. Lett. 2009, 295, 141–147. [Google Scholar] [CrossRef]
- D’Onofrio, A.; Crawford, J.M.; Stewart, E.J.; Witt, K.; Gavrish, E.; Epstein, S.; Clardy, J.; Lewis, K. Siderophores from neighboring organisms promote the growth of uncultured bacteria. Chem. Biol. 2010, 17, 254–264. [Google Scholar] [CrossRef]
- Overmann, J. Principles of enrichment, isolation, cultivation, and preservation of prokaryotes. In The Prokaryotes: Prokaryotic Biology and Symbiotic Associations; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 149–207. [Google Scholar]
- Tripp, H.J.; Kitner, J.B.; Schwalbach, M.S.; Dacey, J.W.H.; Wilhelm, L.J.; Giovannoni, S.J. SAR11 marine bacteria require exogenous reduced sulphur for growth. Nature 2008, 452, 741. [Google Scholar] [CrossRef]
- Available online: https://www.nature.com/articles/nature06776#supplementary-information (accessed on 15 July 2022).
- Nichols, D.; Lewis, K.; Orjala, J.; Mo, S.; Ortenberg, R.; O’Connor, P.; Zhao, C.; Vouros, P.; Kaeberlein, T.; Epstein, S.S. Short peptide induces an “uncultivable” microorganism to grow in vitro. Appl. Environ. Microbiol. 2008, 74, 4889–4897. [Google Scholar] [CrossRef]
- Kaeberlein, T.; Lewis, K.; Epstein, S.S. Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science 2002, 296, 1127–1129. [Google Scholar] [CrossRef]
- Nunoura, T.; Chikaraishi, Y.; Izaki, R.; Suwa, T.; Sato, T.; Harada, T.; Mori, K.; Kato, Y.; Miyazaki, M.; Shimamura, S.; et al. A primordial and reversible TCA cycle in a facultatively chemolithoautotrophic thermophile. Science 2018, 359, 559–563. [Google Scholar] [CrossRef] [PubMed]
- Oliver, J.D. Formation of viable but nonculturable cells. In Starvation in Bacteria; Kjelleberg, S., Ed. Springer: Boston, MA, USA, 1993; pp. 239–272. [Google Scholar]
- Schut, F.; Prins, R.A.; Gottschal, J.C. Oligotrophy and pelagic marine bacteria: Facts and fiction. Aquat. Microb. Ecol. 1997, 12, 177–202. [Google Scholar] [CrossRef][Green Version]
- Bodor, A.; Bounedjoum, N.; Vincze, G.E.; Erdeiné Kis, Á.; Laczi, K.; Bende, G.; Szilágyi, Á.; Kovács, T.; Perei, K.; Rákhely, G. Challenges of unculturable bacteria: Environmental perspectives. Rev. Environ. Sci. Bio/Technol. 2020, 19, 1–22. [Google Scholar] [CrossRef]
- Davis, K.E.; Joseph, S.J.; Janssen, P.H. Effects of growth medium, inoculum size, and incubation time on culturability and isolation of soil bacteria. Appl. Environ. Microbiol. 2005, 71, 826–834. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K.; Epstein, S.; D’Onofrio, A.; Ling, L.L. Uncultured microorganisms as a source of secondary metabolites. J. Antibiot. 2010, 63, 468–476. [Google Scholar] [CrossRef]
- Jung, D.; Machida, K.; Nakao, Y.; Kindaichi, T.; Ohashi, A.; Aoi, Y. Triggering growth via growth initiation factors in Nature: A putative mechanism for in situ cultivation of previously uncultivated microorganisms. Front. Microbiol. 2021, 12, 537194. [Google Scholar] [CrossRef]
- Mu, D.-S.; Ouyang, Y.; Chen, G.-J.; Du, Z.-J. Strategies for culturing active/dormant marine microbes. Mar. Life Sci. Technol. 2021, 3, 121–131. [Google Scholar] [CrossRef]
- Lewis, W.H.; Tahon, G.; Geesink, P.; Sousa, D.Z.; Ettema, T.J.G. Innovations to culturing the uncultured microbial majority. Nat. Rev. Microbiol. 2021, 19, 225–240. [Google Scholar] [CrossRef]
- Steinert, G.; Whitfield, S.; Taylor, M.W.; Thoms, C.; Schupp, P.J. Application of Diffusion Growth Chambers for the Cultivation of Marine Sponge-Associated Bacteria. Mar. Biotechnol. 2014, 16, 594–603. [Google Scholar] [CrossRef]
- Nichols, D.; Cahoon, N.; Trakhtenberg, E.M.; Pham, L.; Mehta, A.; Belanger, A.; Kanigan, T.; Lewis, K.; Epstein, S.S. Use of ichip for high-throughput in situ cultivation of “uncultivable” microbial species. Appl. Environ. Microbiol. 2010, 76, 2445–2450. [Google Scholar] [CrossRef]
