Haemophilus influenzae Carriage among Healthy Children in Portugal, 2015–2019
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Study Design
2.3. Questionnaires
2.4. Sample Collection
2.5. Isolation of Putative Haemophilus influenzae
2.6. Preparation of DNA Templates for Serotyping
2.7. Serotyping
2.8. β-Lactamase Production and Minimum Inhibitory Concentration (MIC) Determination
2.9. DNA Extraction and Whole Genome Sequencing (WGS)
2.10. Whole Genome Sequencing Analysis
2.11. Accession Numbers
2.12. Statistical Analysis
3. Results
3.1. Population Characteristics
3.2. Colonization Rates and Capsular Type
3.3. β-Lactamase Production and Antimicrobial Susceptibility Testing
3.4. Whole Genome Sequencing (WGS)
3.5. Multilocus Sequence Typing (MLST)
3.6. Genetic Determinants of Antimicrobial Resistance
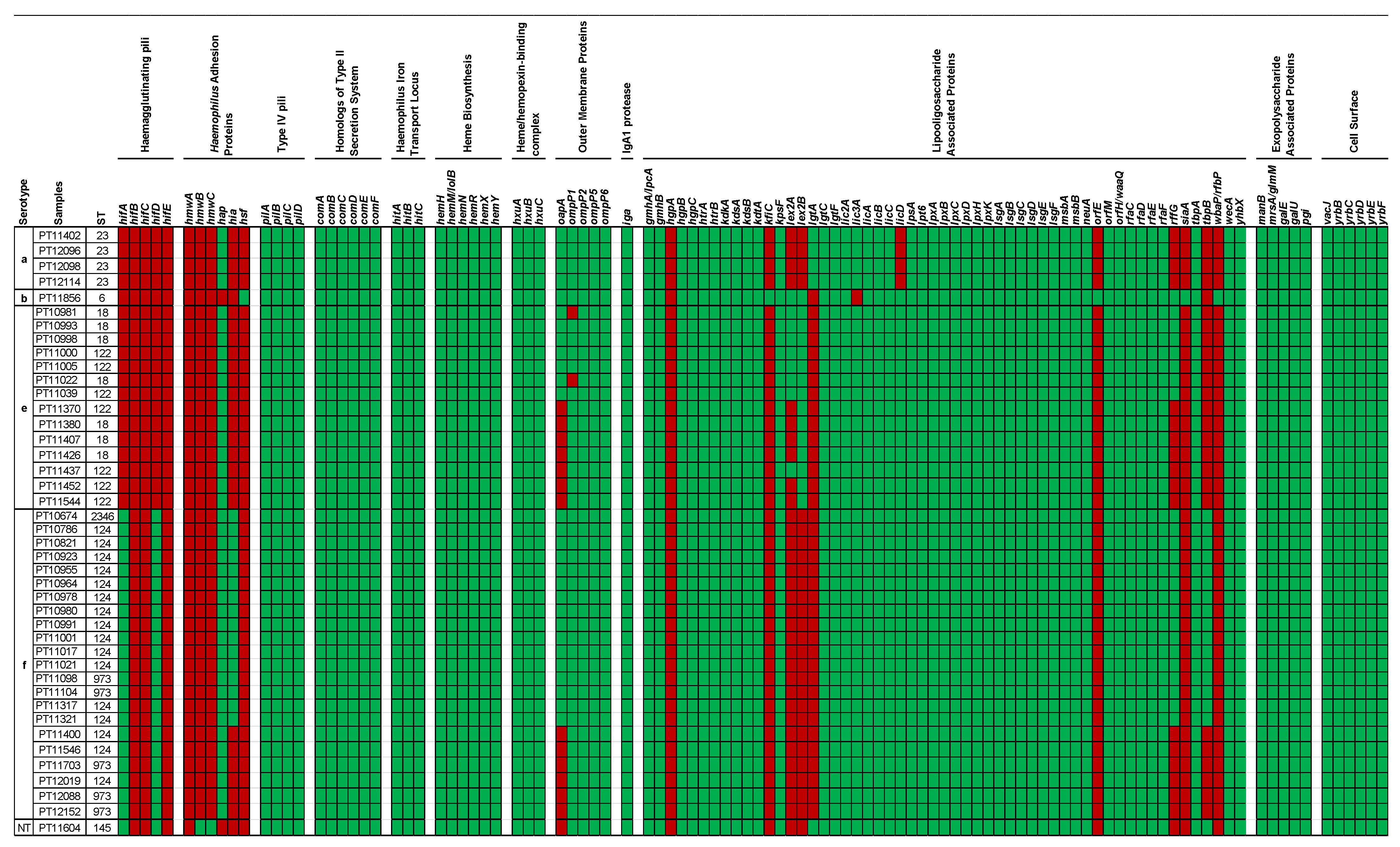
3.7. Virulence Associated Genes
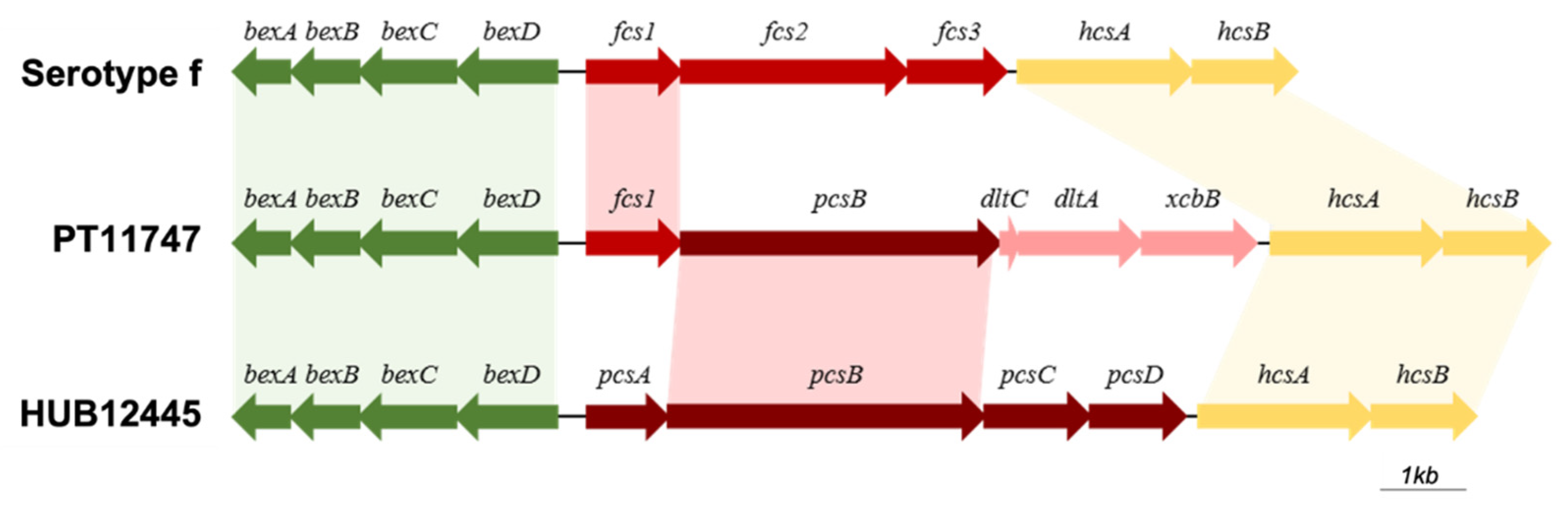
3.8. Characterization of Cap Loci in H. influenzae and H. parainfluenzae
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- De Steenhuijsen Piters, W.A.; Sanders, E.A.; Bogaert, D. The role of the local microbial ecosystem in respiratory health and disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370, 20140294. [Google Scholar] [CrossRef] [PubMed]
- Bogaert, D.; Keijser, B.; Huse, S.; Rossen, J.; Veenhoven, R.; van Gils, E.; Bruin, J.; Montijn, R.; Bonten, M.; Sanders, E. Variability and diversity of nasopharyngeal microbiota in children: A metagenomic analysis. PLoS ONE 2011, 6, e17035. [Google Scholar] [CrossRef] [PubMed]
- Murphy, T.F.; Bakaletz, L.O.; Smeesters, P.R. Microbial interactions in the respiratory tract. Pediatr. Infect. Dis. J. 2009, 28, S121–S126. [Google Scholar] [CrossRef] [PubMed]
- Schenck, L.P.; Surette, M.G.; Bowdish, D.M. Composition and immunological significance of the upper respiratory tract microbiota. FEBS Lett. 2016, 590, 3705–3720. [Google Scholar] [CrossRef]
- García-Rodríguez, J.A.; Fresnadillo Martínez, M.J. Dynamics of nasopharyngeal colonization by potential respiratory pathogens. J. Antimicrob. Chemother. 2002, 50 (Suppl. S2), 59–73. [Google Scholar] [CrossRef] [PubMed]
- Leach, A.J.; Boswell, J.B.; Asche, V.; Nienhuys, T.G.; Mathews, J.D. Bacterial colonization of the nasopharynx predicts very early onset and persistence of otitis media in Australian aboriginal infants. Pediatr. Infect. Dis. J. 1994, 13, 983–989. [Google Scholar] [CrossRef]
- Turk, D.C. The pathogenicity of Haemophilus influenzae. J. Med. Microbiol. 1984, 18, 1–16. [Google Scholar] [CrossRef]
- Pittman, M. Variation and type specificity in the bacterial species Haemophilus influenzae. J. Exp. Med. 1931, 53, 471–492. [Google Scholar] [CrossRef]
- Watts, S.C.; Holt, K.E. hicap: In silico serotyping of the Haemophilus influenzae capsule locus. J. Clin. Microbiol. 2019, 57, e00190-19. [Google Scholar] [CrossRef]
- Satola, S.W.; Schirmer, P.L.; Farley, M.M. Complete sequence of the cap locus of Haemophilus influenzae serotype b and nonencapsulated b capsule-negative variants. Infect. Immun. 2003, 71, 3639–3644. [Google Scholar] [CrossRef]
- Kroll, J.S.; Loynds, B.M.; Moxon, E.R. The Haemophilus influenzae capsulation gene cluster: A compound transposon. Mol. Microbiol. 1991, 5, 1549–1560. [Google Scholar] [CrossRef]
- Park, J.J.; Narayanan, S.; Tiefenbach, J.; Lukšić, I.; Ale, B.M.; Adeloye, D.; Rudan, I. Estimating the global and regional burden of meningitis in children caused by Haemophilus influenzae type b: A systematic review and meta-analysis. J. Glob. Health 2022, 12, 04014. [Google Scholar] [CrossRef]
- Butler, D.F.; Myers, A.L. Changing epidemiology of Haemophilus influenzae in children. Infect. Dis. Clin. 2018, 32, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Bennett, J.V.; Platonov, A.E.; Slack, M.P.; Mala, P.; Burton, A.; Robertson, S.A. Haemophilus influenzae type b (Hib) meningitis in the pre-vaccine era: A global review of incidence, age distributions, and case-fatality rates. Vaccines Biol. 2002, 1, 92. [Google Scholar]
- Dagan, R.; Fraser, D.; Greif, Z.; Keller, N.; Kaufstein, M.; Shazberg, G.; Schlesinger, M. A nationwide prospective surveillance study in Israel to document pediatric invasive infections, with an emphasis on Haemophilus influenzae type b infections. Israeli Pediatric Bacteremia and Meningitis Group. Pediatr. Infect. Dis. J. 1998, 17, S198–S203. [Google Scholar] [CrossRef]