- Angelini, L.M.L.; da Silva, A.R.M.; Rocco, L.d.F.C.; Milagre, C.D.d.F. A high-throughput screening assay for distinguishing nitrile hydratases from nitrilases. Braz. J. Microbiol. 2015, 46, 113–116. [Google Scholar] [CrossRef] [PubMed]
- Baud, D.; Ladkau, N.; Moody, T.S.; Ward, J.M.; Hailes, H.C. A rapid, sensitive colorimetric assay for the high-throughput screening of transaminases in liquid or solid-phase. Chem. Commun. 2015, 51, 17225–17228. [Google Scholar] [CrossRef]
- Rodrigues, C.J.C.; Sanches, J.M.; de Carvalho, C.C.C.R. Determining transaminase activity in bacterial libraries by time-lapse imaging. Chem. Commun. 2019, 55, 13538–13541. [Google Scholar] [CrossRef]
- Pollard, D.J.; Woodley, J.M. Biocatalysis for pharmaceutical intermediates: The future is now. Trends Biotechnol. 2007, 25, 66–73. [Google Scholar] [CrossRef]
- Koutinas, M.; Kiparissides, A.; Pistikopoulos, E.N.; Mantalaris, A. Bioprocess systems engineering: Transferring traditional process engineering principles to industrial biotechnology. Comput. Struct. Biotechnol. J. 2013, 3, e201210022. [Google Scholar] [CrossRef]
- Blesken, C.; Olfers, T.; Grimm, A.; Frische, N. The microfluidic bioreactor for a new era of bioprocess development. Eng. Life Sci. 2016, 16, 190–193. [Google Scholar] [CrossRef]
- Girard, P.; Jordan, M.; Tsao, M.; Wurm, F.M. Small-scale bioreactor system for process development and optimization. Biochem. Eng. J. 2001, 7, 117–119. [Google Scholar] [CrossRef]
- Lattermann, C.; Büchs, J. Microscale and miniscale fermentation and screening. Curr. Opin. Biotechnol. 2015, 35, 1–6. [Google Scholar] [CrossRef]
- Fernandes, P. Miniaturization in Biocatalysis. Int. J. Mol. Sci. 2010, 11, 858–879. [Google Scholar] [CrossRef]
- Marques, M.P.C.; Fernandes, P. Microfluidic Devices: Useful Tools for Bioprocess Intensification. Molecules 2011, 16, 8368. [Google Scholar] [CrossRef]
- Hemmerich, J.; Noack, S.; Wiechert, W.; Oldiges, M. Microbioreactor Systems for Accelerated Bioprocess Development. Biotechnol. J. 2018, 13, e1700141. [Google Scholar] [CrossRef]
- Gruber, P.; Marques, M.P.C.; Szita, N.; Mayr, T. Integration and application of optical chemical sensors in microbioreactors. Lab. Chip 2017, 17, 2693–2712. [Google Scholar] [CrossRef]
- Rodrigues, C.J.C.; de Carvalho, C.C.C.R. Process development for benzyl alcohol production by whole-cell biocatalysis in stirred and packed bed reactors. Microorganisms 2022, 10, 966. [Google Scholar] [CrossRef]
- Rodrigues, C.J.C.; Ferrer, M.; de Carvalho, C.C.C.R. ω-Transaminase-mediated asymmetric synthesis of (S)-1-(4-trifluoromethylphenyl)ethylamine. Catalysts 2021, 11, 307. [Google Scholar] [CrossRef]
- Annamalai, N.; Rajeswari, M.V.; Balasubramanian, T. Enzymatic saccharification of pretreated rice straw by cellulase produced from Bacillus carboniphilus CAS 3 utilizing lignocellulosic wastes through statistical optimization. Biomass Bioenergy 2014, 68, 151–160. [Google Scholar] [CrossRef]
- Vázquez, J.A.; Durán, A.; Nogueira, M.; Menduíña, A.; Antunes, J.; Freitas, A.C.; Gomes, A.M. Production of marine probiotic bacteria in a cost-effective marine media based on peptones obtained from discarded fish by-products. Microorganisms 2020, 8, 1121. [Google Scholar] [CrossRef]
| Enzyme | Marine Bacteria | Sampling Location and/or Organism | Isolation Technique | Enzyme Screening Technique | Activity | Optimal Temperature and pH | Potential Application | Ref. |
|---|---|---|---|---|---|---|---|---|
| Alkaline phosphatases | Cobetia marina | Mussels from Troitza Bay, Japanese Sea | Liquid culture | Standard assay for AP activity | Conversion of p-nitrophenylphosphate | 50 °C; pH 10.3 | Functional studies of nucleic acids and immunologic assays | [54] |
| Alkaline protease | Marine bacterium SD11 | Sea mud, China | Agar plate | Bioactivity screening with agar plate | Hydrolysis of carbohydrates | 60 °C; pH 10 | Synthesis of peptides and detergent formulations in alkaline environments | [55] |
| α-Amylase | Pseudoalteromonas sp. M175 | Antarctic sea ice | Flask culture | Genetic techniques | Cleavage of α-1,4-glycosidic linkages in starch molecules | 25 °C; pH 8.0 | Detergent additive to improve stain removal efficiency | [56] |
| κ-Carrageenase | Pseudoalteromonas porphyrae | Decayed seaweed collected from Yellow Sea, China | Medium containing carrageenan | κ-carrageenase activity | κ-Carrageenan hydrolysis | 55 °C; pH 8.0 | Production of sulfated oligosaccharides | [57] |
| Chitinase | Moritella marina | Sample from a depth of 1200 m, northern Pacific Ocean | Marine agar/broth containing colloidal chitin | Standard chitinase activity | p-nitrophenyl-β-1,4-N,N′-diacetyl-chitobiose | 28 °C; pH 5.0 | Waste management and biocontrol of pathogenic fungi | [58] |
| Dye-decolorizing peroxidase | Actinobacteria strain collection | Shallow water sediments, Trondheim fjord, Norway | Agar plate | Genomic data mining | Oxidization of several phenolic substrates | 25 °C; pH 3–4 | Degradation of natural or artificial dyes, or in chemical synthesis phenolic monomers | [59] |
| Esterase | Thalassospira sp. GB04J01 | Sea fan, northern Norway | Flask culture | Genetic techniques | Hydrolysis of simple esters | 45 °C; pH 8.5 | Additives in laundry detergents | [60] |
| Fibrinolytic enzymes | Bacillus subtilis ICTF-1 | Western seacoast of Maharashtra, India | Agar plate | Activity screening in microplate | Conversion of N-succinyl-Ala-Ala-Pro-Phe-p-nitroanilide | 50 °C; pH 9.0 | Drugs for the treatment of thrombosis | [61] |
| Inulinase (whole-cell) | Bacillus subtilis | Intertidal shallow water thermal vent, Azores | Agar plate | Bioactivity screening with agar plate | Hydrolysis of inulin to reducing sugars | 40 °C; pH 8.5 | Food industry | [25] |
| Lactase | Pseudoalteromonas sp. KNOUC808 | Antarctic polar sea | Enrichments cultures | Hydrolysis of ONPG | Hydrolysis of o-nitrophenyl β-D-galactopyranoside | 20 °C; pH 7.8 | Detergent formulations, dairy industry, biosensors | [62] |
| Lipase | Pseudomonas sp. LSK25 | Signy Island, Antarctica | Growth in LB broth at 4 °C | Tributyrin agar plates | Colorimetric assay with olive oil as substrate | 10 °C; pH 7.0 | Detergent, textile, and food industries | [63] |
| Sulfatase | Pseudoalteromonas sp. | Sea cucumber gut, Vietnam | Agar plate | Bioactivity screening with agar plate | Hydrolytic cleavage of sulfate ester groups on carbohydrates | 68 °C; pH 6.5 | Production of partially desulphated fucoidan oligosaccharides | [64] |
| Xylose isomerase | Fulvimarina pelagi | Sargasso Sea | Dilution-to-extinction | Genetic techniques | Interconversion of d-xylose and d-xylulose | 35 °C; pH 7 | Biofuels production | [65] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodrigues, C.J.C.; de Carvalho, C.C.C.R. Marine Bioprospecting, Biocatalysis and Process Development. Microorganisms 2022, 10, 1965. https://doi.org/10.3390/microorganisms10101965
Rodrigues CJC, de Carvalho CCCR. Marine Bioprospecting, Biocatalysis and Process Development. Microorganisms. 2022; 10(10):1965. https://doi.org/10.3390/microorganisms10101965
Chicago/Turabian StyleRodrigues, Carlos J. C., and Carla C. C. R. de Carvalho. 2022. "Marine Bioprospecting, Biocatalysis and Process Development" Microorganisms 10, no. 10: 1965. https://doi.org/10.3390/microorganisms10101965
APA StyleRodrigues, C. J. C., & de Carvalho, C. C. C. R. (2022). Marine Bioprospecting, Biocatalysis and Process Development. Microorganisms, 10(10), 1965. https://doi.org/10.3390/microorganisms10101965