- Jordens, J.Z.; Slack, M.P. Haemophilus influenzae: Then and now. Eur. J. Clin. Microbiol. Infect. Dis. 1995, 14, 935–948. [Google Scholar] [CrossRef] [PubMed]
- Whittaker, R.; Economopoulou, A.; Dias, J.G.; Bancroft, E.; Ramliden, M.; Celentano, L.P. Epidemiology of invasive Haemophilus influenzae disease, Europe, 2007–2014. Emerg. Infect. Dis. 2017, 23, 396–404. [Google Scholar] [CrossRef] [PubMed]
- Ladhani, S.N. Two decades of experience with the Haemophilus influenzae serotype b conjugate vaccine in the United Kingdom. Clin. Ther. 2012, 34, 385–399. [Google Scholar] [CrossRef]
- Campos, J.; Hernando, M.; Román, F.; Pérez-Vázquez, M.; Aracil, B.; Oteo, J.; Lázaro, E.; de Abajo, F.; Group of Invasive Haemophilus Infections of the Autonomous Community of Madrid, Spain. Analysis of invasive Haemophilus influenzae infections after extensive vaccination against H. influenzae type b. J. Clin. Microbiol. 2004, 42, 524–529. [Google Scholar] [CrossRef] [PubMed]
- Heath, P.T.; Booy, R.; Azzopardi, H.J.; Slack, M.P.; Fogarty, J.; Moloney, A.C.; Ramsay, M.E.; Moxon, E.R. Non-type b Haemophilus influenzae disease: Clinical and epidemiologic characteristics in the Haemophilus influenzae type b vaccine era. Pediatr. Infect. Dis. J. 2001, 20, 300–305. [Google Scholar] [CrossRef]
- Peltola, H. Worldwide Haemophilus influenzae type b disease at the beginning of the 21st century: Global analysis of the disease burden 25 years after the use of the polysaccharide vaccine and a decade after the advent of conjugates. Clin. Microbiol. Rev. 2000, 13, 302–317. [Google Scholar] [CrossRef] [PubMed]
- Adegbola, R.A.; Usen, S.O.; Weber, M.; Lloyd-Evans, N.; Jobe, K.; Mulholland, K.; McAdam, K.P.; Greenwood, B.M.; Milligan, P.J. Haemophilus influenzae type b meningitis in The Gambia after introduction of a conjugate vaccine. Lancet 1999, 354, 1091–1092. [Google Scholar] [CrossRef]
- Slack, M.P.; Azzopardi, H.J.; Hargreaves, R.M.; Ramsay, M.E. Enhanced surveillance of invasive Haemophilus influenzae disease in England, 1990 to 1996: Impact of conjugate vaccines. Pediatr. Infect. Dis. J. 1998, 17, S204–S207. [Google Scholar] [CrossRef] [PubMed]
- Adams, W.G.; Deaver, K.A.; Cochi, S.L.; Plikaytis, B.D.; Zell, E.R.; Broome, C.V.; Wenger, J.D. Decline of childhood Haemophilus influenzae type b (Hib) disease in the Hib vaccine era. JAMA 1993, 269, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Carrera-Salinas, A.; González-Díaz, A.; Calatayud, L.; Mercado-Maza, J.; Puig, C.; Berbel, D.; Càmara, J.; Tubau, F.; Grau, I.; Domínguez, M.; et al. Epidemiology and population structure of Haemophilus influenzae causing invasive disease. Microb. Genom. 2021, 7, 000723. [Google Scholar] [CrossRef]
- Fuji, N.; Pichichero, M.; Kaur, R. Haemophilus influenzae prevalence, proportion of capsulated strains and antibiotic susceptibility during colonization and acute otitis media in children, 2019–2020. Pediatr. Infect. Dis. J. 2021, 40, 792–796. [Google Scholar] [CrossRef] [PubMed]
- Heliodoro, C.I.M.; Bettencourt, C.R.; Bajanca-Lavado, M.P. Molecular epidemiology of invasive Haemophilus influenzae disease in Portugal: An update of the post-vaccine period, 2011–2018. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 1471–1480. [Google Scholar] [CrossRef] [PubMed]
- Giufrè, M.; Fabiani, M.; Cardines, R.; Riccardo, F.; Caporali, M.G.; D’Ancona, F.; Pezzotti, P.; Cerquetti, M. Increasing trend in invasive non-typeable Haemophilus influenzae disease and molecular characterization of the isolates, Italy, 2012–2016. Vaccine 2018, 36, 6615–6622. [Google Scholar] [CrossRef] [PubMed]
- Puig, C.; Grau, I.; Marti, S.; Tubau, F.; Calatayud, L.; Pallares, R.; Liñares, J.; Ardanuy, C. Clinical and molecular epidemiology of Haemophilus influenzae causing invasive disease in adult patients. PLoS ONE 2014, 9, e112711. [Google Scholar] [CrossRef] [PubMed]
- DGS. Programa Nacional de Vacinação—2017. Available online: https://www.dgs.pt/em-destaque/novo-programa-nacional-de-vacinacao-pdf.aspx (accessed on 21 March 2020).
- Bajanca, P.; Caniça, M.; Group, M.S. Emergence of nonencapsulated and encapsulated non-b-type invasive Haemophilus influenzae isolates in Portugal (1989–2001). J. Clin. Microbiol. 2004, 42, 807–810. [Google Scholar] [CrossRef] [PubMed]
- Bajanca-Lavado, M.P.; Simões, A.S.; Betencourt, C.R.; Sá-Leão, R. Characteristics of Haemophilus influenzae invasive isolates from Portugal following routine childhood vaccination against H. influenzae serotype b (2002–2010). Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, F.; Foster, D.; Nicoli, E.; Trotter, C.; Vipond, B.; Muir, P.; Gonçalves, G.; Januário, L.; Finn, A. Relationships between rhinitis symptoms, respiratory viral infections and nasopharyngeal colonization with Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus in children attending daycare. Pediatr. Infect. Dis. J. 2013, 32, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Sá-Leão, R.; Nunes, S.; Brito-Avô, A.; Alves, C.R.; Carriço, J.A.; Saldanha, J.; Almeida, J.S.; Sanches, I.S.; de Lencastre, H. High rates of transmission of and colonization by Streptococcus pneumoniae and Haemophilus influenzae within a day care center revealed in a longitudinal study. J. Clin. Microbiol. 2008, 46, 225–234. [Google Scholar] [CrossRef] [PubMed]
- De Lencastre, H.; Kristinsson, K.G.; Brito-Avô, A.; Sanches, I.S.; Sá-Leão, R.; Saldanha, J.; Sigvaldadottir, E.; Karlsson, S.; Oliveira, D.; Mato, R.; et al. Carriage of respiratory tract pathogens and molecular epidemiology of Streptococcus pneumoniae colonization in healthy children attending day care centers in Lisbon, Portugal. Microb. Drug Resist. 1999, 5, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Neto, A.S.; Lavado, P.; Flores, P.; Dias, R.; Pessanha, M.A.; Sousa, E.; Palminha, J.M.; Caniça, M.; Esperança-Pina, J. Risk factors for the nasopharyngeal carriage of respiratory pathogens by Portuguese children: Phenotype and antimicrobial susceptibility of Haemophilus influenzae and Streptococcus pneumoniae. Microb. Drug Resist. 2003, 9, 99–108. [Google Scholar] [CrossRef]
- Ortiz-Romero, M.D.M.; Espejo-García, M.P.; Alfayate-Miguelez, S.; Ruiz-López, F.J.; Zapata-Hernandez, D.; Gonzalez-Pacanowska, A.J. Epidemiology of nasopharyngeal carriage by Haemophilus influenzae in healthy children: A study in the mediterranean coast region. Pediatr. Infect. Dis. J. 2017, 36, 919–923. [Google Scholar] [CrossRef]
- Peerbooms, P.G.; Engelen, M.N.; Stokman, D.A.; van Benthem, B.H.; van Weert, M.L.; Bruisten, S.M.; van Belkum, A.; Coutinho, R.A. Nasopharyngeal carriage of potential bacterial pathogens related to day care attendance, with special reference to the molecular epidemiology of Haemophilus influenzae. J. Clin. Microbiol. 2002, 40, 2832–2836. [Google Scholar] [CrossRef]
- Dabernat, H.; Plisson-Sauné, M.A.; Delmas, C.; Séguy, M.; Faucon, G.; Pélissier, R.; Carsenti, H.; Pradier, C.; Roussel-Delvallez, M.; Leroy, J.; et al. Haemophilus influenzae carriage in children attending French day care centers: A molecular epidemiological study. J. Clin. Microbiol. 2003, 41, 1664–1672. [Google Scholar] [CrossRef]
- Bonifácio Da Silva, M.E.N.; Da Silva, P.; Medeiros, M.I.C.; Neme, S.N.; Macedo, C.; Marin, J.M. Nasopharyngeal colonization by Haemophilus influenzae in children attending day-care centers, in Ribeirão Preto, State of São Paulo, Brazil. Braz. J. Microbiol. 2006, 37, 33–38. [Google Scholar] [CrossRef][Green Version]
- Medeiros, A.A.; Levesque, R.; Jacoby, G.A. An animal source for the ROB-1 beta-lactamase of Haemophilus influenzae type b. Antimicrob. Agents Chemother. 1986, 29, 212–215. [Google Scholar] [CrossRef]
- Khan, W.; Ross, S.; Rodriguez, W.; Controni, G.; Saz, A.K. Haemophilus influenzae type B resistant to ampicillin. A report of two cases. JAMA 1974, 229, 298–301. [Google Scholar] [CrossRef] [PubMed]
- Mathies, A.W. Penicillins in the treatment of bacterial meningitis. J. R. Coll. Physicians Lond. 1972, 6, 139–146. [Google Scholar] [PubMed]
- Dabernat, H.; Delmas, C.; Seguy, M.; Pelissier, R.; Faucon, G.; Bennamani, S.; Pasquier, C. Diversity of beta-lactam resistance-conferring amino acid substitutions in penicillin-binding protein 3 of Haemophilus influenzae. Antimicrob. Agents Chemother. 2002, 46, 2208–2218. [Google Scholar] [CrossRef] [PubMed]
- Ubukata, K.; Shibasaki, Y.; Yamamoto, K.; Chiba, N.; Hasegawa, K.; Takeuchi, Y.; Sunakawa, K.; Inoue, M.; Konno, M. Association of amino acid substitutions in penicillin-binding protein 3 with beta-lactam resistance in beta-lactamase-negative ampicillin-resistant Haemophilus influenzae. Antimicrob. Agents Chemother. 2001, 45, 1693–1699. [Google Scholar] [CrossRef]
- Tristram, S.; Jacobs, M.R.; Appelbaum, P.C. Antimicrobial resistance in Haemophilus influenzae. Clin. Microbiol. Rev. 2007, 20, 368–389. [Google Scholar] [CrossRef] [PubMed]
- Doern, G.V.; Brueggemann, A.B.; Pierce, G.; Holley, H.P.; Rauch, A. Antibiotic resistance among clinical isolates of Haemophilus influenzae in the United States in 1994 and 1995 and detection of beta-lactamase-positive strains resistant to amoxicillin-clavulanate: Results of a national multicenter surveillance study. Antimicrob. Agents Chemother. 1997, 41, 292–297. [Google Scholar] [CrossRef]
- Nunes, S.; Félix, S.; Valente, C.; Simões, A.S.; Tavares, D.A.; Almeida, S.T.; Paulo, A.C.; Brito-Avô, A.; de Lencastre, H.; Sá-Leão, R. The impact of private use of PCV7 in 2009 and 2010 on serotypes and antimicrobial resistance of Streptococcus pneumoniae carried by young children in Portugal: Comparison with data obtained since 1996 generating a 15-year study prior to PCV13 introduction. Vaccine 2016, 34, 1648–1656. [Google Scholar] [CrossRef]
- Satzke, C.; Turner, P.; Virolainen-Julkunen, A.; Adrian, P.V.; Antonio, M.; Hare, K.M.; Henao-Restrepo, A.M.; Leach, A.J.; Klugman, K.P.; Porter, B.D.; et al. Standard method for detecting upper respiratory carriage of Streptococcus pneumoniae: Updated recommendations from the World Health Organization Pneumococcal Carriage Working Group. Vaccine 2013, 32, 165–179. [Google Scholar] [CrossRef]
- Falla, T.J.; Crook, D.W.; Brophy, L.N.; Maskell, D.; Kroll, J.S.; Moxon, E.R. PCR for capsular typing of Haemophilus influenzae. J. Clin. Microbiol. 1994, 32, 2382–2386. [Google Scholar] [CrossRef]
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 12.0. 2022. Available online: http://www.eucast.org (accessed on 21 March 2020).
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Chen, L.; Yang, J.; Yu, J.; Yao, Z.; Sun, L.; Shen, Y.; Jin, Q. VFDB: A reference database for bacterial virulence factors. Nucleic Acids Res. 2005, 33, D325–D328. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.M.; Akerley, B.J. Genome-scale approaches to identify genes essential for Haemophilus influenzae pathogenesis. Front. Cell. Infect. Microbiol. 2012, 2, 23. [Google Scholar] [CrossRef] [PubMed]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; De Pascale, G.; Ejim, L.; et al. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef]
- González-Díaz, A.; Tubau, F.; Pinto, M.; Sierra, Y.; Cubero, M.; Càmara, J.; Ayats, J.; Bajanca-Lavado, P.; Ardanuy, C.; Marti, S. Identification of polysaccharide capsules among extensively drug-resistant genitourinary Haemophilus parainfluenzae isolates. Sci. Rep. 2019, 9, 4481. [Google Scholar] [CrossRef]
- Agrawal, A.; Murphy, T.F. Haemophilus influenzae infections in the H. influenzae type b conjugate vaccine era. J. Clin. Microbiol. 2011, 49, 3728–3732. [Google Scholar] [CrossRef]
- De Carvalho, C.X.; Kipnis, A.; Thörn, L.; de Andrade, J.G.; Pimenta, F.; Brandileone, M.C.; Zanella, R.C.; Flannery, B.; Sgambatti, S.; Andrade, A.L. Carriage of Haemophilus influenzae among Brazilian children attending day care centers in the era of widespread Hib vaccination. Vaccine 2011, 29, 1438–1442. [Google Scholar] [CrossRef]
- Puig, C.; Marti, S.; Fleites, A.; Trabazo, R.; Calatayud, L.; Liñares, J.; Ardanuy, C. Oropharyngeal colonization by nontypeable Haemophilus influenzae among healthy children attending day care centers. Microb. Drug Resist. 2014, 20, 450–455. [Google Scholar] [CrossRef]
- Dunne, E.M.; Murad, C.; Sudigdoadi, S.; Fadlyana, E.; Tarigan, R.; Indriyani, S.A.K.; Pell, C.L.; Watts, E.; Satzke, C.; Hinds, J.; et al. Carriage of Streptococcus pneumoniae, Haemophilus influenzae, Moraxella catarrhalis, and Staphylococcus aureus in Indonesian children: A cross-sectional study. PLoS ONE 2018, 13, e0195098. [Google Scholar] [CrossRef]
- Canada. Public Health Agency & Centre for Communicable Diseases and Infection Control. Routine Practices and Additional Precautions for Preventing the Transmission of Infection in Healthcare Settings. 2016. Available online: https://www.canada.ca/en/public-health/services/publications/diseases-conditions/routine-practices-precautions-healthcare-associated-infections.html; (accessed on 21 March 2020).
- Zanella, R.C.; Brandileone, M.C.; Andrade, A.L.; Ogassavara, C.T.; Fiório, C.E.; Brandão, A.P.; Almeida, S.C.; Lemos, A.P.; Gorla, M.C.; Carvalhanas, T.R.; et al. Evaluation of Haemophilus influenzae type b carrier status among children 10 years after the introduction of Hib vaccine in Brazil. Mem. Inst. Oswaldo Cruz 2015, 110, 755–759. [Google Scholar] [CrossRef][Green Version]
- Barbosa-Cesnik, C.; Farjo, R.S.; Patel, M.; Gilsdorf, J.; McCoy, S.I.; Pettigrew, M.M.; Marrs, C.; Foxman, B. Predictors for Haemophilus influenzae colonization, antibiotic resistance and for sharing an identical isolate among children attending 16 licensed day-care centers in Michigan. Pediatr. Infect. Dis. J. 2006, 25, 219–223. [Google Scholar] [CrossRef]
- Schumacher, S.K.; Marchant, C.D.; Loughlin, A.M.; Bouchet, V.; Stevenson, A.; Pelton, S.I. Prevalence and genetic diversity of nontypeable Haemophilus influenzae in the respiratory tract of infants and primary caregivers. Pediatr. Infect. Dis. J. 2012, 31, 145–149. [Google Scholar] [CrossRef] [PubMed]
- Kovács, E.; Sahin-Tóth, J.; Tóthpál, A.; van der Linden, M.; Tirczka, T.; Dobay, O. Co-carriage of Staphylococcus aureus, Streptococcus pneumoniae, Haemophilus influenzae and Moraxella catarrhalis among three different age categories of children in Hungary. PLoS ONE 2020, 15, e0229021. [Google Scholar] [CrossRef] [PubMed]
- Janapatla, R.P.; Chang, H.J.; Hsu, M.H.; Hsieh, Y.C.; Lin, T.Y.; Chiu, C.H. Nasopharyngeal carriage of Streptococcus pneumoniae, Haemophilus influenzae, Moraxella catarrhalis, and Alloiococcus otitidis in young children in the era of pneumococcal immunization, Taiwan. Scand. J. Infect. Dis. 2011, 43, 937–942. [Google Scholar] [CrossRef] [PubMed]
- Giufrè, M.; Daprai, L.; Cardines, R.; Bernaschi, P.; Ravà, L.; Accogli, M.; Raponi, M.; Garlaschi, M.L.; Ciofi degli Atti, M.L.; Cerquetti, M. Carriage of Haemophilus influenzae in the oropharynx of young children and molecular epidemiology of the isolates after fifteen years of H. influenzae type b vaccination in Italy. Vaccine 2015, 33, 6227–6234. [Google Scholar] [CrossRef]
- Brown, V.M.; Madden, S.; Kelly, L.; Jamieson, F.B.; Tsang, R.S.; Ulanova, M. Invasive Haemophilus influenzae disease caused by non-type b strains in Northwestern Ontario, Canada, 2002–2008. Clin. Infect. Dis. 2009, 49, 1240–1243. [Google Scholar] [CrossRef]
- Giufrè, M.; Dorrucci, M.; Lo Presti, A.; Farchi, F.; Cardines, R.; Camilli, R.; Pimentel de Araujo, F.; Mancini, F.; Ciervo, A.; Corongiu, M.; et al. Nasopharyngeal carriage of Haemophilus influenzae among adults with co-morbidities. Vaccine 2022, 40, 826–832. [Google Scholar] [CrossRef]
- Giufrè, M.; Cardines, R.; Accogli, M.; Pardini, M.; Cerquetti, M. Identification of Haemophilus influenzae clones associated with invasive disease a decade after introduction of H. influenzae serotype b vaccination in Italy. Clin. Vaccine Immunol. 2013, 20, 1223–1229. [Google Scholar] [CrossRef] [PubMed]
- Smith-Vaughan, H.C.; Sriprakash, K.S.; Leach, A.J.; Mathews, J.D.; Kemp, D.J. Low genetic diversity of Haemophilus influenzae type b compared to nonencapsulated H. influenzae in a population in which H. influenzae is highly endemic. Infect. Immun. 1998, 66, 3403–3409. [Google Scholar] [CrossRef]
- Pinto, M.; González-Díaz, A.; Machado, M.P.; Duarte, S.; Vieira, L.; Carriço, J.A.; Marti, S.; Bajanca-Lavado, M.P.; Gomes, J.P. Insights into the population structure and pan-genome of Haemophilus influenzae. Infect. Genet. Evol. 2019, 67, 126–135. [Google Scholar] [CrossRef]
- Cherkaoui, A.; Gaïa, N.; Baud, D.; Leo, S.; Fischer, A.; Ruppe, E.; François, P.; Schrenzel, J. Molecular characterization of fluoroquinolones, macrolides, and imipenem resistance in Haemophilus influenzae: Analysis of the mutations in QRDRs and assessment of the extent of the AcrAB-TolC-mediated resistance. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 2201–2210. [Google Scholar] [CrossRef]
- Shoji, H.; Shirakura, T.; Fukuchi, K.; Takuma, T.; Hanaki, H.; Tanaka, K.; Niki, Y. A molecular analysis of quinolone-resistant Haemophilus influenzae: Validation of the mutations in Quinolone Resistance-Determining Regions. J. Infect. Chemother. 2014, 20, 250–255. [Google Scholar] [CrossRef]
- Puig, C.; Tirado-Vélez, J.M.; Calatayud, L.; Tubau, F.; Garmendia, J.; Ardanuy, C.; Marti, S.; de la Campa, A.G.; Liñares, J. Molecular characterization of fluoroquinolone resistance in nontypeable Haemophilus influenzae clinical isolates. Antimicrob. Agents Chemother. 2015, 59, 461–466. [Google Scholar] [CrossRef]
- Leflon-Guibout, V.; Speldooren, V.; Heym, B.; Nicolas-Chanoine, M. Epidemiological survey of amoxicillin-clavulanate resistance and corresponding molecular mechanisms in Escherichia coli isolates in France: New genetic features of bla(TEM) genes. Antimicrob. Agents Chemother. 2000, 44, 2709–2714. [Google Scholar] [CrossRef] [PubMed]
- Chaïbi, E.B.; Sirot, D.; Paul, G.; Labia, R. Inhibitor-resistant TEM beta-lactamases: Phenotypic, genetic and biochemical characteristics. J. Antimicrob. Chemother. 1999, 43, 447–458. [Google Scholar] [CrossRef] [PubMed]
- Tristam, S.G.; Burdach, J.G. Effect of cloned inhibitor-resistant TEM beta-lactamases on the susceptibility of Haemophilus influenzae to amoxicillin/clavulanate. J. Antimicrob. Chemother. 2007, 60, 1151–1154. [Google Scholar] [CrossRef] [PubMed]
- García-Cobos, S.; Campos, J.; Lázaro, E.; Román, F.; Cercenado, E.; García-Rey, C.; Pérez-Vázquez, M.; Oteo, J.; de Abajo, F. Ampicillin-resistant non-beta-lactamase-producing Haemophilus influenzae in Spain: Recent emergence of clonal isolates with increased resistance to cefotaxime and cefixime. Antimicrob. Agents Chemother. 2007, 51, 2564–2573. [Google Scholar] [CrossRef] [PubMed]
- Sierra, Y.; Tubau, F.; González-Díaz, A.; Carrera-Salinas, A.; Moleres, J.; Bajanca-Lavado, P.; Garmendia, J.; Domínguez, M.; Ardanuy, C.; Martí, S. Assessment of trimethoprim-sulfamethoxazole susceptibility testing methods for fastidious Haemophilus spp. Clin. Microbiol. Infect. 2020, 26, 944.e941–944.e947. [Google Scholar] [CrossRef]
- Li, X.X.; Xiao, S.Z.; Gu, F.F.; He, W.P.; Ni, Y.X.; Han, L.Z. Molecular epidemiology and antimicrobial resistance of Haemophilus influenzae in adult patients in shanghai, china. Front. Public Health 2020, 8, 95. [Google Scholar] [CrossRef]
- Li, J.P.; Hua, C.Z.; Sun, L.Y.; Wang, H.J.; Chen, Z.M.; Shang, S.Q. Epidemiological features and antibiotic resistance patterns of Haemophilus influenzae originating from respiratory tract and vaginal specimens in pediatric patients. J. Pediatr. Adolesc. Gynecol. 2017, 30, 626–631. [Google Scholar] [CrossRef]
| Total | Year, n (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Characteristic | 2015 | 2016 | 2018 | 2019 | ||||||
| Participants | 1524 | 315 | 318 | 427 | 464 | |||||
| Age group (months) a | ||||||||||
| 0–12 | 122 | (8.0) | 21 | (6.7) | 30 | (9.6) | 28 | (6.6) | 43 | (9.4) |
| 13–24 | 252 | (16.6) | 47 | (14.9) | 49 | (15.6) | 86 | (20.2) | 70 | (15.2) |
| 25–36 | 287 | (19.0) | 55 | (17.5) | 59 | (18.8) | 80 | (18.8) | 93 | (20.2) |
| 37–48 | 362 | (23.9) | 88 | (27.9) | 64 | (20.4) | 98 | (23.1) | 112 | (24.3) |
| 49–60 | 272 | (18.0) | 60 | (19.0) | 69 | (29) | 67 | (15.8) | 76 | (16.5) |
| ≥61 | 219 | (14.5) | 44 | (14.0) | 43 | (13.7) | 66 | (15.5) | 66 | (14.4) |
| Gender | ||||||||||
| Male | 817 | (53.6) | 163 | (51.8) | 171 | (53.8) | 234 | (54.8) | 249 | (53.7) |
| Antimicrobial use | ||||||||||
| At sampling b | 63 | (4.3) | 14 | (4.5) | 11 | (3.7) | 21 | (5.0) | 17 | (3.8) |
| In previous month c | 246 | (17.6) | 45 | (15.6) | 48 | (17.8) | 73 | (18.1) | 80 | (18.5) |
| In previous six months d | 503 | (36.1) | 94 | (33.0) | 90 | (31.4) | 167 | (42.8) | 152 | (35.3) |
| H. influenzae Carriage | Total | Year, % (n/Total) | |||
|---|---|---|---|---|---|
| 2015 | 2016 | 2018 | 2019 | ||
| Global | 84.1 (1282/1524) | 88.6 (279/315) | 75.2 (239/318) | 90.6 (387/427) | 81.3 (377/464) |
| by age group (months) a | |||||
| 0–12 | 83.6 (102/122) | 81.0 (17/21) | 73.3 (22/30) | 92.9 (26/28) | 86.0 (37/43) |
| 13–24 | 92.1 (232/252) | 91.5 (43/47) | 89.8 (44/49) | 95.3 (82/86) | 90.0 (63/70) |
| 25–36 | 86.4 (248/287) | 92.7 (51/59) | 81.4 (48/59) | 91.3 (73/80) | 81.7 (76/93) |
| 37–48 | 84.3 (305/362) | 92.0 (81/88) | 73.4 (47/64) | 89.8 (88/98) | 79.5 (89/112) |
| 49–60 | 82.4 (224/272) | 86.7 (52/60) | 76.8 (53/69) | 86.6 (58/67) | 80.3 (61/76) |
| ≥61 | 75.3 (165/219) | 79.5 (35/44) | 51.2 (22/43) | 90.9 (60/66) | 72.7 (48/66) |
| by serotype | |||||
| serotype a | 0.3 (4/1282) | 0.0 (0/279) | 0.0 (0/239) | 0.3 (1/387) | 0.8 (3/377) |
| serotype b | 0.1(1/1282) | 0.0 (0/279) | 0.0 (0/239) | 0.0 (0/387) | 0.3 (1/377) |
| serotype e | 1.1(14/1282) | 0.0 (0/279) | 2.9 (7/239) | 1.8 (7/387) | 0.0 (0/377) |
| serotype f | 1.8 (23/1282) | 0.7 (2/279) | 5.9 (14/239) | 0.8 (3/387) | 1.0 (4/377) |
| nonencapsulated | 96.7 (1240/1282) | 99.3 (277/279) | 91.2 (218/239) | 97.1 (376/387) | 97.9 (369/377) |
| Antimicrobial Agent | β-Lactamase Producers (n = 96) a | Capsulated (n = 42) a | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S (%) | I (%) | R (%) | S (%) | I (%) | R (%) | |||||||
| Ampicillin | 0 | (0.0) | 0 | (0.0) | 96 | (100.0) | 37 | (88.1) | 0 | (0.0) | 5 | (11.9) |
| Amoxicillin-clavulanic acid | 95 | (99.0) | 0 | (0.0) | 1 | (1.0) | 42 | (100.0) | 0 | (0.0) | 0 | (0.0) |
| Cefepime | 93 | (96.9) | 0 | (0.0) | 3 | (3.1) | 41 | (97.6) | 0 | (0.0) | 1 | (2.4) |
| Cefotaxime | 96 | (100.0) | 0 | (0.0) | 0 | (0.0) | 42 | (100.0) | 0 | (0.0) | 0 | (0.0) |
| Cefuroxime | 0 | (0.0) | 87 | (90.6) | 9 | (9.4) | 0 | (0.0) | 40 | (95.2) | 2 | (4.8) |
| Ciprofloxacin | 95 | (99.0) | 0 | (0.0) | 1 | (1.0) | 42 | (100.0) | 0 | (0.0) | 0 | (0.0) |
| Chloramphenicol | 93 | (96.9) | 0 | (0.0) | 3 | (3.1) | 42 | (100.0) | 0 | (0.0) | 0 | (0.0) |
| Meropenem | 96 | (100.0) | 0 | (0.0) | 0 | (0.0) | 42 | (100.0) | 0 | (0.0) | 0 | (0.0) |
| Tetracycline | 94 | (97.9) | 0 | (0.0) | 2 | (2.1) | 42 | (100.0) | 0 | (0.0) | 0 | (0.0) |
| Trimethoprim-sulfamethoxazole | 63 | (65.6) | 0 | (0.0) | 33 | (34.4) | 40 | (95.2) | 0 | (0.0) | 2 | (4.8) |
| Rifampicin | 96 | (100.0) | 0 | (0.0) | 0 | (0.0) | 42 | (100.0) | 0 | (0.0) | 0 | (0.0) |
| Strain Reference | Sampling Year | Genome Size (Mbp) | Genome Coverage | Number of Contigs | Serotype | Multilocus Sequence Typing | Phenotype Resistance Pattern | Antimicrobial Resistance Determinants | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| adk | atpG | frdB | fucK | mdh | pgi | recA | ST | Resistance Genotype | bla Gene | ftsI Allele | PBP3 Mutations a | folA Mutations a | folP Mutations a | |||||||
| PT12096 | 2019 | 1.76 | 352 | 32 | a | 13 | 16 | 5 | 2 | 3 | 11 | 7 | 23 | - | - | nd | 46 | - | ||
| PT12098 | 2019 | 1.76 | 323 | 30 | a | 13 | 16 | 5 | 2 | 3 | 11 | 7 | 23 | - | - | nd | 46 | - | ||
| PT12114 | 2019 | 1.76 | 292 | 33 | a | 13 | 16 | 5 | 2 | 3 | 11 | 7 | 23 | - | - | nd | 46 | - | ||
| PT11402 | 2018 | 1.76 | 27 | 46 | a | 13 | 16 | 5 | 2 | 3 | 11 | 7 | 23 | - | - | nd | 46 | - | ||
| PT11856 | 2019 | 1.87 | 217 | 46 | b | 10 | 14 | 4 | 5 | 4 | 7 | 8 | 6 | - | - | nd | 10 | - | ||
| PT10981 | 2016 | 1.85 | 243 | 33 | e | 18 | 6 | 3 | 7 | 10 | 28 | 12 | 18 | Amp, Cfx(I),SXT, BLP | BLP | bla TEM-1 | 55 | D350N | F154S | P64E |
| PT10993 | 2016 | 1.84 | 287 | 37 | e | 18 | 6 | 3 | 7 | 10 | 28 | 12 | 18 | - | - | nd | 55 | D350N | ||
| PT10998 | 2016 | 1.84 | 291 | 37 | e | 18 | 6 | 3 | 7 | 10 | 28 | 12 | 18 | - | - | nd | 55 | D350N | ||
| PT11000 | 2016 | 1.84 | 307 | 31 | e | 18 | 6 | 3 | 18 | 10 | 28 | 12 | 122 | Cfx (I) | BLNAR IIb | nd | 2 | D350N M377I A502V N526K | ||
| PT11005 | 2016 | 1.84 | 308 | 29 | e | 18 | 6 | 3 | 18 | 10 | 28 | 12 | 122 | Cfx | BLNAR IIb | nd | 2 | D350N M377I A502V N526K | ||
| PT11022 | 2016 | 1.84 | 101 | 36 | e | 18 | 6 | 3 | 7 | 10 | 28 | 12 | 18 | Amp, Cfx(I),SXT, BLP | BLP | bla TEM-1 | 55 | D350N | F154S | P64E |
| PT11039 | 2016 | 1.84 | 190 | 33 | e | 18 | 6 | 3 | 18 | 10 | 28 | 12 | 122 | Cfx | BLNAR IIb | nd | 2 | D350N M377I A502V N526K | ||
| PT11370 | 2018 | 1.90 | 52 | 41 | e | 18 | 6 | 3 | 18 | 10 | 28 | 12 | 122 | Amp, Cfx(I), BLP | BLPACR | bla TEM-82 | 2 | D350N M377I A502V N526K | ||
| PT11380 | 2018 | 1.84 | 43 | 32 | e | 18 | 6 | 3 | 7 | 10 | 28 | 12 | 18 | - | - | nd | 55 | D350N | ||
| PT11407 | 2018 | 1.84 | 27 | 40 | e | 18 | 6 | 3 | 7 | 10 | 28 | 12 | 18 | - | - | nd | 55 | D350N | ||
| PT11426 | 2018 | 1.84 | 32 | 31 | e | 18 | 6 | 3 | 7 | 10 | 28 | 12 | 18 | - | - | nd | 55 | D350N | ||
| PT11437 | 2018 | 1.84 | 35 | 35 | e | 18 | 6 | 3 | 18 | 10 | 28 | 12 | 122 | Cfx (I) | BLNAR IIb | nd | 2 | D350N M377I A502V N526K | ||
| PT11452 | 2018 | 1.84 | 25 | 43 | e | 18 | 6 | 3 | 18 | 10 | 28 | 12 | 122 | Cfx (I) | BLNAR IIb | nd | 2 | D350N M377I A502V N526K | ||
| PT11544 | 2018 | 1.84 | 33 | 39 | e | 18 | 6 | 3 | 18 | 10 | 28 | 12 | 122 | Cfx (I) | BLNAR IIb | nd | 2 | D350N M377I A502V N526K | ||
| PT10674 | 2015 | 1.81 | 262 | 21 | f | 22 | 19 | 11 | 11 | 86 | 19 | 15 | 2346 | Cfx (I), Cpe | - | nd | 6 | D350N | ||
| PT10786 | 2015 | 1.81 | 222 | 23 | f | 22 | 19 | 11 | 11 | 22 | 19 | 15 | 124 | - | - | nd | 6 | D350N | ||
| PT10821 | 2016 | 1.81 | 290 | 20 | f | 22 | 19 | 11 | 11 | 22 | 19 | 15 | 124 | - | - | nd | 6 | D350N | ||
| PT10923 | 2016 | 1.81 | 195 | 21 | f | 22 | 19 | 11 | 11 | 22 | 19 | 15 | 124 | - | - | nd | 6 | D350N | ||
| PT10955 | 2016 | 1.81 | 266 | 22 | f | 22 | 19 | 11 | 11 | 22 | 19 | 15 | 124 | - | - | nd | 6 | D350N | ||
| PT10964 | 2016 | 1.81 | 319 | 25 | f | 22 | 19 | 11 | 11 | 22 | 19 | 15 | 124 | - | - | nd | 6 | D350N | ||
| PT10978 | 2016 | 1.81 | 335 | 22 | f | 22 | 19 | 11 | 11 | 22 | 19 | 15 | 124 | - | - | nd | 6 | D350N | ||
| PT10980 | 2016 | 1.81 | 236 | 24 | f | 22 | 19 | 11 | 11 | 22 | 19 | 15 | 124 | - | - | nd | 6 | D350N | ||
| PT10991 | 2016 | 1.81 | 238 | 23 | f | 22 | 19 | 11 | 11 | 22 | 19 | 15 | 124 | - | - | nd | 6 | D350N | ||
| PT11001 | 2016 | 1.81 | 324 | 25 | f | 22 | 19 | 11 | 11 | 22 | 19 | 15 | 124 | - | - | nd | 6 | D350N | ||
| PT11017 | 2016 | 1.81 | 258 | 22 | f | 22 | 19 | 11 | 11 | 22 | 19 | 15 | 124 | - | - | nd | 6 | D350N | ||
| PT11021 | 2016 | 1.81 | 91 | 21 | f | 22 | 19 | 11 | 11 | 22 | 19 | 15 | 124 | - | - | nd | 6 | D350N | ||
| PT11098 | 2016 | 1.81 | 78 | 24 | f | 22 | 19 | 140 | 11 | 22 | 19 | 15 | 973 | - | - | nd | 6 | D350N | ||
| PT11104 | 2016 | 1.81 | 75 | 24 | f | 22 | 19 | 140 | 11 | 22 | 19 | 15 | 973 | - | - | nd | 6 | D350N | ||
| PT11317 | 2016 | 1.81 | 73 | 24 | f | 22 | 19 | 11 | 11 | 22 | 19 | 15 | 124 | - | - | nd | 6 | D350N | ||
| PT11321 | 2016 | 1.81 | 66 | 21 | f | 22 | 19 | 11 | 11 | 22 | 19 | 15 | 124 | - | - | nd | 6 | D350N | ||
| PT11400 | 2018 | 1.81 | 53 | 21 | f | 22 | 19 | 11 | 11 | 22 | 19 | 15 | 124 | - | - | nd | 6 | D350N | ||
| PT11546 | 2018 | 1.81 | 37 | 29 | f | 22 | 19 | 11 | 11 | 22 | 19 | 15 | 124 | - | - | nd | 6 | D350N | ||
| PT11703 | 2018 | 1.81 | 156 | 20 | f | 22 | 19 | 140 | 11 | 22 | 19 | 15 | 973 | Cip | - | nd | 6 | D350N | ||
| PT12019 | 2019 | 1.81 | 308 | 23 | f | 22 | 19 | 11 | 11 | 22 | 19 | 15 | 124 | Amp, Cfx(I), BLP | BLP | bla TEM-1 | 6 | D350N | ||
| PT12088 | 2019 | 1.81 | 286 | 22 | f | 22 | 19 | 140 | 11 | 22 | 19 | 15 | 973 | - | - | nd | 6 | D350N | ||
| PT12152 | 2019 | 1.81 | 298 | 21 | f | 22 | 19 | 140 | 11 | 22 | 19 | 15 | 973 | - | - | nd | 6 | D350N | ||
| PT11604 | 2018 | 1.84 | 84 | 101 | NT | 1 | 8 | 1 | 14 | 22 | 14 | 13 | 145 | Amp, Aug, Cfx, Cpe, SXT, BLP | BLPACR | bla TEM-1 | 24 | D350N A502T N526K | F154S | P64E |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bajanca-Lavado, M.P.; Cavaco, L.; Fernandes, M.; Touret, T.; Candeias, C.; Simões, A.S.; Sá-Leão, R. Haemophilus influenzae Carriage among Healthy Children in Portugal, 2015–2019. Microorganisms 2022, 10, 1964. https://doi.org/10.3390/microorganisms10101964
Bajanca-Lavado MP, Cavaco L, Fernandes M, Touret T, Candeias C, Simões AS, Sá-Leão R. Haemophilus influenzae Carriage among Healthy Children in Portugal, 2015–2019. Microorganisms. 2022; 10(10):1964. https://doi.org/10.3390/microorganisms10101964
Chicago/Turabian StyleBajanca-Lavado, Maria Paula, Luís Cavaco, Mariana Fernandes, Tiago Touret, Catarina Candeias, Alexandra S. Simões, and Raquel Sá-Leão. 2022. "Haemophilus influenzae Carriage among Healthy Children in Portugal, 2015–2019" Microorganisms 10, no. 10: 1964. https://doi.org/10.3390/microorganisms10101964
APA StyleBajanca-Lavado, M. P., Cavaco, L., Fernandes, M., Touret, T., Candeias, C., Simões, A. S., & Sá-Leão, R. (2022). Haemophilus influenzae Carriage among Healthy Children in Portugal, 2015–2019. Microorganisms, 10(10), 1964. https://doi.org/10.3390/microorganisms10101964






